Transgenic Xenopus laevis Line for In Vivo Labeling of Nephrons within the Kidney
Abstract
:1. Introduction
2. Materials and Methods
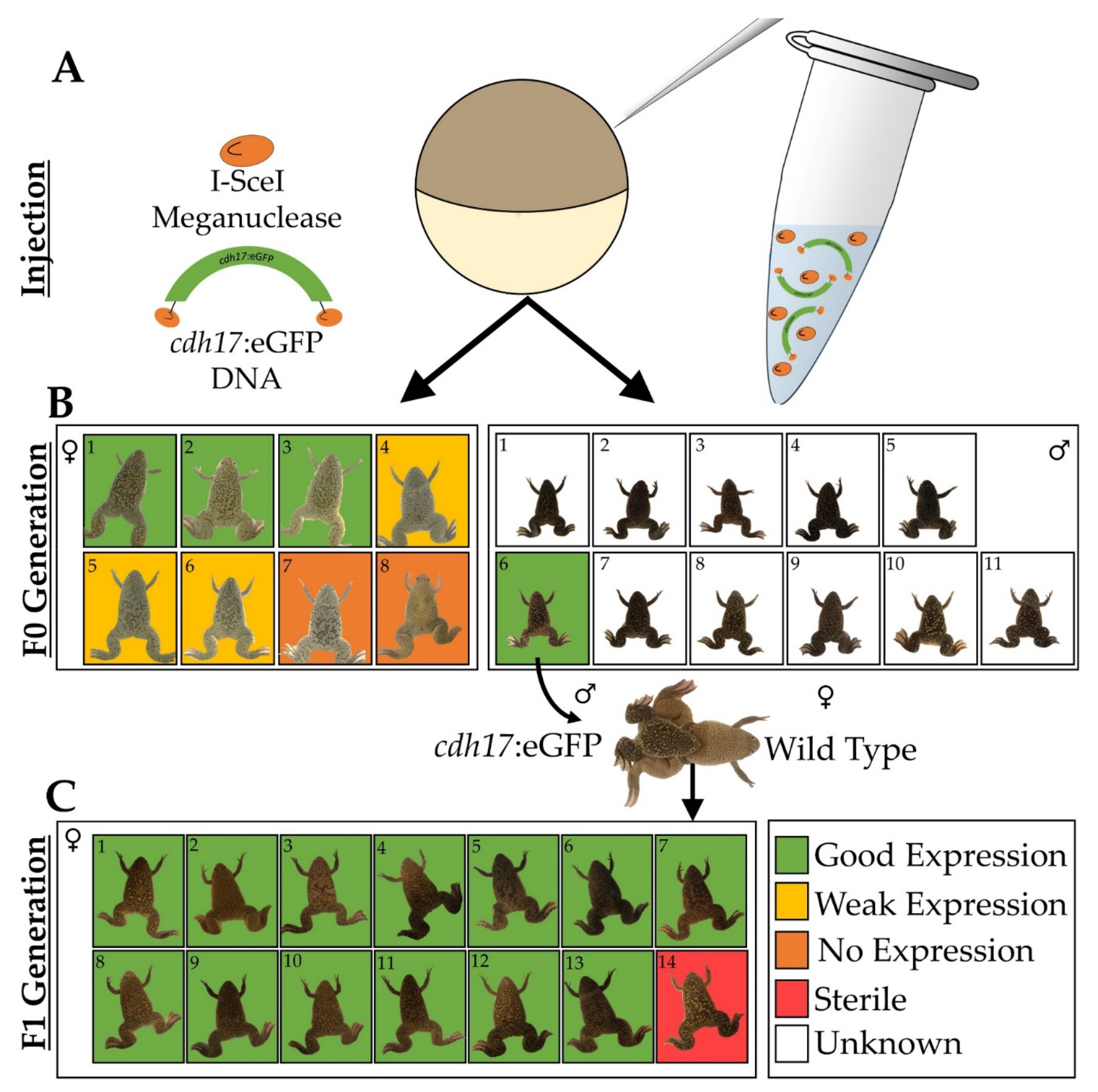
2.1. Transgenic Animal Generation and Embryo Culture
2.2. Immunostaining
2.3. Imaging
2.4. Secretion Assay
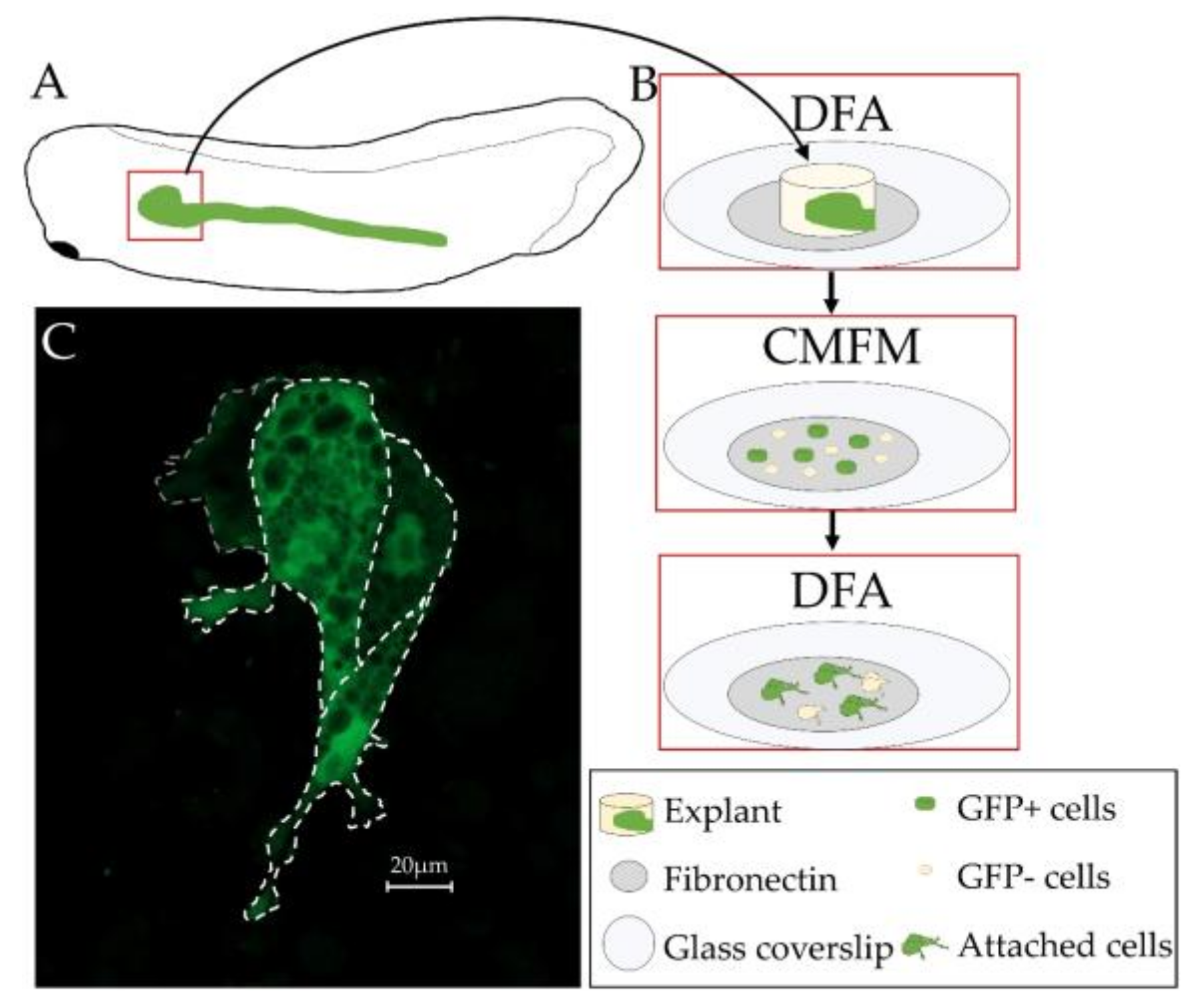
2.5. Kidney Cell Primary Culture
2.6. Microinjection
3. Results
3.1. Generation of Xenopus laevis cdh17:eGFP Line
3.2. cdh17:eGFP Colocalizes with Kidney Markers
3.3. In Vivo Characterization of cdh17:eGFP Transgenic Line
3.4. Assessment of Kidney Function in cdh17:eGFP Embryos
3.5. Primary Culture of Xenopus laevis Kidney Cells
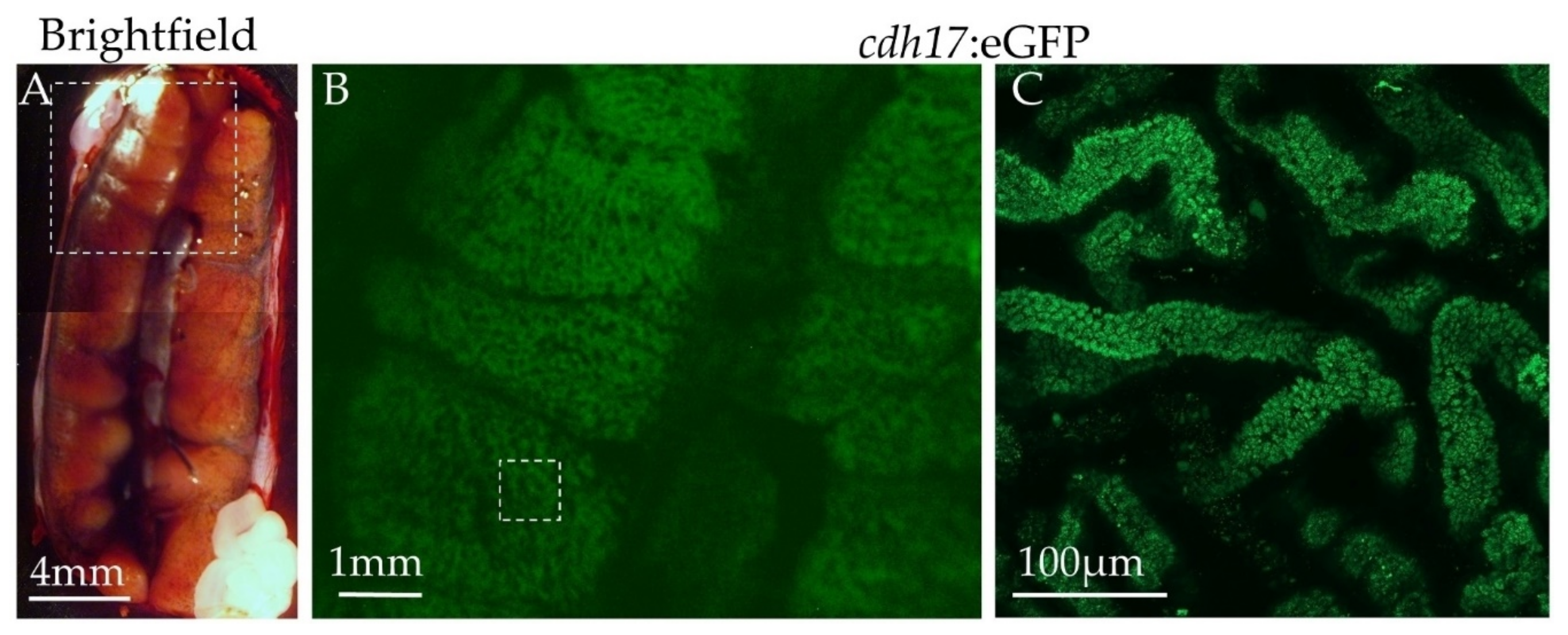
3.6. Expression of Green Fluorescent Protein in Adult Animals
3.7. Utility of cdh17:eGFP for Identifying Genes Involved in Kidney Diseases and Development
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Tomlinson, M.L.; Field, R.A.; Wheeler, G.N. Xenopus as a model organism in developmental chemical genetic screens. Mol. BioSyst. 2005, 1, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Lienkamp, S.S. Vertebrate kidney tubules elongate using a planar cell polarity-dependent rosette-based mechanism of convergent extension. Nat. Genet. 2012, 44, 1382–1387. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, G.N.; Brändli, A.W. Simple vertebrate models for chemical genetics and drug discovery screens: Lessons from zebrafish and Xenopus. Dev. Dyn. 2009, 238, 1287–1308. [Google Scholar] [CrossRef] [PubMed]
- Karpinka, J.B.; Fortriede, J.D.; Burns, K.A.; James-Zorn, C.; Ponferrada, V.G.; Lee, J.; Karimi, K.; Zorn, A.M.; Vize, P.D. Xenbase, the Xenopus model organism database; new virtualized system, data types and genomes. Nucleic Acids Res. 2015, 43, D756–D763. [Google Scholar] [CrossRef] [PubMed]
- Bauer, D.V.; Huang, S.; Moody, S.A. The cleavage stage origin of Spemann’s Organizer: Analysis of the movements of blastomere clones before and during gastrulation in Xenopus. Development 1994, 120, 1179–1189. [Google Scholar] [PubMed]
- Moody, S.A. Fates of the blastomeres of the 16-cell stage Xenopus embryo. Dev. Biol. 1987, 119, 560–578. [Google Scholar] [CrossRef]
- Moody, S.A. Fates of the blastomeres of the 32-cell-stage Xenopus embryo. Dev. Biol. 1987, 122, 300–319. [Google Scholar] [CrossRef]
- Moody, S.A.; Kline, M.J. Segregation of fate during cleavage of frog (Xenopus laevis) blastomeres. Anat. Embryol. 1990, 182, 347–362. [Google Scholar] [CrossRef] [PubMed]
- DeLay, B.D.; Krneta-Stankic, V.; Miller, R.K. Technique to target microinjection to the developing Xenopus kidney. J. Vis. Exp. 2016. [Google Scholar] [CrossRef] [PubMed]
- Vize, P.D.; Seufert, D.W.; Carroll, T.J.; Wallingford, J.B. Model systems for the study of kidney development: Use of the pronephros in the analysis of organ induction and patterning. Dev. Biol. 1997, 188, 189–204. [Google Scholar] [CrossRef] [PubMed]
- Dressler, G.R. The cellular basis of kidney development. Annu. Rev. Cell Dev. Biol. 2006, 22, 509–529. [Google Scholar] [CrossRef] [PubMed]
- Carroll, T.; Vize, P.D. Synergism between pax-8 and lim-1 in embryonic kidney development. Dev. Biol. 1999, 214, 46–59. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Vize, P.D. Proximo-distal specialization of epithelial transport processes within the Xenopus pronephric tubules. Dev. Biol. 2004, 271, 322–338. [Google Scholar] [CrossRef] [PubMed]
- Raciti, D.; Reggiani, L.; Geffers, L.; Jiang, Q.; Bacchion, F.; Subrizi, A.E.; Clements, D.; Tindal, C.; Davidson, D.R.; Kaissling, B.; et al. Organization of the pronephric kidney revealed by large-scale gene expression mapping. Genome Biol. 2008, 9, R84. [Google Scholar] [CrossRef] [PubMed]
- Brandli, A.W. Towards a molecular anatomy of the Xenopus pronephric kidney. Int. J. Dev. Biol. 1999, 43, 381–395. [Google Scholar] [PubMed]
- Hensey, C.; Dolan, V.; Brady, H.R. The Xenopus pronephros as a model system for the study of kidney development and pathophysiology. Nephrol. Dial. Transplant. 2002, 17, 73–74. [Google Scholar] [CrossRef] [PubMed]
- Droz, S.T.; McLaughlin, K.A. Use of Xenopus Frogs to study Renal Development/Repair. In Kidney Development and Disease; Kubiak, J.Z., Kloc, M., Eds.; Springer: Berlin, Germany, 2017; Volume 60, Chapter 4. [Google Scholar]
- Vize, P.; Carroll, T.; Wallingford, J. Induction, Development, and Physiology of the Pronephric Tubules The Kidney: From Normal Development to Congenital Disease; Elsevier: Amsterdam, The Netherlands, 2003. [Google Scholar]
- Nieuwkoop, P.D.; Faber, J. Normal Table of Xenopus laevis (Daudin): A Systematical & Chronological Survey of the Development from the Fertilized Egg Till the End of Metamorphosis; Garland Publishing, Inc.: New York, NY, USA, 1994. [Google Scholar]
- Buisson, I.; le Bouffant, R.; Futel, M.; Riou, J.F.; Umbhauer, M. Pax8 and Pax2 are specifically required at different steps of Xenopus pronephros development. Dev. Biol. 2015, 397, 175–190. [Google Scholar] [CrossRef] [PubMed]
- Lienkamp, S.S. Using Xenopus to study genetic kidney diseases. Semin. Cell Dev. Biol. 2016, 51, 117–124. [Google Scholar] [CrossRef] [PubMed]
- DeLay, B.D.; Corkins, M.E.; Hanania, H.L.; Salanga, M.; Deng, J.M.; Sudou, N.; Taira, M.; Horb, M.E.; Miller, R.K. Tissue-specific gene inactivation in Xenopus laevis: Knockout of lhx1 in the kidney with CRISPR/Cas9. Genetics 2018, 208, 673–686. [Google Scholar] [CrossRef] [PubMed]
- Barth, L.G.; Barth, L.J. Differentiation of cells of the Rana pipiens gastrula in unconditioned medium. J. Embryol. Exp. Morphol. 1959, 7, 210–222. [Google Scholar] [PubMed]
- Krneta-Stankic, V.; DeLay, B.D.; Miller, R.K. Xenopus: Leaping forward in kidney organogenesis. Pediatr. Nephrol. 2017, 32, 547–555. [Google Scholar] [CrossRef] [PubMed]
- Pan, F.C.; Chen, Y.; Loeber, J.; Henningfeld, K.; Pieler, T. I-SceI meganuclease-mediated transgenesis in Xenopus. Dev. Dyn. 2006, 235, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Ogino, H.; McConnell, W.B.; Grainger, R.M. High-throughput transgenesis in Xenopus using I-SceI meganuclease. Nat. Protoc. 2006, 1, 1703–1710. [Google Scholar] [CrossRef] [PubMed]
- Diep, C.Q.; Ma, D.; Deo, R.C.; Holm, T.M.; Naylor, R.W.; Arora, N.; Wingert, R.A.; Bollig, F.; Djordjevic, G.; Lichman, B.; et al. Identification of adult nephron progenitors capable of kidney regeneration in zebrafish. Nature 2011, 470, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Boucher, R.C.; Bollig, F.; Englert, C.; Hildebrandt, F. Characterization of mesonephric development and regeneration using transgenic zebrafish. Am. J. Physiol. Renal Physiol. 2010, 299, F1040–F1047. [Google Scholar] [CrossRef] [PubMed]
- Sive, H.L.; Grainger, R.M.; Harland, R.M. Early Development of Xenopus laevis: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2000. [Google Scholar]
- Lyons, J.P.; Miller, R.K.; Zhou, X.; Weidinger, G.; Deroo, T.; Park, J.I.; Ji, H.; Hong, J.Y.; Li, A.; Moon, R.T.; et al. Requirement of Wnt/β-catenin signaling in pronephric kidney development. Mech. Dev. 2009, 126, 142–159. [Google Scholar] [CrossRef] [PubMed]
- Vize, P.D.; Jones, E.A.; Pfister, R. Development of the Xenopus pronephric system. Dev. Biol. 1995, 171, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Wallingford, J.B. Preparation of fixed Xenopus embryos for confocal imaging. Cold Spring Harb. Protoc. 2010. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.D.; Davidson, L.A. Live-cell imaging and quantitative analysis of embryonic epithelial cells in Xenopus laevis. J. Vis. Exp. 2010. [Google Scholar] [CrossRef] [PubMed]
- Sive, H.L.; Grainger, R.M.; Harland, R.M. Xenopus laevis Keller Explants. Cold Spring Harb. Protoc. 2007. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.K.; Canny, S.G.; Jang, C.W.; Cho, K.; Ji, H.; Wagner, D.S.; Jones, E.A.; Habas, R.; McCrea, P.D. Pronephric tubulogenesis requires Daam1-mediated planar cell polarity signaling. J. Am. Soc. Nephrol. 2011, 22, 1654–1664. [Google Scholar] [CrossRef] [PubMed]
- Horsfield, J.; Ramachandran, A.; Reuter, K.; LaVallie, E.; Collins-Racie, L.; Crosier, K.; Crosier, P. Cadherin-17 is required to maintain pronephric duct integrity during zebrafish development. Mech. Dev. 2002, 115, 15–26. [Google Scholar] [CrossRef]
- Miller, R.K.; Lee, M.; McCrea, P.D. The Xenopus Pronephros: A Kidney Model Making Leaps toward Understanding Tubule Development. In Xenopus Development; Wiley Blackwell: Hoboken, NJ, USA, 2014; Chapter 12. [Google Scholar]
- Tandon, P.; Showell, C.; Christine, K.; Conlon, F.L. Morpholino injection in Xenopus. Methods Mol. Biol. 2012, 843, 29–46. [Google Scholar] [PubMed]
- Tandon, P.; Conlon, F.; Furlow, J.D.; Horb, M.E. Expanding the genetic toolkit in Xenopus: Approaches and opportunities for human disease modeling. Dev. Biol. 2017, 426, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Hellsten, U.; Harland, R.M.; Gilchrist, M.J.; Hendrix, D.; Jurka, J.; Kapitonov, V.; Ovcharenko, I.; Putnam, N.H.; Shu, S.; Taher, L.; et al. The genome of the Western clawed frog Xenopus tropicalis. Science 2010, 328, 633–636. [Google Scholar] [CrossRef] [PubMed]
- Mizell, M.; Tulane University; Cancer Association of Greater New Orleans; National Cancer Institute (U.S.). Biology of Amphibian Tumors (Recent Results in Cancer Research Special Supplement); Springer: New York, NY, USA, 1969; 484p.
- Shiina, N.; Tsukita, S. Mutations at phosphorylation sites of Xenopus microtubule-associated protein 4 affect its microtubule-binding ability and chromosome movement during mitosis. Mol. Biol. Cell 1999, 10, 597–608. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Gordillo, D.; Trujillo-Provencio, C.; Knight, V.B.; Serrano, E.E. Optimization of gene delivery methods in Xenopus laevis kidney (A6) and Chinese hamster ovary (CHO) cell lines for heterologous expression of Xenopus inner ear genes. In Vitro Cell. Dev. Biol. Anim. 2011, 47, 640–652. [Google Scholar] [CrossRef] [PubMed]
- Fox, M.A.; Nieuwesteeg, M.A.; Willson, J.A.; Cepeda, M.; Damjanovski, S. Knockdown of Pex11β reveals its pivotal role in regulating peroxisomal genes, numbers, and ROS levels in Xenopus laevis A6 cells. In Vitro Cell. Dev. Biol. Anim. 2014, 50, 340–349. [Google Scholar] [CrossRef] [PubMed]






© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corkins, M.E.; Hanania, H.L.; Krneta-Stankic, V.; DeLay, B.D.; Pearl, E.J.; Lee, M.; Ji, H.; Davidson, A.J.; Horb, M.E.; Miller, R.K. Transgenic Xenopus laevis Line for In Vivo Labeling of Nephrons within the Kidney. Genes 2018, 9, 197. https://doi.org/10.3390/genes9040197
Corkins ME, Hanania HL, Krneta-Stankic V, DeLay BD, Pearl EJ, Lee M, Ji H, Davidson AJ, Horb ME, Miller RK. Transgenic Xenopus laevis Line for In Vivo Labeling of Nephrons within the Kidney. Genes. 2018; 9(4):197. https://doi.org/10.3390/genes9040197
Chicago/Turabian StyleCorkins, Mark E., Hannah L. Hanania, Vanja Krneta-Stankic, Bridget D. DeLay, Esther J. Pearl, Moonsup Lee, Hong Ji, Alan J. Davidson, Marko E. Horb, and Rachel K. Miller. 2018. "Transgenic Xenopus laevis Line for In Vivo Labeling of Nephrons within the Kidney" Genes 9, no. 4: 197. https://doi.org/10.3390/genes9040197
APA StyleCorkins, M. E., Hanania, H. L., Krneta-Stankic, V., DeLay, B. D., Pearl, E. J., Lee, M., Ji, H., Davidson, A. J., Horb, M. E., & Miller, R. K. (2018). Transgenic Xenopus laevis Line for In Vivo Labeling of Nephrons within the Kidney. Genes, 9(4), 197. https://doi.org/10.3390/genes9040197





