From Chromosomes to Genome: Insights into the Evolutionary Relationships and Biogeography of Old World Knifefishes (Notopteridae; Osteoglossiformes)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals, Chromosome Preparations, and Bandings
2.2. Probes and Fluorescence In Situ Hybridization.
2.3. Image Processing
2.4. DNA Extraction and Genotyping by Sequencing
2.5. Analysis of Genetic Diversity between Species
2.6. Feature Annotation of Allele Sequences
3. Results
3.1. Karyotypes and Chromosome Bandings
3.2. Telomere (TTAGGG)n and Ribosomal DNA (5S and 18S Ribosomal DNA) Sequence Mapping
3.3. Chromosome Mapping of Microsatellite Motif Sequences
3.4. Feature Annotation of Diversity Arrays Technology Sequencing Markers
3.5. Comparative Analyses Using Diversity Arrays Technology Sequencing Data
4. Discussion
4.1. Chromosomal Evolution in Notopteridae
4.2. Genetic Variability among Notopterids
4.3. Hypotheses on Biogeographical History of Notopteridae in Light of Our Data
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bănărescu, P. Zoogeography of Fresh Waters. General Distribution and Dispersal of Freshwater Animals, 1st ed.; Aula-Verlag: Wiesbaden, Germany, 1990; ISBN 3891044801. [Google Scholar]
- Greenwood, P.H.; Wilson, M.V.; Paxton, J.R.; Eschmeyer, W.N. Encyclopedia of Fishes; Academic Press: San Diego, CA, USA, 1998. [Google Scholar]
- Hilton, E.J. Comparative osteology and phylogenetic systematics of fossil and living bony-tongue fishes (Actinopterygii, Teleostei, Osteoglossomorpha). Zool. J. Linn. Soc. 2002, 137, 1–100. [Google Scholar] [CrossRef] [Green Version]
- Lavoué, S. Was Gondwanan breakup the cause of the intercontinental distribution of Osteoglossiformes? A time-calibrated phylogenetic test combining molecular, morphological, and paleontological evidence. Mol. Phylogenet. Evol. 2016, 99, 34–43. [Google Scholar] [PubMed]
- Vidthayanon, C. Thailand Red Data: Fishes; Office of Natural Resources and Environmental Policy and Planning: Bangkok, Thailand, 2005; ISBN 974992987X.
- Wilson, M.V.H.; Murray, A.M. Osteoglossomorpha: Phylogeny, biogeography, and fossil record and the significance of key African and Chinese fossil taxa. Geol. Soc. Lond. Spec. Publ. 2008, 295, 185–219. [Google Scholar] [CrossRef]
- Roberts, T.R. Systematic revision of the old world freshwater fish family Notopteridae. Ichthyol. Explor. Freshw. 1992, 2, 361–383. [Google Scholar]
- Nelson, J.S.; Grande, T.C.; Wilson, M.V.H. Fishes of the World; John Wiley & Sons: Hoboken, NJ, USA, 2016; ISBN 111834233X. [Google Scholar]
- Lavoué, S.; Sullivan, J.P. Simultaneous analysis of five molecular markers provides a well-supported phylogenetic hypothesis for the living bony-tongue fishes (Osteoglossomorpha: Teleostei). Mol. Phylogenet. Evol. 2004, 33, 171–185. [Google Scholar] [CrossRef] [PubMed]
- Inoue, J.G.; Kumazawa, Y.; Miya, M.; Nishida, M. The historical biogeography of the freshwater knifefishes using mitogenomic approaches: A Mesozoic origin of the Asian notopterids (Actinopterygii: Osteoglossomorpha). Mol. Phylogenet. Evol. 2009, 51, 486–499. [Google Scholar] [CrossRef] [PubMed]
- Taverne, L.; Maisey, J.G. A Notopterid Skull (Teleostei, Osteoglossomorpha) from the Continental Early Cretaceous of Southern Morocco; American Museum Novitates No. 3260; American Museum of Natural History: New York, NY, USA, 1999. [Google Scholar]
- Cavin, L.; Forey, P.L. Osteology and systematic affinities of Palaeonotopterus greenwoodi Forey 1997 (Teleostei: Osteoglossomorpha). Zool. J. Linn. Soc. 2001, 133, 25–52. [Google Scholar] [CrossRef]
- DeConto, R.M.; Wold, C.N.; Wilson, K.M.; Voigt, S.; Schulz, M.; Wold, A.R.; Dullo, W.-C.; Ronov, A.B.; Balukhovsky, A.N.; Soding, E. Alternative global Cretaceous paleogeography. Evol. Cretac. Ocean.-Clim. Syst. 1999, 332, 1–435. [Google Scholar] [CrossRef]
- Bănărescu, P. Volume 2: Distribution and dispersal of freshwater animals in North America and Eurasia. In Zoogeography of Fresh Waters; Aula-Verlag: Wiesbaden, Germany, 1991; ISBN 3891044828. [Google Scholar]
- Rögl, F. Palaeogeographic considerations for Mediterranean and Paratethys seaways (Oligocene to Miocene). Ann. Naturhistorischen Mus. 1997, 99A, 279–310. [Google Scholar]
- Myers, G.S. Salt-tolerance of fresh-water fish groups in relation to zoogeographical problems. Bijdr. Dierkd. 1949, 28, 315–322. [Google Scholar]
- Sanders, M. Die Fossilen Fische der Alttertiären Süsswasserablagerungen aus Mittel-Sumatra; Mouton: Berlin, Germany, 1934. [Google Scholar]
- Kumazawa, Y.; Nishida, M. Molecular phylogeny of osteoglossoids: A new model for Gondwanian origin and plate tectonic transportation of the Asian arowana. Mol. Biol. Evol. 2000, 17, 1869–1878. [Google Scholar] [CrossRef] [PubMed]
- Ráb, P.; Yano, C.F.; Lavoué, S.; Jegede, O.I.; Bertollo, L.A.C.; Ezaz, T.; Majtánová, Z.; de Oliveira, E.A.; Cioffi, M.B. Karyotype and mapping of repetitive DNAs in the African butterfly fish Pantodon buchholzi, the sole species of the family Pantodontidae. Cytogenet. Genome Res. 2016, 149, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Canitz, J.; Kirschbaum, F.; Tiedemann, R. Karyotype description of the African weakly electric fish Campylomormyrus compressirostris in the context of chromosome evolution in Osteoglossiformes. J. Physiol. 2016, 110, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Nayyar, R.P. Karyotype studies in the genus Notopterus (Lacepede). the occurrence and fate of univalent chromosomes in spermatocytes of N. Chitala. Genetica 1965, 36, 398–406. [Google Scholar] [PubMed]
- Uyeno, T. A comparative study of chromosomes in the teleostean fish order Osteoglossiformes. Jpn. J. Ichthyol. 1973, 20, 211–217. [Google Scholar]
- Takai, A.; Ojima, Y. C-banded karyotype and nucleolus organizer regions of a notopterid fish, Notopterus chitala (Notopteridae, Osteoglossiformes). Chromosome Sci. 1998, 2, 35–38. [Google Scholar]
- Rishi, K.K.; Singh, J. Chromosomes of Notopterus notopterus (Pallas) (Notopteridae: Clupeiformes). Chromosome Inf. Serv. 1983, 34, 9–10. [Google Scholar]
- Srivastava, M.D.; Kaur, P. The structure and behaviour of chromosomes in six freshwater Teleosts. Cellule 1964, 65, 93–107. [Google Scholar] [PubMed]
- Urushido, T. Karyotype of three species of fishes in the order Osteoglossiformes. Chromosome Inf. Serv. 1975, 18, 20–22. [Google Scholar]
- Donsakul, T.; Magtoon, W. A chromosome study on three species of featherbacks, Notopterus chitala (Hamilton), N. bland D’Aubenton and N. notopterus (Pallas), from Thailand. In Proceedings of the 28th Kasetsart University Conference, Bangkok, Thailand, 29–31 January 1990; pp. 29–31. [Google Scholar]
- Silawong, K.; Aoki, S.; Supiwong, W.; Tanomtong, A.; Khakhong, S.; Sanoamuang, L. The first chromosomal characteristics of nucleolar organizer regions (NORs) in grey featherback fish, Notopterus notopterus (Osteoglossiformes, Notopteridae) by conventional and Ag-NOR staining techniques. Cytologia 2012, 77, 279–285. [Google Scholar] [CrossRef]
- Supiwong, W.; Tanomtong, A.; Khakhong, S.; Silawong, K.; Aoki, S.; Sanoamuang, L. The first chromosomal characteristics of nucleolar organizer regions and karyological analysis of clown knife fish, Chitala ornata (Osteoglossiformes, Notopteridae) by T-lymphocyte cell culture. Cytologia 2012, 77, 393–399. [Google Scholar] [CrossRef]
- Bertollo, L.A.C.; Cioffi, M.B.; Moreira-Filho, O. Direct chromosome preparation from Freshwater Teleost Fishes. In Fish Cytogenetic Techniques (Chondrichthyans and Teleosts); Ozouf-Costaz, C., Pisano, E., Foresti, F., Almeida Toledo, L.F., Eds.; Enfield/CRC Press: Boca Raton, FL, USA, 2015. [Google Scholar]
- Sumner, A.T. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 1972, 75, 204–206. [Google Scholar] [CrossRef]
- Howell, W.M.; Black, D.A. Controlled silver staining of nucleolus organizer regions with a protective colloidal developer: A 1-step method. Experientia 1980, 36, 1014–1015. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M. Chromosome banding in Amphibia. IV. Differentiation of GC-and AT-rich chromosome regions in Anura. Chromosoma 1980, 77, 83–103. [Google Scholar] [CrossRef] [PubMed]
- Martins, C.; Ferreira, I.A.; Oliveira, C.; Foresti, F.; Galetti, P.M. A tandemly repetitive centromeric DNA sequence of the fish Hoplias malabaricus (Characiformes: Erythrinidae) is derived from 5S rDNA. Genetica 2006, 127, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Cioffi, M.B.; Martins, C.; Bertollo, L.A.C. Comparative chromosome mapping of repetitive sequences. Implications for genomic evolution in the fish, Hoplias malabaricus. BMC Genet. 2009, 10, 34. [Google Scholar] [CrossRef] [PubMed]
- Kubat, Z.; Hobza, R.; Vyskot, B.; Kejnovsky, E. Microsatellite accumulation in the Y chromosome of Silene latifolia. Genome 2008, 51, 350–356. [Google Scholar] [CrossRef] [PubMed]
- Ijdo, J.W.; Wells, R.A.; Baldini, A.; Reeders, S.T. Improved telomere detection using a telomere repeat probe (TTAGGG)n generated by PCR. Nucleic Acids Res. 1991, 19, 4780. [Google Scholar] [CrossRef] [PubMed]
- Pinkel, D.; Straume, T.; Gray, J. Cytogenetic analysis using quantitative, high sensitivity, fluorescence hybridization. Proc. Natl. Acad. Sci. USA 1986, 83, 2934–2938. [Google Scholar] [CrossRef] [PubMed]
- Levan, A.; Fredga, K.; Sandberg, A.A. Nomenclature for centromeric position on chromosomes. Hereditas 1964, 52, 201–220. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning, A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Grewe, P.M.; Feutry, P.; Hill, P.L.; Gunasekera, R.M.; Schaefer, K.M.; Itano, D.G.; Fuller, D.W.; Foster, S.D.; Davies, C.R. Evidence of discrete yellowfin tuna (Thunnus albacares) populations demands rethink of management for this globally important resource. Sci. Rep. 2015, 5, 16916. [Google Scholar] [CrossRef] [PubMed]
- Kilian, A.; Wenzl, P.; Huttner, E.; Carling, J.; Xia, L.; Blois, H.; Caig, V.; Heller-Uszynska, K.; Jaccoud, D.; Hopper, C. Diversity arrays technology: A generic genome profiling technology on open platforms. In Data Production and Analysis in Population Genomics; Springer: Berlin/Heidelberg, Germany, 2012; pp. 67–89. [Google Scholar]
- Lambert, M.R.; Skelly, D.K.; Ezaz, T. Sex-linked markers in the North American green frog (Rana clamitans) developed using DArTseq provide early insight into sex chromosome evolution. BMC Genom. 2016, 17, 844. [Google Scholar] [CrossRef] [PubMed]
- Dice, L.R. Measures of the amount of ecologic association between species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R package for multivariate analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Suzuki, R.; Shimodaira, H. Pvclust: An R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 2006, 22, 1540–1542. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [PubMed]
- Rambaut, A. FigTree Version 1.3.1. Computer Program. Available online: http//tree.bio.ed.ac.uk/software/figtree/ (accessed on 15 August 2009).
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Ribeiro, J.M.C.; Alarcon-Chaidez, F.; Francischetti, I.M.B.; Mans, B.J.; Mather, T.N.; Valenzuela, J.G.; Wikel, S.K. An annotated catalog of salivary gland transcripts from Ixodes scapularis ticks. Insect Biochem. Mol. Biol. 2006, 36, 111–129. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2015, 44, D733–D745. [Google Scholar] [CrossRef] [PubMed]
- Obermiller, L.E.; Pfeiler, E. Phylogenetic relationships of elopomorph fishes inferred from mitochondrial ribosomal DNA sequences. Mol. Phylogenet. Evol. 2003, 26, 202–214. [Google Scholar] [CrossRef]
- Chen, W.; Lavoué, S.; Mayden, R.L. Evolutionary origin and early biogeography of otophysan fishes (Ostariophysi: Teleostei). Evolution 2013, 67, 2218–2239. [Google Scholar] [CrossRef] [PubMed]
- Sallan, L.C. Major issues in the origins of ray-finned fish (Actinopterygii) biodiversity. Biol. Rev. 2014, 89, 950–971. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S.; Atkin, N.B. Comparative DNA values and chromosome complements of eight species of fishes. Chromosoma 1966, 18, 455–466. [Google Scholar] [CrossRef] [PubMed]
- López-Flores, I.; Garrido-Ramos, M.A. Repetitive DNA; Garrido-Ramos, M.A., Ed.; Karger: Basel, Switzerland, 2012. [Google Scholar]
- Gornung, E. Twenty years of physical mapping of major ribosomal RNA genes across the teleosts: A review of research. Cytogenet. Genome Res. 2013, 141, 90–102. [Google Scholar] [CrossRef] [PubMed]
- Marques, D.K.; Venere, P.C.; Galetti Junior, P.M. Chromosomal characterization of the bonytongue Arapaima gigas (Osteoglossiformes: Arapaimidae). Neotrop. Ichthyol. 2006, 4, 215–218. [Google Scholar] [CrossRef]
- Da Rosa, R.; Rubert, M.; Caetano-Filho, M.; Giuliano-Caetano, L. Conserved cytogenetic features in the Amazonian arapaima, Arapaima gigas (Schinz 1822) from Jamari river, Rondonia-Brazil. Open Biol. J. 2009, 2, 91–94. [Google Scholar] [CrossRef]
- Ozouf-Costaz, C.; Coutanceau, J.-P.; BOnillO, C.; Belkadi, L.; Fermon, Y.; Agnèse, J.-F.; Guidi-Rontani, C.; Paugy, D. First insights into karyotype evolution within the family Mormyridae. Cybium 2015, 39, 227–236. [Google Scholar]
- Majtánová, Z.; Symonová, R.; Arias-Rodriguez, L.; Sallan, L.; Ráb, P. “Holostei versus Halecostomi” problem: Insight from cytogenetics of ancient nonteleost actinopterygian fish, bowfin Amia calva. J. Exp. Zool. Part B Mol. Dev. Evol. 2017, 328, 620–628. [Google Scholar] [CrossRef] [PubMed]
- Symonová, R.; Majtánová, Z.; Arias-Rodriguez, L.; Mořkovský, L.; Kořínková, T.; Cavin, L.; Pokorná, M.J.; Doležálková, M.; Flajšhans, M.; Normandeau, E.; et al. Genome compositional organization in gars shows more similarities to mammals than to other ray-finned fish. J. Exp. Zool. Part B Mol. Dev. Evol. 2017, 328, 607–619. [Google Scholar]
- Mayr, B.; Kalat, M.; Ràb, P. Localization of NORs and counterstain enhanced fluorescence studies in Perca fluviatilis (Pisces, Percidae). Genetica 1985, 67, 51–56. [Google Scholar] [CrossRef]
- Amemiya, C.T.; Gold, J.R. Chromomycin A 3 stains nucleolus organizer regions of fish chromosomes. Copeia 1986, 1986, 226–231. [Google Scholar] [CrossRef]
- Schmid, M.; Guttenbach, M. Evolutionary diversity of reverse (R) fluorescent chromosome bands in vertebrates. Chromosoma 1988, 97, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Fontana, F.; Lanfredi, M.; Congiu, L.; Tagliavini, J.; Rossi, R. Fluorescent in situ hybridization with rDNA probes on chromosomes of Acipenser ruthenus and Acipenser naccarii (Osteichthyes Acipenseriformes). Genome 1999, 42, 1008–1012. [Google Scholar]
- Inafuku, J.; Nabeyama, M.; Kikuma, Y.; Saitoh, J.; Kubota, S.; Kohno, S. Chromosomal location and nucleotide sequences of 5S ribosomal DNA of two cyprinid species (Osteichthyes, Pisces). Chromosome Res. 2000, 8, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Fontana, F.; Lanfredi, M.; Congiu, L.; Leis, M.; Chicca, M.; Rossi, R. Chromosomal mapping of 18S–28S and 5S rRNA genes by two-colour fluorescent in situ hybridization in six sturgeon species. Genome 2003, 46, 473–477. [Google Scholar] [CrossRef] [PubMed]
- Tigano, C.; Rocco, L.; Ferrito, V.; Costagliola, D.; Pappalardo, A.M.; Stingo, V. Chromosomal mapping and molecular characterization of ribosomal RNA genes in Lebias fasciata (Teleostei, Cyprinodontidae). Genetica 2004, 121, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Cioffi, M.B.; Martins, C.; Vicari, M.R.; Rebordinos, L.; Bertollo, L.A.C. Differentiation of the XY sex chromosomes in the fish Hoplias malabaricus (Characiformes, Erythrinidae): Unusual accumulation of repetitive sequences on the X chromosome. Sex. Dev. 2010, 4, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Martins, N.F.; Bertollo, L.A.C.; Troy, W.P.; Feldberg, E.; de Souza Valentin, F.C.; de Bello Cioffi, M. Differentiation and evolutionary relationships in Erythrinus erythrinus (Characiformes, Erythrinidae): Comparative chromosome mapping of repetitive sequences. Rev. Fish Biol. Fish. 2013, 23, 261–269. [Google Scholar] [CrossRef]
- Marquioni, V.; Bertollo, L.A.C.; Diniz, D.; de Bello Cioffi, M. Comparative chromosomal mapping in Triportheus fish species. Analysis of synteny between ribosomal genes. Micron 2013, 45, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Yano, C.F.; Bertollo, L.A.C.; Ezaz, T.; Trifonov, V.; Sember, A.; Liehr, T.; Cioffi, M.B. Highly conserved Z and molecularly diverged W chromosomes in the fish genus Triportheus (Characiformes, Triportheidae). Heredity 2017, 118, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Kidwell, M.G. Transposable elements and the evolution of genome size in eukaryotes. Genetica 2002, 115, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Cioffi, M.B.; Bertollo, L.A.C. Chromosomal distribution and evolution of repetitive DNAs in fish. In Repetitive DNAs; Garrido, R., Ed.; Karger: Basel, Switzerland, 2012. [Google Scholar]
- Scotese, C.R. Atlas of Early Cretaceous Paleogeographic Maps, PALEOMAP Atlas for ArcGIS. The Cretaceous, Maps 23–31; Mollweide Projection: Evanston, IL, USA, 2014; Volume 2. [Google Scholar]
- Scotese, C.R. Atlas of Late Cretaceous Paleogeographic Maps, PALEOMAP Atlas for ArcGIS. The Cretaceous, Maps 16–22; Mollweide Projection: Evanston, IL, USA, 2014; Volume 2. [Google Scholar]
- Molina, W.F. Chromosomal changes and stasis in marine fish groups. In Fish Cytogenetics; Taylor Francis Group: Boca Raton, FL, USA, 2007; pp. 69–110. [Google Scholar]
- Steane, D.A.; Nicolle, D.; Sansaloni, C.P.; Petroli, C.D.; Carling, J.; Kilian, A.; Myburg, A.A.; Grattapaglia, D.; Vaillancourt, R.E. Population genetic analysis and phylogeny reconstruction in Eucalyptus (Myrtaceae) using high-throughput, genome-wide genotyping. Mol. Phylogenet. Evol. 2011, 59, 206–224. [Google Scholar] [CrossRef] [PubMed]
- Resende, M.D.V.; Resende, M.F.R.; Sansaloni, C.P.; Petroli, C.D.; Missiaggia, A.A.; Aguiar, A.M.; Abad, J.M.; Takahashi, E.K.; Rosado, A.M.; Faria, D.A. Genomic selection for growth and wood quality in Eucalyptus: Capturing the missing heritability and accelerating breeding for complex traits in forest trees. New Phytol. 2012, 194, 116–128. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Sevilla, J.F.; Horvath, A.; Botella, M.A.; Gaston, A.; Folta, K.; Kilian, A.; Denoyes, B.; Amaya, I. Diversity Arrays Technology (DArT) marker platforms for diversity analysis and linkage mapping in a complex crop, the octoploid cultivated strawberry (Fragaria × ananassa). PLoS ONE 2015, 10, e0144960. [Google Scholar] [CrossRef] [PubMed]
- Brandolini, A.; Volante, A.; Heun, M. Geographic differentiation of domesticated einkorn wheat and possible Neolithic migration routes. Heredity 2016, 117, 135–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, J.R.; Aitchison, J.C. Gondwana to Asia: Plate tectonics, paleogeography and the biological connectivity of the Indian sub-continent from the Middle Jurassic through latest Eocene (166–35 Ma). Earth-Sci. Rev. 2008, 88, 145–166. [Google Scholar]
- Agnarsson, I.; Kuntner, M. The generation of a biodiversity hotspot: Biogeography and phylogeography of the western Indian Ocean islands. In Current Topics in Phylogenetics and Phylogeography of Terrestrial and Aquatic Systems; InTech: Rijeka, Croatia, 2012. [Google Scholar]
- Vences, M.; Freyhof, J.; Sonnenberg, R.; Kosuch, J.; Veith, M. Reconciling fossils and molecules: Cenozoic divergence of cichlid fishes and the biogeography of Madagascar. J. Biogeogr. 2001, 28, 1091–1099. [Google Scholar] [CrossRef] [Green Version]
- Yoder, A.D.; Nowak, M.D. Has vicariance or dispersal been the predominant biogeographic force in Madagascar? Only time will tell. Annu. Rev. Ecol. Evol. Syst. 2006, 37, 405–431. [Google Scholar] [CrossRef]
- Kuntner, M.; Agnarsson, I. Phylogeography of a successful aerial disperser: The golden orb spider Nephila on Indian Ocean islands. BMC Evol. Biol. 2011, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Kuntner, M.; Agnarsson, I. Biogeography and diversification of hermit spiders on Indian Ocean islands (Nephilidae: Nephilengys). Mol. Phylogenet. Evol. 2011, 59, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Masters, J.C.; De Wit, M.J.; Asher, R.J. Reconciling the origins of Africa, India and Madagascar with vertebrate dispersal scenarios. Folia Primatol. 2006, 77, 399–418. [Google Scholar] [CrossRef] [PubMed]
- Schatz, G.E. Malagasy/Indo-Australo-Malesian Phytogeographic Connections; ORSTOM: Paris, France, 1996. [Google Scholar]
- Van Steenis, C.G.G.J. The land-bridge theory in botany with particular reference to tropical plants. Blumea Biodivers. Evol. Biogeogr. Plants 1962, 11, 235–372. [Google Scholar]
- Rage, J.-C. Relationships of the Malagasy fauna during the Late Cretaceous: Northern or Southern routes? Acta Palaeontol. Pol. 2003, 48, 661–662. [Google Scholar]
- Aitchison, J.C.; Ali, J.R.; Davis, A.M. When and where did India and Asia collide? J. Geophys. Res. Solid Earth 2007, 112. [Google Scholar] [CrossRef]
- Daniels, S.R. Reconstructing the colonisation and diversification history of the endemic freshwater crab (Seychellum alluaudi) in the granitic and volcanic Seychelles Archipelago. Mol. Phylogenet. Evol. 2011, 61, 534–542. [Google Scholar] [CrossRef] [PubMed]
- Arai, R. Fish Karyotypes: A Check List; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2011; ISBN 4431538771. [Google Scholar]
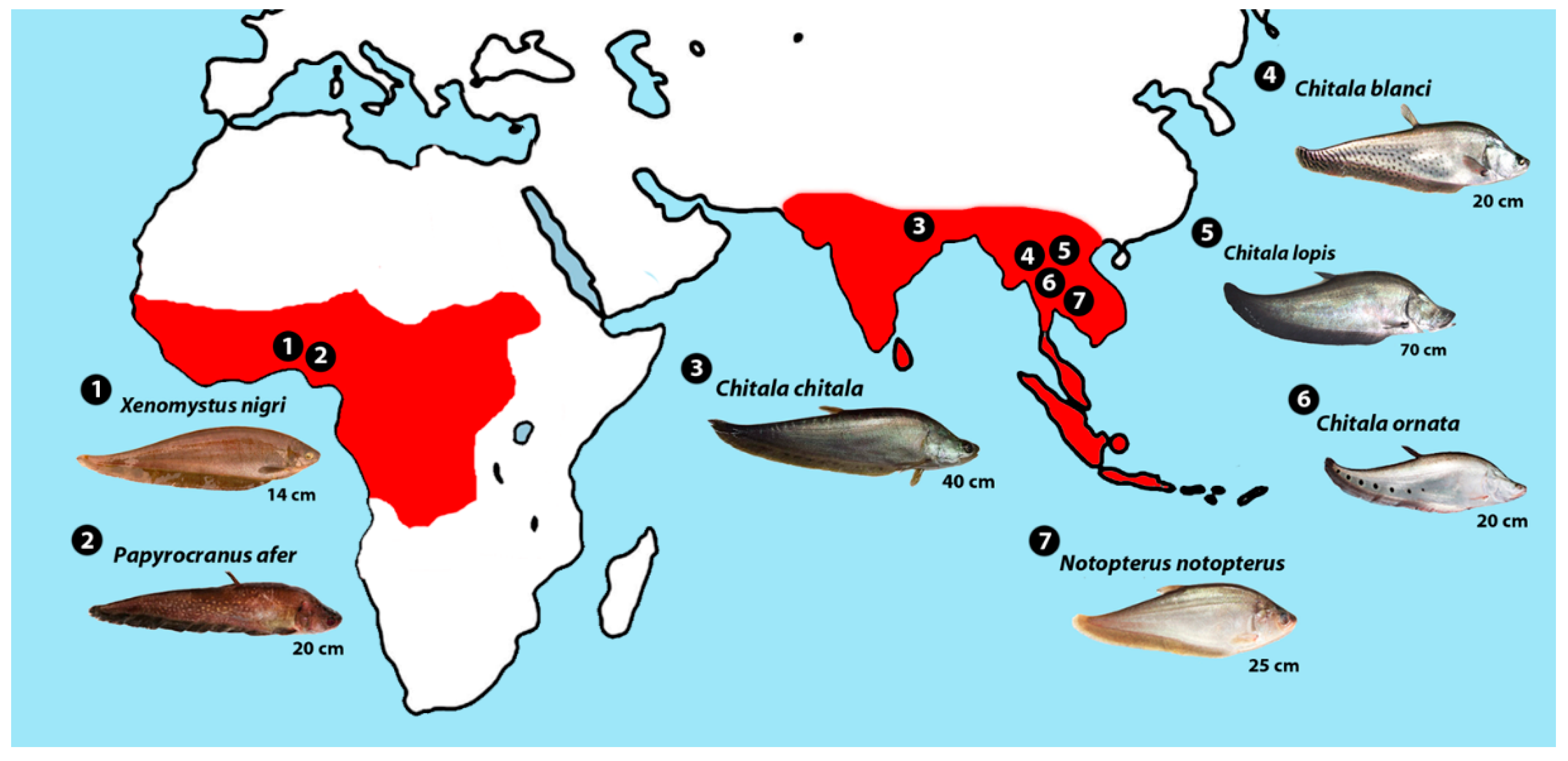









| Species | 2n | Karyotype | FN | Reference | Locality |
|---|---|---|---|---|---|
| Chitala chitala | 42 | 42a | 42 | Present study | Song Khram basin, Thailand |
| C. chitala | 48 | 12m + 36a | 60 | [21] | India, Delhi, River Jumna |
| C. chitala | 42 | 42a | 42 | [22] | Not known |
| C. chitala | 42 | 42a | 42 | [23] | Aquarium trade |
| Notopterus notopterus | 42 | 42a | 42 | Present study | Chi and Mekong basins, Thailand |
| N. notopterus | 42 | 42a | 42 | [24] | India, Kurukshetra |
| N. notopterus | 42 | 42a | 42 | [25] | Thailand |
| N. notopterus | 48 | 12m + 36a | 60 | [21] | India, Delhi, River Jumna |
| N. notopterus | 42 | 42a | 42 | [26] | Not known |
| N. notopterus | 42 | 42a | 42 | [27] | Thailand |
| N. notopterus | 42 | 42a | 42 | [28] | Thailand, Chi Basin. |
| Chitala ornata | 42 | 42a | 42 | Present Study | Chi and Mekong basins, Thailand |
| C. ornata | 42 | 42a | 42 | [27] | Thailand, Chi Basin |
| C. ornata | 42 | 42a | 42 | [29] | Thailand, Chi Basin |
| C. blanci | 42 | 42a | 42 | Present Study | Song Khram basin, Thailand |
| C. blanci | 42 | 42a | 42 | [27] | Thailand, Chi Basin |
| C. lopis | 38 | 38a | 38 | Present study | Song Khram basin, Thailand |
| Papyrocranus afer | 34 | 4sm + 30a | 38 | [22] | Africa |
| P. afer | 50 | 2m + 2sm + 36a | 54 | Present study | Oluwa River, Nigeria |
| Xenomystus nigri | 42 | 42a | 42 | [22] | Africa |
| X. nigri | 42 | 42a | 42 | Present Study | Oluwa River, Nigeria |
| Species | Sampling Sites | N |
|---|---|---|
| Chitala blanci | Song Khram basin, Thailand | (04 ♀; 04 ♂) |
| Chitala chitala | Ganges river, India | (05 ♀; 04 ♂) |
| Chitala lopis | Song Khram basin, Thailand | (12 ♀; 06 ♂) |
| Chitala ornate | Chi and Mekong basins, Thailand | (09 ♀; 07 ♂) |
| Notopterus notopterus | Chi and Mekong basins, Thailand | (06 ♀; 04 ♂) |
| Papyrocranus afer | Oluwa River, Nigeria | (19 ♀; 21 ♂) |
| Xenomystus nigri | Oluwa River, Nigeria | (13 ♀; 24 ♂) |
| Sequence Collections Retrieved from NCBI | Number of Representative Species | Number of Sequences c | Number of DArTseq Alleles with BLAST Hits a |
|---|---|---|---|
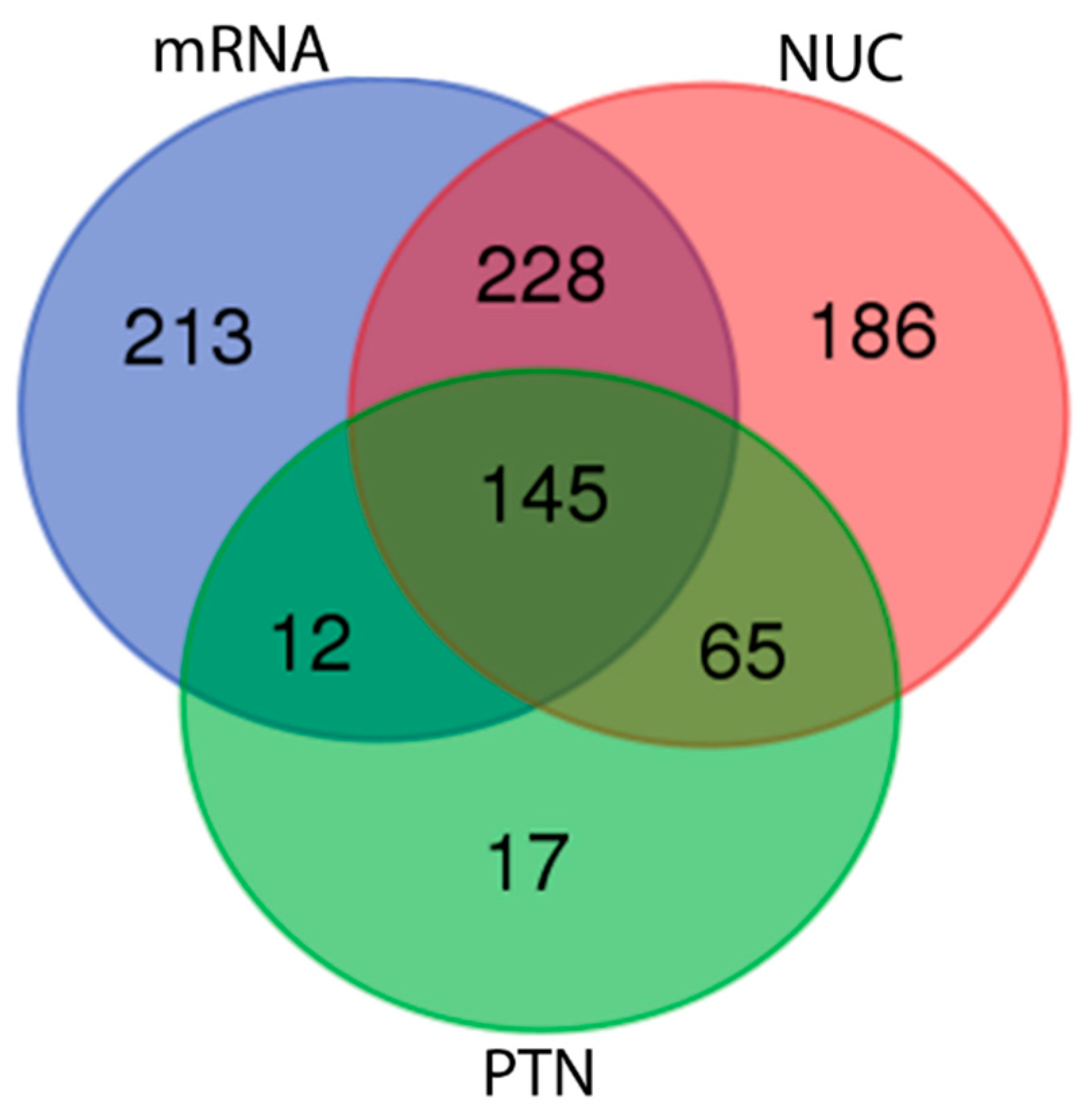
| Teleostei RefSeq mRNA | 6 b | 373,153 | 619 (40%) |
| Osteoglossiformes RefSeq Nucleotide | 23 d | 48,195 | 625 (41%) |
| Osteoglossiformes RefSeq Protein | 239 e | 41,731 | 238 (16%) |
| Type of Mutation | Count | Percentage |
|---|---|---|
| Transition | 882 | 57% |
| Transversion | 655 | 42% |
| Unique SNP in an allele sequence | 1201 | 88% |
| Multiple SNP in an allele sequence | ||
| 2 SNP | 143 | 10.50% |
| 3 SNP | 10 | 0.70% |
| 4 SNP | 1 | 0.07% |
| 5 SNP | 1 | 0.07% |
| 11 SNP | 1 | 0.07% |
| Alleles found in Heterozygosity | 287 | 19% |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barby, F.F.; Ráb, P.; Lavoué, S.; Ezaz, T.; Bertollo, L.A.C.; Kilian, A.; Maruyama, S.R.; Aguiar de Oliveira, E.; Artoni, R.F.; Santos, M.H.; et al. From Chromosomes to Genome: Insights into the Evolutionary Relationships and Biogeography of Old World Knifefishes (Notopteridae; Osteoglossiformes). Genes 2018, 9, 306. https://doi.org/10.3390/genes9060306
Barby FF, Ráb P, Lavoué S, Ezaz T, Bertollo LAC, Kilian A, Maruyama SR, Aguiar de Oliveira E, Artoni RF, Santos MH, et al. From Chromosomes to Genome: Insights into the Evolutionary Relationships and Biogeography of Old World Knifefishes (Notopteridae; Osteoglossiformes). Genes. 2018; 9(6):306. https://doi.org/10.3390/genes9060306
Chicago/Turabian StyleBarby, Felipe Faix, Petr Ráb, Sébastien Lavoué, Tariq Ezaz, Luiz Antônio Carlos Bertollo, Andrzej Kilian, Sandra Regina Maruyama, Ezequiel Aguiar de Oliveira, Roberto Ferreira Artoni, Mateus Henrique Santos, and et al. 2018. "From Chromosomes to Genome: Insights into the Evolutionary Relationships and Biogeography of Old World Knifefishes (Notopteridae; Osteoglossiformes)" Genes 9, no. 6: 306. https://doi.org/10.3390/genes9060306
APA StyleBarby, F. F., Ráb, P., Lavoué, S., Ezaz, T., Bertollo, L. A. C., Kilian, A., Maruyama, S. R., Aguiar de Oliveira, E., Artoni, R. F., Santos, M. H., Ilesanmi Jegede, O., Hatanaka, T., Tanomtong, A., Liehr, T., & Cioffi, M. D. B. (2018). From Chromosomes to Genome: Insights into the Evolutionary Relationships and Biogeography of Old World Knifefishes (Notopteridae; Osteoglossiformes). Genes, 9(6), 306. https://doi.org/10.3390/genes9060306







