16S rRNA Gene-Based Profiling of the Microbial Community in an Acid Mine Drainage Fe Precipitate at Libiola Mine (Liguria, Italy)
Abstract
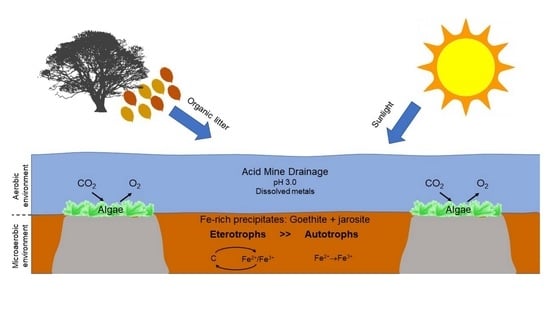
:1. Introduction
2. Materials and Methods
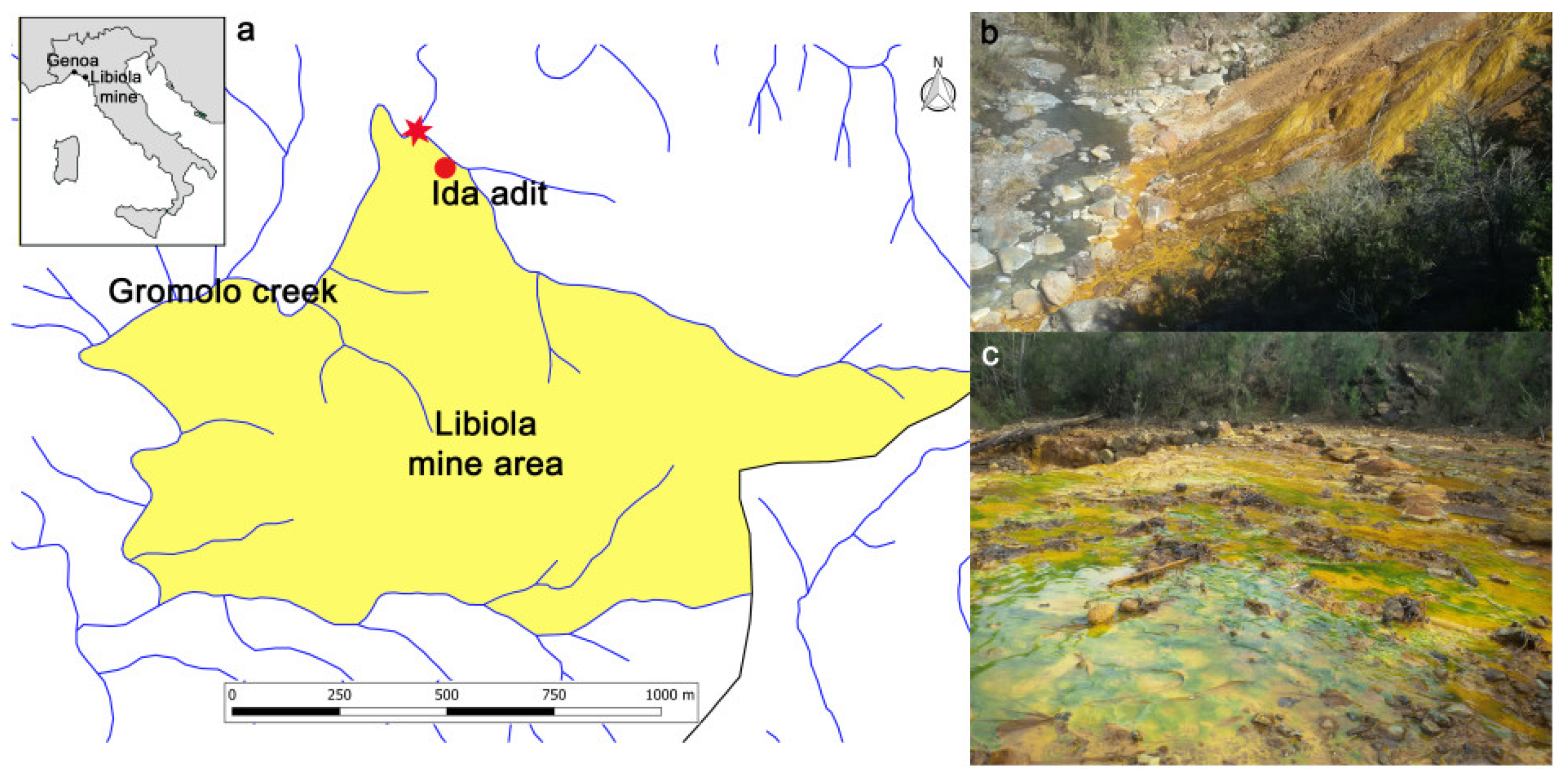
2.1. Study Area and Sampling
2.2. DNA Extraction and PCR
2.3. Sequence Analyses
3. Results
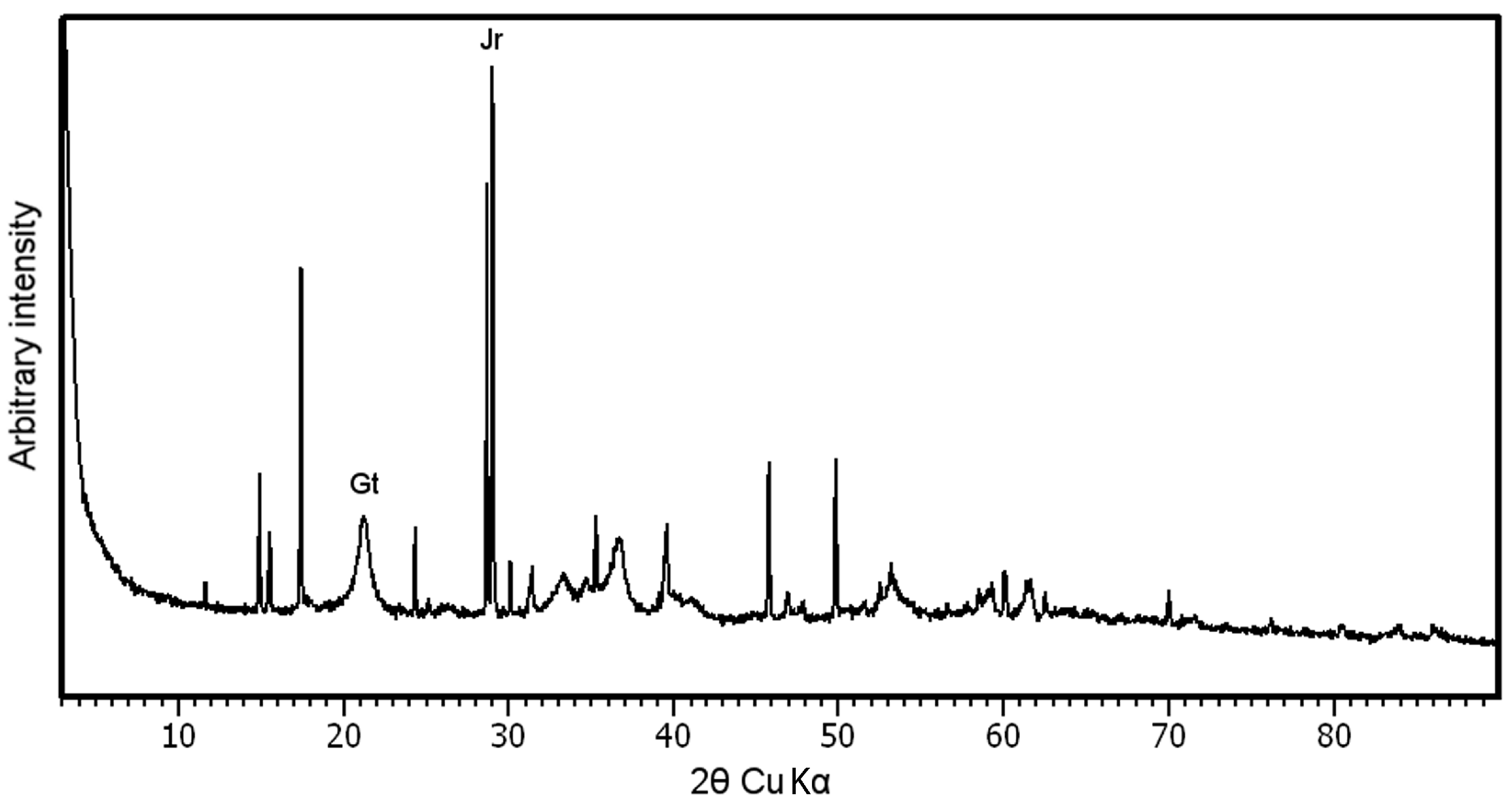
3.1. Mineralogical Characterization of the Sample
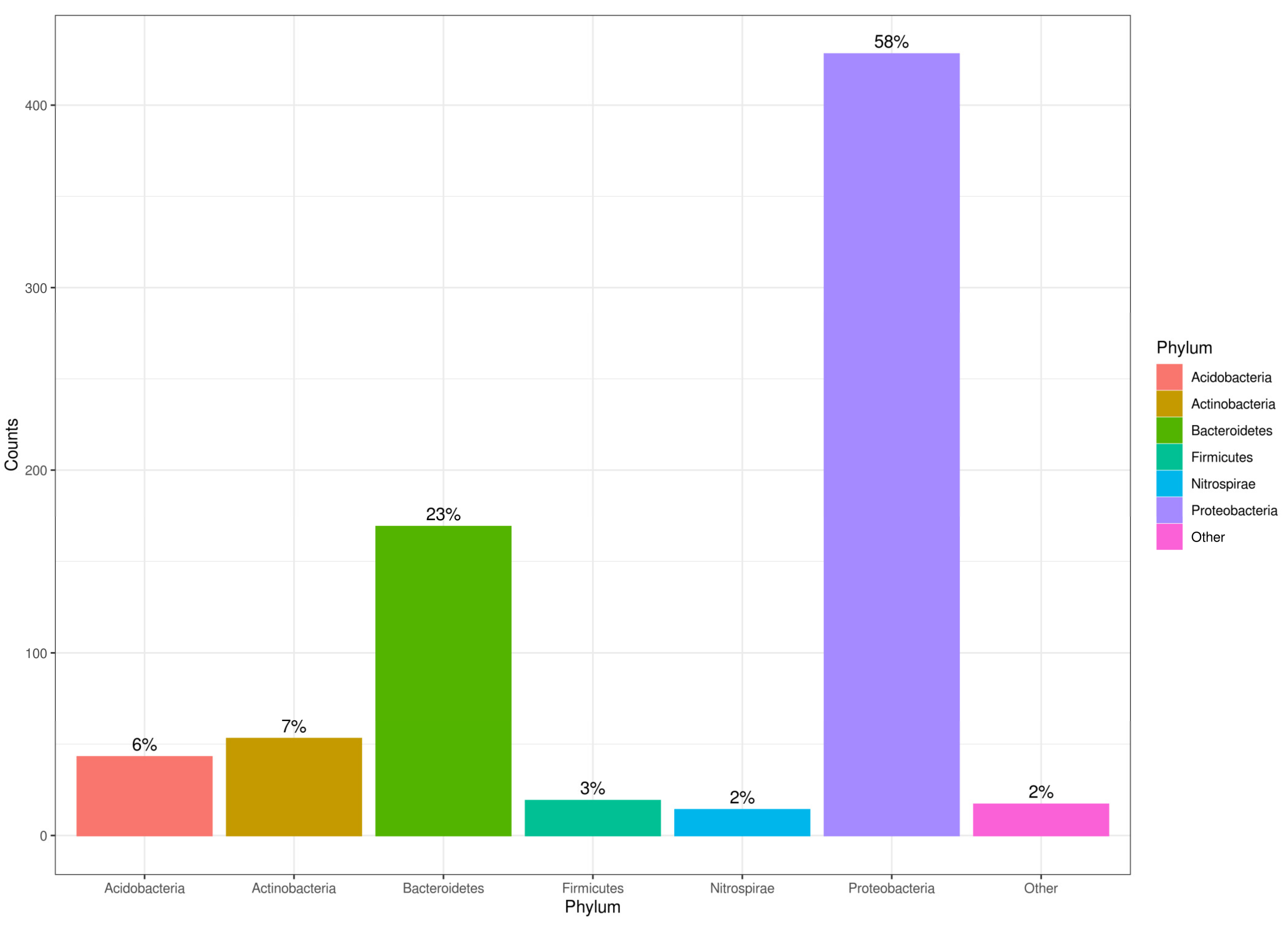
3.2. Genomic DNA Extraction, Microbial Diversity, and Richness in AMD Precipitates
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blowes, D.W.; Ptacek, C.J.; Jambor, J.L.; Weisener, C.G.; Paktunc, D.; Gould, W.D.; Johnson, D.B. The geochemistry of acid mine drainage. In Treatise of Geochemistry, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2014; Volume 11, pp. 131–190. [Google Scholar]
- Nordstrom, D.K.; Alpers, C.N. Geochemistry of Acid Mine Waters. In The Environmental Geochemistry of Mineral Deposits, Part A; Plumlee, G.S., Longdon, M.J., Eds.; Society of Economic Geologists Inc.: Littleton, CO, USA, 1999; pp. 133–157. [Google Scholar]
- Ritchie, A.I.M. The waste-rock environment. In Short Course Handbook on Environmental Geochemistry of Sulfide Mine-Waste; Jambor, J.L., Blowes, D.W., Eds.; Mineralogical Association of Canada: Ottawa, QC, Canada, 1994; pp. 133–162. [Google Scholar]
- Singer, P.C.; Stumm, W. Acid mine drainage-rate determining step. Science 1970, 167, 1121–1123. [Google Scholar] [CrossRef] [PubMed]
- Stumm, W.; Morgan, J.J. Aquatic Chemistry, 3rd ed.; Wiley: New York, NY, USA, 1996; p. 1040. [Google Scholar]
- Hallberg, K.B.; Johnson, D.B. Novel acidophiles isolated from moderately acidic mine drainage waters. Hydrometallurgy 2003, 71, 139–148. [Google Scholar] [CrossRef]
- Hallberg, K.B. New perspectives in acid mine drainage microbiology. Hydrometallurgy 2010, 104, 448–453. [Google Scholar] [CrossRef]
- Korehi, H.; Blothe, M.; Schippers, A. Microbial at the moderate acidic stage in three different sulfidic mine tailings dumps generating acid mine drainage. Res. Microbiol. 2014, 165, 713–718. [Google Scholar] [CrossRef]
- Bomberg, M.; Makinen, J.; Salo, M.; Kinnunen, P. High diversity in iron cycling microbial communities in acidic iron-rich water of Pyhasalmi mine, Finland. Geofluids 2019, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Johnson, D.B. Extremophiles: Acidic Environments. In Encyclopedia of Microbiology, 3rd ed.; Schaechter, M., Ed.; Academic Press: Cambridge, MA, USA; Elsevier: Amsterdam, The Netherlands, 2009; pp. 107–126. [Google Scholar]
- Yang, Y.; Li, Y.; Sun, Q. Archaeal and bacterial communities in acid mine drainage from metal-rich abandoned tailing ponds, Tongling, China. Trans. Nonferr. Met. Soc. China 2014, 24, 3332–3342. [Google Scholar] [CrossRef]
- Johnson, D.B.; Hallberg, K.B. Carbon, iron and sulfur metabolism in acidophilic micro-organisms. Adv. Microb. Physiol. 2008, 54, 201–255. [Google Scholar]
- Rodrigues, M.L.M.; Lopes, K.C.S.; Leôncio, H.C.; Silva, L.A.M.; Leão, V.A. Bioleaching of fluoride-bearing secondary copper sulphides: Column experiments with Acidithiobacillus ferrooxidans. Chem. Eng. J. 2016, 284, 1279–1286. [Google Scholar] [CrossRef]
- Orell, A.; Navarro, C.A.; Arancibia, R.; Mobarec, J.C.; Jerez, C.A. Life in blue: Copper resistance mechanisms of bacteria and Archaea used in industrial biomining of minerals. Biotech. Adv. 2010, 28, 839–848. [Google Scholar]
- Watling, H.R. Chalcopyrite hydrometallurgy at atmospheric pressure: 1. Review of acidic sulfate, sulphate-chloride and sulphate-nitrate process options. Hydrometallurgy 2013, 140, 163–180. [Google Scholar] [CrossRef]
- Funari, V.; Mäkinen, J.; Salminen, J.; Braga, R.; Dinelli, E.; Revitzer, H. Metal removal from Municipal Solid Waste Incineration fly ash: A comparison between chemical leaching and bioleaching. Waste Manag. 2017, 60, 397–406. [Google Scholar] [CrossRef]
- Rastegar, S.O.; Mousavi, S.M.; Shojaosadati, S.A.; Sarraf Mamoory, R. Bioleaching of V, Ni, and Cu from residual produced in oil fired furnaces using Acidithiobacillus ferrooxidans. Hydrometallurgy 2015, 157, 50–59. [Google Scholar] [CrossRef]
- Gomes, H.I.; Funari, V.; Mayes, W.M.; Rogerson, M.; Prior, T.J. Recovery of Al, Cr and V from steel slag by bioleaching: Batch and column experiments. J. Environ. Manag. 2018, 222, 30–36. [Google Scholar] [CrossRef]
- Di Piazza, S.; Cecchi, G.; Cardinale, A.M.; Carbone, C.; Mariotti, M.G.; Giovine, M.; Zotti, M. Penicillium expansum Link strain for a biometallurgical method to recover REEs from WEEE. Waste Manag. 2017, 60, 596–600. [Google Scholar] [CrossRef] [PubMed]
- Carbone, C.; Di Benedetto, F.; Marescotti, P.; Martinelli, A.; Sangregorio, C.; Cipriani, C.; Lucchetti, G.; Romanelli, M. Genetic evolution of nanocrystalline Fe oxide and oxyhydroxide assemblages from the Libiola Mine (eastern Liguria, Italy): Structural and microstructural investigations. Eur. J. Mineral. 2005, 17, 785–795. [Google Scholar] [CrossRef]
- Maggi, R.; Pearce, M. Mid fourth-millennium copper mining in Liguria, north-west Italy: The earliest known copper mines in Western Europe. Antiquity 2005, 79, 66–77. [Google Scholar] [CrossRef]
- Garuti, G.; Zaccarini, F. Minerals of Au, Ag, and U in volcanic rock-associated massive sulfide deposits of the Northern Apennine ophiolite, Italy. Can. Mineral. 2005, 43, 935–950. [Google Scholar] [CrossRef]
- Zaccarini, F.; Garuti, G. Mineralogy and chemical composition of VMS deposits of northern Apennine ophiolites, Italy: Evidence for the influence of country rock type on ore composition. Mineral. Petrol. 2008, 94, 61–83. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; Wiley: Hoboken, NJ, USA, 1991; pp. 115–175. [Google Scholar]
- Nadkarni, M.A.; Martin, F.E.; Jacques, N.A.; Hunter, N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 2002, 48, 257–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects (accessed on 24 January 2020).
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate illumina paired-end reAd mergeR. Bioinformatics 2013, 30, 614–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria. 2020. Available online: https://www.R-project.org/ (accessed on 24 January 2020).
- Oksanen, J.F.; Blanchet, G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package Version 2.4-4. 2017. Available online: https://CRAN.R-project.org/package=vegan (accessed on 25 January 2020).
- McMurdy, P.J.; Holmes, S. Phyloseq:an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar]
- Carbone, C.; Dinelli, E.; Marescotti, P.; Gasparotto, G.; Lucchetti, G. The role of AMD secondary minerals in controlling environmental pollution: Indications from bulk leaching tests. J. Geochem. Explor. 2013, 132, 188–200. [Google Scholar] [CrossRef]
- Consani, S.; Ianni, M.C.; Dinelli, E.; Capello, M.; Cutroneo, L.; Carbone, C. Assessment of metal distribution in different Fe precipitates related to Acid Mine Drainage through two sequential extraction procedures. J. Geochem. Explor. 2019, 196, 247–258. [Google Scholar] [CrossRef]
- Rowe, O.F.; Sánchez-España, J.; Hallberg, K.B.; Johnson, D.B. Microbial communities and geochemical dynamics in an extremely acidic, metal-rich stream at an abandoned sulfide mine (Huelva, Spain) underpinned by two functional primary production systems. Environ. Microbiol. 2007, 9, 1761–1771. [Google Scholar] [CrossRef]
- An, X.; Baker, P.; Li, H.; Su, J.; Yu, C.; Cai, C. The patterns of bacterial community and relationships between sulfate-reducing bacteria and hydrochemistry in sulfate-polluted groundwater of Baogang rare earth tailings. Environ. Sci. Pollut. Res. 2016, 23, 21766–21779. [Google Scholar] [CrossRef] [Green Version]
- Amils, R. Lessons learned from thirty years of geomicrobiological studies of Río Tinto. Res. Microbiol 2016, 167, 539–545. [Google Scholar] [CrossRef]
- Baker, B.J.; Banfield, J.F. Microbial communities in acid mine drainage. FEMS Microbiol. Ecol. 2003, 44, 139–152. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Sun, X.; Li, B.; Xu, R.; Young, L.Y.; Dong, Y.; Zhang, M.; Kong, T.; Xiao, E.; Wang, Q. Bacterial response to sharp geochemical gradients caused by acid mine drainage intrusion in a terrace: Relevance of C, N, and S cycling and metal resistance. Environ. Int. 2020, 138, 105601. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.B.; Bridge, T.A.M. Reduction of ferric iron by acidophilic heterotrophic bacteria: Evidence for constitutive and inducible enzyme systems in Acidiphilium spp. J. Appl. Microbiol. 2002, 92, 315–321. [Google Scholar] [CrossRef]
- Xu, R.; Li, B.; Xiao, E.; Young, L.Y.; Sun, X.; Kong, T.; Dong, Y.; Wang, Q.; Yang, Z.; Chen, L.; et al. Uncovering microbial responses to sharp geochemical gradients in a terrace contaminated by acid mine drainage. Environ. Pollut. 2020, 261, 114226. [Google Scholar] [CrossRef]
- Druschel, G.K.; Baker, B.J.; Gihring, T.; Banfield, J.F. Acid mine drainage biogeochemistry at Iron Mountain, California. Geochem. Trans. 2004, 5, 13–32. [Google Scholar] [CrossRef] [Green Version]
- Coupland, K. A Study of the Geomicrobiology of Acid Mine Drainage Impacted Environments. Ph.D. Thesis, University of Wales, Bangor, UK, 2006. [Google Scholar]
- Xu, R.; Sun, X.; Häggblom, M.M.; Dong, Y.; Zhang, M.; Xiao, E.; Xiao, T.; Gao, P.; Li, B.; Sun, W. Metabolic potentials of members of the class Acidobacteriia in metal-contaminated soils revealed by metagenomic analysis. Environ. Microbiol. 2021, 23. [Google Scholar] [CrossRef]
- Kimura, S.; Bryan, C.G.; Hallberg, K.B.; Johnson, D.B. Biodiversity and geochemistry of an extremely acidic, low-temperature subterranean environment sustained by chemolithotrophy. Environ. Microbiol. 2011, 13, 2092–2104. [Google Scholar] [CrossRef]
- Foster, A.L.; Munk, L.; Koski, R.A.; Shanks, W.C., III; Stillings, L.L. Relationships between microbial communities and environmental parameters at sites impacted by mining of volcanogenic massive sulphide deposits, Prince William Sound, Alaska. Appl. Geochem. 2008, 23, 279–307. [Google Scholar] [CrossRef]
- Paustian, T.; Roberts, G. Deinococcus and Cytophaga. In Through the Microscope: A Look at All Things Small. 2007. Available online: http://www.microbiologytext.com/6th_ed/ (accessed on 17 April 2020).
| Element | (wt%) | Element | (mg·kg−1) |
|---|---|---|---|
| Fe | 50.69 | Cu | 2018 |
| S | n.a. | Zn | 41 |
| Al | 0.06 | Mn | 77 |
| Mg | 0.03 | Ni | 11 |
| Ca | 0.04 | Cr | 96 |
| Ti | 0.02 | V | 64 |
| Na | 0.01 | As | 4.30 |
| K | 0.05 | Pb | 2 |
| P | 0.01 | Co | 2 |
| Genus | Relative Abundance (%) |
|---|---|
| Flavobacterium | 5.11 |
| undetermined_Comamonadaceae | 5.11 |
| undetermined_Acidobacteriaceae | 4.44 |
| Unclassified | 3.90 |
| undetermined_Acetobacteraceae | 3.77 |
| undetermined_Acidimicrobiales | 3.63 |
| undetermined_Sphingobacteriales | 3.50 |
| Novosphingobium | 3.10 |
| undetermined_Oxalobacteraceae | 3.10 |
| Pedobacter | 2.96 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Consani, S.; Ghignone, S.; Pozzolini, M.; Giovine, M.; Vezzulli, L.; Carbone, C. 16S rRNA Gene-Based Profiling of the Microbial Community in an Acid Mine Drainage Fe Precipitate at Libiola Mine (Liguria, Italy). Minerals 2021, 11, 1064. https://doi.org/10.3390/min11101064
Consani S, Ghignone S, Pozzolini M, Giovine M, Vezzulli L, Carbone C. 16S rRNA Gene-Based Profiling of the Microbial Community in an Acid Mine Drainage Fe Precipitate at Libiola Mine (Liguria, Italy). Minerals. 2021; 11(10):1064. https://doi.org/10.3390/min11101064
Chicago/Turabian StyleConsani, Sirio, Stefano Ghignone, Marina Pozzolini, Marco Giovine, Luigi Vezzulli, and Cristina Carbone. 2021. "16S rRNA Gene-Based Profiling of the Microbial Community in an Acid Mine Drainage Fe Precipitate at Libiola Mine (Liguria, Italy)" Minerals 11, no. 10: 1064. https://doi.org/10.3390/min11101064
APA StyleConsani, S., Ghignone, S., Pozzolini, M., Giovine, M., Vezzulli, L., & Carbone, C. (2021). 16S rRNA Gene-Based Profiling of the Microbial Community in an Acid Mine Drainage Fe Precipitate at Libiola Mine (Liguria, Italy). Minerals, 11(10), 1064. https://doi.org/10.3390/min11101064