New Features of Acidophilic Bacteria of the Genus Sulfobacillus: Polysaccharide Biosynthesis and Degradation Pathways
Abstract
:1. Introduction
2. Materials and Methods
2.1. Objectives of Research, Culture Media, and Conditions of Cultivation
2.2. Analytical Techniques
2.3. Enzyme Assays
2.4. Protein Alignments
2.5. Protein Characterization and Pathway Mapping
2.6. Phylogenetic Analysis of Proteins
3. Results and Discussion
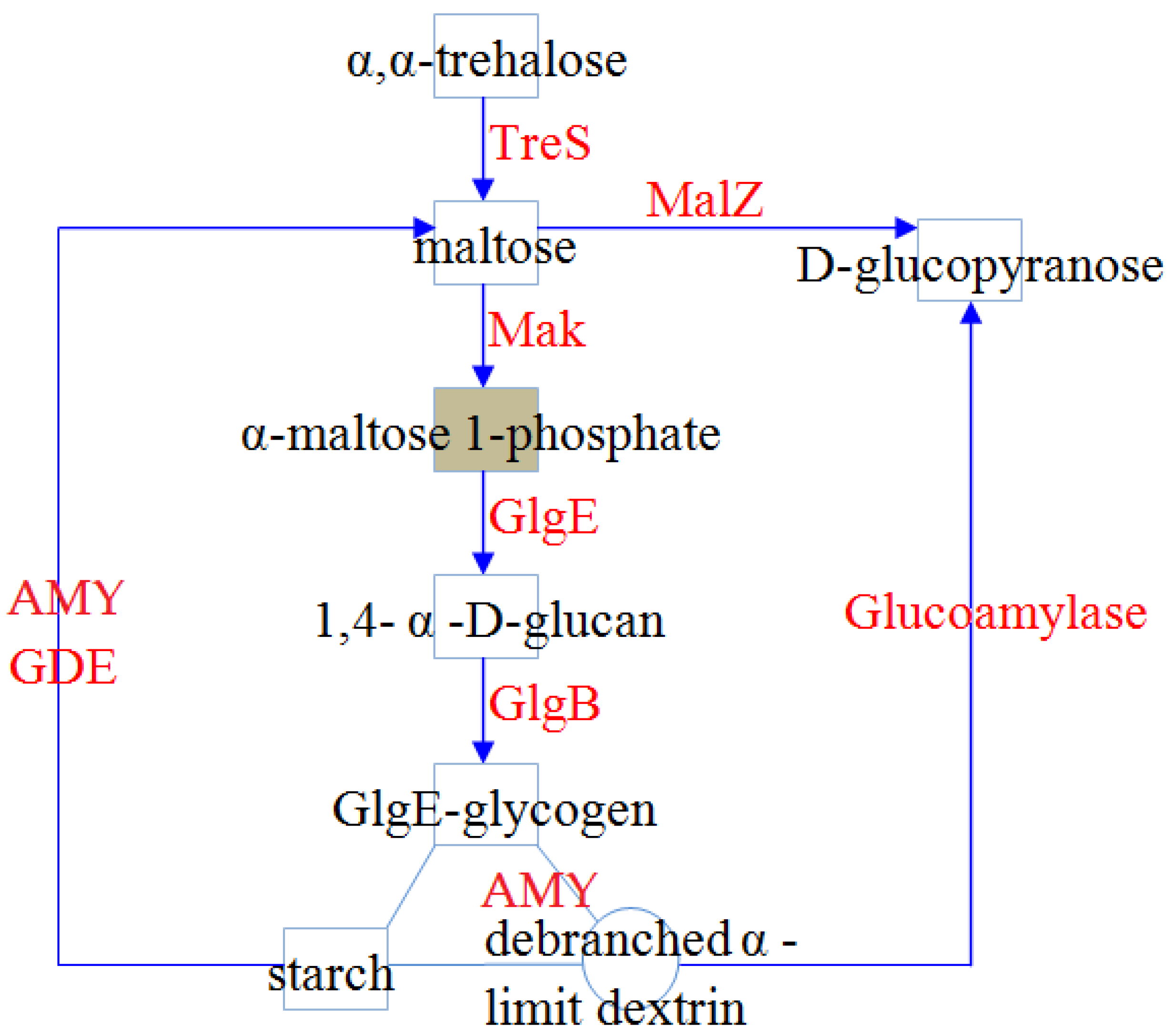
3.1. α-Glucan metabolism in Sulfobacillus Species: Biosynthesis and Degradation of Polysaccharides
3.1.1. Biosynthesis of α-Glucans in Sulfobacillus Species
3.1.2. α-Glucan Degradation Pathways in Bacteria of the Genus Sulfobacillus
3.2. Microbial Growth and Polysaccharide Oxidation
3.3. Activities of Enzymes Degrading α-Glucans
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kaksonen, A.H.; Deng, X.; Bohu, T.; Zea, L.; Khaleque, H.N.; Gumulya, Y.; Boxall, N.J.; Morris, C.; Cheng, K.Y. Prospective directions for biohydrometallurgy. Hydrometallurgy 2020, 195, 105376. [Google Scholar] [CrossRef]
- Wills, B.A.; Finch, J.A. Wills’ Mineral Processing Technology: An Introduction to the Practical Aspects of Ore Treatment and Mineral Recovery, 8th ed.; Butterworth-Heinemann: Oxford, UK, 2015. [Google Scholar]
- Muravyov, M.; Panyushkina, A.; Fomchenko, N. Bulk flotation followed by selective leaching with biogenic ferric iron is a promising solution for eco-friendly processing of complex sulfidic ores. J. Environ. Manag. 2022, 318, 115587. [Google Scholar] [CrossRef]
- Kondrat’eva, T.F.; Pivovarova, T.A.; Tsaplina, I.A.; Fomchenko, N.V.; Zhuravleva, A.E.; Murav’ev, M.I.; Melamud, V.S.; Bulayev, A.G. Diversity of the communities of acidophilic chemolithotrophic microorganisms in natural and technogenic ecosystems. Microbiology 2012, 81, 1–24. [Google Scholar] [CrossRef]
- Hedrich, S.; Schippers, A. Distribution of acidophilic microorganisms in natural and man-made acidic environments. Curr. Issues Mol. Biol. 2021, 40, 25–48. [Google Scholar] [CrossRef]
- Panyushkina, A.; Bulaev, A.; Belyi, A.V. Unraveling the central role of sulfur-oxidizing Acidiphilium multivorum LMS in industrial bioprocessing of gold-bearing sulfide concentrates. Microorganisms 2021, 9, 984. [Google Scholar] [CrossRef] [PubMed]
- Panyushkina, A.; Muravyov, M.; Fomchenko, N. A Case of predominance of Alicyclobacillus tolerans in microbial community during bioleaching of pentlandite-chalcopyrite concentrate. Minerals 2022, 12, 396. [Google Scholar] [CrossRef]
- Rawlings, D.E.; Johnson, D.B. The microbiology of biomining: Development and optimization of mineral-oxidizing microbial consortia. Microbiology 2007, 153, 315–324. [Google Scholar] [CrossRef]
- Panyushkina, A.E.; Babenko, V.V.; Nikitina, A.S.; Selezneva, O.V.; Tsaplina, I.A.; Letarova, M.A.; Kostryukova, E.S.; Letarov, A.V. Sulfobacillus thermotolerans: New insights into resistance and metabolic capacities of acidophilic chemolithotrophs. Sci. Rep. 2019, 9, 15069. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Hedrich, S.; Jin, D.; Breuker, A.; Schippers, A. Sulfobacillus harzensis sp. nov., an acidophilic bacterium inhabiting mine tailings from a polymetallic mine. Int. J. Syst. Evol. Microbiol. 2021, 71, 004871. [Google Scholar] [CrossRef]
- Muravyov, M.; Panyushkina, A. Distinct roles of acidophiles in complete oxidation of high-sulfur ferric leach product of zinc sulfide concentrate. Microorganisms 2020, 8, 386. [Google Scholar] [CrossRef] [Green Version]
- Tsaplina, I.A.; Bogdanova, T.I.; Sayakin, D.D.; Karavaiko, G.I. Effects of organic substances on the growth of Sulfobacillus thermosulfidooxidans and pyrite oxidation. Microbiology 1992, 60, 686–692. [Google Scholar]
- Watling, H.R. The bioleaching of nickel-copper sulfides. Hydrometallurgy 2008, 91, 70–88. [Google Scholar] [CrossRef]
- Collinson, D.M.; Usher, K.M.; Nichols, P.D.; Watling, H.R. Habituation of Sulfobacillus thermosulfidooxidans to 4-nonylphenol in ferrous ion growth medium. Process Biochem. 2011, 46, 108–115. [Google Scholar] [CrossRef]
- Zhuravleva, A.E.; Ismailov, A.D.; Tsaplina, I.A. Electron donors at oxidative phosphorylation in bacteria of the genus Sulfobacillus. Microbiology 2009, 78, 811–814. [Google Scholar] [CrossRef]
- Tsaplina, I.A.; Krasil’nikova, E.N.; Zakharchuk, L.M.; Egorova, M.A.; Bogdanova, T.I.; Karavaiko, G.I. Carbon metabolism in Sulfobacillus thermosulfidooxidans subsp. asporogenes, strain 41. Microbiology 2000, 69, 271–276. [Google Scholar] [CrossRef]
- Tsaplina, I.A.; Zhuravleva, A.E.; Egorova, M.A.; Bogdanova, T.I.; Krasil’nikova, E.N.; Zakharchuk, L.M.; Kondrat’eva, T.F. Response to oxygen limitation in bacteria of the genus Sulfobacillus. Microbiology 2010, 79, 13–22. [Google Scholar] [CrossRef]
- Hedrich, S.; Johnson, D.B. Aerobic and anaerobic oxidation of hydrogen by acidophilic bacteria. FEMS Microbiol. Lett. 2013, 349, 40–45. [Google Scholar] [CrossRef]
- Panyushkina, A.; Matyushkina, D.; Pobeguts, O. Understanding stress response to high-arsenic gold-bearing sulfide concentrate in extremely metal-resistant acidophile Sulfobacillus thermotolerans. Microorganisms 2020, 8, 1076. [Google Scholar] [CrossRef] [PubMed]
- Zhuravleva, A.E.; Tsaplina, I.A.; Kondrat’eva, T.F. Specific characteristics of the strains isolated from a thermoacidophilic microbial community oxidizing antimony sulfide ore. Microbiology 2011, 80, 70–81. [Google Scholar] [CrossRef]
- Vardanyan, A.; Vyrides, I. Acidophilic bioleaching at high dissolved organic compounds: Inhibition and strategies to counteract this. Miner. Eng. 2019, 143, 105943. [Google Scholar] [CrossRef]
- Merino, M.P.; Andrews, B.A.; Parada, P.; Asenjo, J.A. Characterization of Ferroplasma acidiphilum growing in pure and mixed culture with Leptospirillum ferriphilum. Biotechnol. Prog. 2016, 32, 1390–1396. [Google Scholar] [CrossRef]
- Bogdanova, T.I.; Tsaplina, I.A.; Kondrat’eva, T.F.; Duda, V.I.; Suzina, N.E.; Melamud, V.S.; Tourova, T.P.; Karavaiko, G.I. Sulfobacillus thermotolerans sp. nov., a thermotolerant, chemolithotrophic bacterium. Int. J. Syst. Evol. Microbiol. 2006, 56, 1039–1042. [Google Scholar] [CrossRef] [PubMed]
- Galleguillos, P.A.; Grail, B.M.; Hallberg, K.B.; Demergasso, C.S.; Johnson, D.B. Identification of trehalose as a compatible solute in different species of acidophilic bacteria. J. Microbiol. 2018, 56, 727–733. [Google Scholar] [CrossRef]
- Justice, N.B.; Norman, A.; Brown, C.T.; Singh, A.; Thomas, B.C.; Banfield, J.F. Comparison of environmental and isolate Sulfobacillus genomes reveals diverse carbon, sulfur, nitrogen, and hydrogen metabolisms. BMC Genom. 2014, 15, 1107. [Google Scholar] [CrossRef] [PubMed]
- Wilson, W.A.; Roach, P.J.; Montero, M.; Baroja-Fernández, E.; Muñoz, F.J.; Eydallin, G.; Viale, A.M.; Pozueta-Romero, J. Regulation of glycogen metabolism in yeast and bacteria. FEMS Microbiol. Rev. 2010, 34, 952–985. [Google Scholar] [CrossRef]
- Guo, X.; Yin, H.; Liang, Y.; Hu, Q.; Zhou, X.; Xiao, Y.; Ma, L.; Zhang, X.; Qiu, G.; Liu, X. Comparative genome analysis reveals metabolic versatility and environmental adaptations of Sulfobacillus thermosulfidooxidans strain ST. PLoS ONE 2014, 9, e99417. [Google Scholar] [CrossRef]
- Matzke, J.; Schwermann, B.; Bakker, E.P. Acidostable and acidophilic proteins: The example of the α-amylase from Alicyclobacillus acidocaldarius. Comp. Biochem. Physiol. A Physiol. 1997, 118, 475–479. [Google Scholar] [CrossRef]
- Karavaiko, G.I.; Bogdanova, T.I.; Tourova, T.P.; Kondrat’eva, T.F.; Tsaplina, I.A.; Egorova, M.A.; Krasil’nikova, E.N.; Zakharchuk, L.M. Reclassification of ‘Sulfobacillus thermosulfidooxidans subsp. thermotolerans’ strain K1 as Alicyclobacillus tolerans sp. nov. and Sulfobacillus disulfidooxidans Dufresne et al. 1996 as Alicyclobacillus disulfidooxidans comb. nov., and emended description of the genus Alicyclobacillus. Int. J. Syst. Evol. Microbiol. 2005, 55, 941–947. [Google Scholar] [PubMed]
- Panyushkina, A.E.; Tsaplina, I.A.; Grigor’eva, N.V.; Kondrat’eva, T.F. Thermoacidophilic microbial community oxidizing the gold-bearing flotation concentrate of a pyrite-arsenopyrite ore. Microbiology 2014, 83, 539–549. [Google Scholar] [CrossRef]
- Silverman, M.P.; Lundgren, D.G. Studies on the chemoautotrophic iron bacterium Ferrobacillus ferrooxidans. I. An improved medium and a harvesting procedure for securing high cell yields. J. Bacteriol. 1959, 77, 642–647. [Google Scholar] [CrossRef]
- Nelson, N. A photometric adaptation of the Somogyi method for the determination of glucose. J. Biol. Chem. 1944, 153, 375–380. [Google Scholar] [CrossRef]
- Somogyi, M. Notes on sugar determination. J. Biol. Chem. 1952, 195, 19–23. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randal, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA 11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef]
- Caspi, R.; Billington, R.; Ferrer, L.; Foerster, H.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Mueller, L.A.; et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2016, 44, D471–D480. [Google Scholar] [CrossRef]
- Paley, S.; O’Maille, P.E.; Weaver, D.; Karp, P.D. Pathway collages: Personalized multi-pathway diagrams. BMC Bioinform. 2016, 17, 529. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Zuckerkandl, E.; Pauling, L. Evolutionary divergence and convergence in proteins. In Evolving Genes and Proteins; Bryson, V., Vogel, H.J., Eds.; Academic Press: New York, USA, 1965; pp. 97–166. [Google Scholar]
- Kalscheuer, R.; Syson, K.; Veeraraghavan, U.; Weinrick, B.; Biermann, K.E.; Liu, Z.; Sacchettini, J.C.; Besra, G.; Bornemann, S.; Jacobs, W.R., Jr. Self-poisoning of Mycobacterium tuberculosis by targeting GlgE in an alpha-glucan pathway. Nat. Chem. Biol. 2010, 6, 376–384. [Google Scholar] [CrossRef]
- Fraga, J.; Maranha, A.; Mendes, V.; Pereira, P.J.; Empadinhas, N.; Macedo-Ribeiro, S. Structure of mycobacterial maltokinase, the missing link in the essential GlgE-pathway. Sci. Rep. 2015, 5, 8026. [Google Scholar] [CrossRef]
- Suzuki, R.; Koide, K.; Hayashi, M.; Suzuki, T.; Sawada, T.; Ohdan, T.; Takahashi, H.; Nakamura, Y.; Fujita, N.; Suzuki, E. Functional characterization of three (GH13) branching enzymes involved in cyanobacterial starch biosynthesis from Cyanobacterium sp. NBRC 102756. Biochim. et Biophys. Acta (BBA) Proteins Proteom. 2015, 1854, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Marín-Navarro, J.; Polaina, J. Glucoamylases: Structural and biotechnological aspects. Appl. Microbiol. Biotechnol. 2011, 89, 1267–1273. [Google Scholar] [CrossRef] [PubMed]
- Senyushkin, A.A.; Severina, L.O.; Mityushina, L.L. Capsule formation by Sulfobacillus thermosulfidooxidans cells growing under oligotrophic and mixotrophic conditions. Microbiology 1997, 66, 455–461. [Google Scholar]
- Tsaplina, I.A.; Krasil’nikova, E.N.; Zhuravleva, A.E.; Egorova, M.A.; Zakharchuk, L.M.; Suzina, N.E.; Duda, V.I.; Bogdanova, T.I.; Stadnichuk, I.N.; Kondrat’eva, T.F. Phenotypic properties of Sulfobacillus thermotolerans: Comparative aspects. Microbiology 2008, 77, 654–664. [Google Scholar] [CrossRef]
- Zakharchuk, L.M.; Tsaplina, I.A.; Krasil’nikova, E.N.; Bogdanova, T.I.; Karavaiko, G.I. Carbon Metabolism in Sulfobacillus thermosulfidooxidans. Microbiology 1994, 63, 573–580. [Google Scholar]
- Tsaplina, I.A.; Zhuravleva, A.E.; Ismailov, A.D.; Zakharchuk, L.M.; Krasil’nikova, E.N.; Bogdanova, T.I.; Karavaiko, G.I. The dependence of intracellular ATP level on the nutrition mode of the acidophilic bacteria Sulfobacillus thermotolerans and Alicyclobacillus tolerans. Microbiology 2007, 76, 654–662. [Google Scholar] [CrossRef]
- Donati, E.R.; Sand, W. (Eds.) Microbial Processing of Metal Sulfides; Springer: Dordrecht, The Netherlands, 2007. [Google Scholar]
- Bai, Y.; Huang, H.; Meng, K.; Shi, P.; Yang, P.; Luo, H.; Luo, C.; Feng, Y.; Zhang, W.; Yao, B. Identification of an acidic α-amylase from Alicyclobacillus sp. A4 and assessment of its application in the starch industry. Food Chem. 2012, 131, 1473–1478. [Google Scholar] [CrossRef]
- Li, Q.; Sand, W. Mechanical and chemical studies on eps from Sulfobacillus thermosulfidooxidans: From planktonic to biofilm cells. Colloids Surf. B Biointerfaces 2017, 153, 34–40. [Google Scholar] [CrossRef]

| Strain, ORF ID | KO 1 | Protein Size (a. a.) | Protein Similarity (Coverage), % | Putative Homolog | ||
|---|---|---|---|---|---|---|
| Kr1, BXT84_ | 1269, SAMN00768000_ | Kr1 | 1269 | |||
| 11955 | 3318 | K16147 | 655 | 661 | 63 (98) | GlgE, α-1,4-glucan:maltose-1-phosphate maltosyltransferase (EC 2.4.99.16) |
| 11960 | 3317 | K05343 | 1082 | 1081 | 73 (99) | TreS-Mak/AmyA, fused trehalose synthase (EC 5.4.99.16)/maltokinase (EC 2.7.1.175)/α-amylase (EC 3.2.1.1) |
| 11965 | 3316 | K00700 | 632 | 630 | 68 (99) | GlgB, 1,4-α-glucan branching enzyme (EC 2.4.1.18) |
| 16100 | 1445 | - | 746 | 1020 | 53 (72) | GDE; amylo-α-1,6-glucosidase (glycogen debranching enzyme) (EC 3.2.1.133) |
| 08005 | 0408 | K01178 | 801 | 789 | 70 (98) | Glucoamylase, maltase-glucoamylase, (glucan 1,4-α-glucosidase) (EC 3.2.1.3) |
| 12195 | 2946 | - | 652 | 654 | 84 (99.7) | Glucoamylase, glycoside hydrolase family 15 (EC 3.2.1.3) |
| 16125 | 1977 | - | 601 | 614 | 44 (96) | |
| 13535 | 1977 | - | 601 | 614 | 74 (97) | |
| 05115 | 2343 | K15922 | 795 | 806 | 53 (96) | Maltase-glucoamylase, α-glucosidase (EC 3.2.1.20) |
| Substrate | Product | Putative Enzyme(s) | Enzyme Activity, µmol/(min∙mg Protein) 1 | |
|---|---|---|---|---|
| Kr1 | 1269 | |||
| Starch | Maltose | α-Amylase (EC 3.2.1.1) | 0.54 ± 0.05 | 0.66 ± 0.05 |
| Glycogen | Maltose | α-Amylase (EC 3.2.1.1) | 0.62 ± 0.03 | 0.71± 0.04 |
| Starch | Glucose | γ-Amylase/maltase-glucoamylase (EC 3.2.1.3) + α-amylase (EC 3.2.1.1) + α-glucosidase (EC 3.2.1.20) | 1.31 ± 0.04 | 1.40 ± 0.05 |
| Glycogen | Glucose | γ-Amylase/maltase-glucoamylase (EC 3.2.1.3) + α-amylase (EC 3.2.1.1) + α-glucosidase (EC 3.2.1.20) | 1.87 ± 0.06 | 1.93 ± 0.06 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panyushkina, A.; Muravyov, M. New Features of Acidophilic Bacteria of the Genus Sulfobacillus: Polysaccharide Biosynthesis and Degradation Pathways. Minerals 2023, 13, 255. https://doi.org/10.3390/min13020255
Panyushkina A, Muravyov M. New Features of Acidophilic Bacteria of the Genus Sulfobacillus: Polysaccharide Biosynthesis and Degradation Pathways. Minerals. 2023; 13(2):255. https://doi.org/10.3390/min13020255
Chicago/Turabian StylePanyushkina, Anna, and Maxim Muravyov. 2023. "New Features of Acidophilic Bacteria of the Genus Sulfobacillus: Polysaccharide Biosynthesis and Degradation Pathways" Minerals 13, no. 2: 255. https://doi.org/10.3390/min13020255
APA StylePanyushkina, A., & Muravyov, M. (2023). New Features of Acidophilic Bacteria of the Genus Sulfobacillus: Polysaccharide Biosynthesis and Degradation Pathways. Minerals, 13(2), 255. https://doi.org/10.3390/min13020255








