Seasonal Microbial Population Shifts in a Bioremediation System Treating Metal and Sulfate-Rich Seepage
Abstract
:1. Introduction
2. Results
2.1. Pore Water Chemistry Surrounding the Biosolids Samples
2.2. Organic Material Characteristics
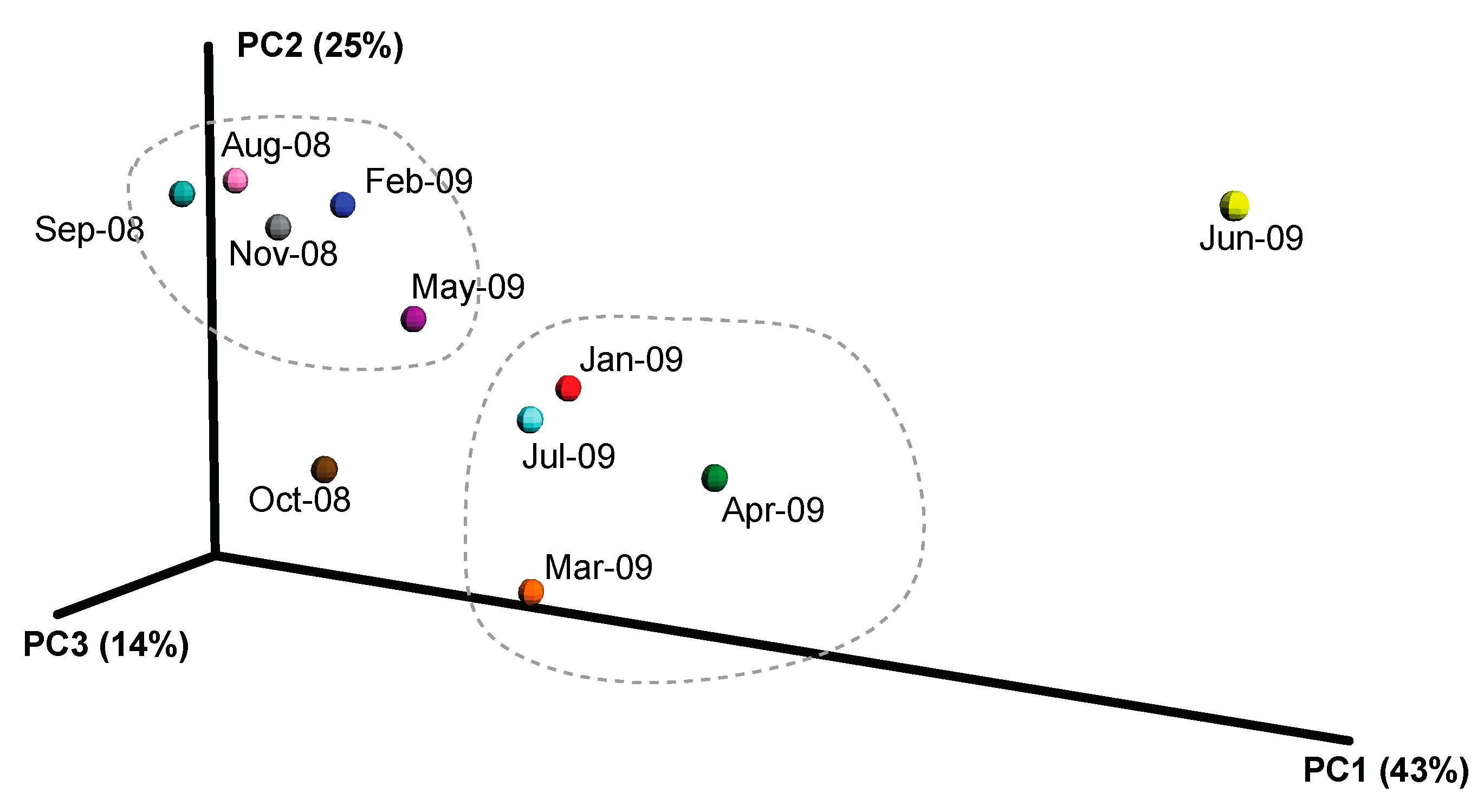
2.3. Overall Microbial Population Composition
2.4. Diversity of Sulfate-Reducing Bacteria
2.5. Quantification of Bacterial Genes
3. Discussion
3.1. Environmental Factors Influencing Microbial Population Structure
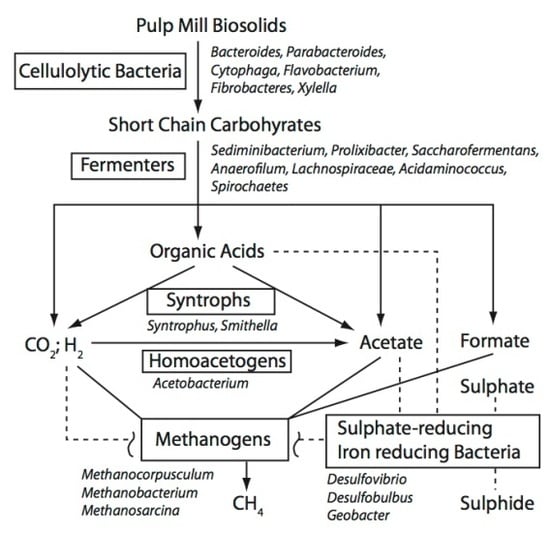
3.2 Possible Functional Roles for the Taxonomic Groups Found in the BCR Biosolids
3.2.1. Cellulose-Degrading Bacteria
3.2.2. Fermenters
3.2.3. Others
3.3. Co-Occurrence of Taxa in the BCR Biosolids
4. Materials and Methods
4.1. Experimental Design
4.2. Chemistry Measurements
4.3. Characteristics of the Organic Matrix
4.4. Deoxyribonucleic Acid (DNA) Extraction
4.5. Pyrotag Sequencing, Phylogenetic Analysis and Statistics
4.6. Quantitative Polymerase Chain Reaction
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| BCR | Biochemical reactor |
| DO | Dissolved oxygen |
| Fe | Iron |
| HC | Hydrocarbons |
| ORP | Oxidation-reduction potential |
| OTU | Operational taxonomic unit |
| NCBI | National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/ |
| PC | Pyrolysable carbon |
| PD | Phylogenetic diversity |
| rRNA | ribosomal ribonucleic acid |
| S1 | Hydrocarbons released during pyrolysis at 300 °C, mg-HC/g-solids |
| S2 | Hydrocarbons released during pyrolysis from 300–650 °C, mg-HC/g-solids |
| S3 | CO released during pyrolysis, mg-CO2/g-solids |
| SSU | Small sub-unit |
| TOC | Total organic carbon |
References
- Zinck, J.; Griffith, W. Review of Mine Drainage Treatment and Sludge Management Operations; Natural Resources Canada Report CANMET-MMSL 10-058(CR); Natural Resources Canada: Ottawa, ON, Canada, 2013.
- Gadd, G.M. Metals, minerals and microbes: Geomicrobiology and bioremediation. Microbiology 2010, 156, 609–643. [Google Scholar] [CrossRef] [PubMed]
- Gusek, J.J. Passive Treatment 101: An overview of the technologies. In Proceedings of the U.S. EPA/National Groundwater Association’s Remediation of Abandoned Mine Lands Conference, Denver, CO, USA, 2–3 October 2008; pp. 1–13.
- Jalali, K.; Baldwin, S.A. The role of sulphate reducing bacteria in copper removal from aqueous sulphate solutions. Water Res. 2000, 34, 797–806. [Google Scholar] [CrossRef]
- Ziemkiewicz, P.F.; Skousen, J.G.; Simmons, J. Long-term performance of passive acid mine drainage treatment systems. Mine Water Environ. 2003, 22, 118–129. [Google Scholar] [CrossRef]
- Hiibel, S.R.; Pereyra, L.P.; Breazeal, M.V.R.; Reisman, D.J.; Reardon, K.F.; Pruden, A. Effect of organic substrate on the microbial community structure in pilot-scale sulfate-reducing biochemical reactors treating mine drainage. Environ. Eng. Sci. 2011, 28, 563–572. [Google Scholar] [CrossRef]
- Hiibel, S.R.; Pereyra, L.P.; Inman, L.Y.; Tischer, A.; Reisman, D.J.; Reardon, K.F.; Pruden, A. Microbial community analysis of two field-scale sulfate-reducing bioreactors treating mine drainage. Environ. Microbiol. 2008, 10, 2087–2097. [Google Scholar] [CrossRef] [PubMed]
- Edwards, K.J.; Gihring, T.M.; Banfield, J.F. Seasonal variations in microbial populations and environmental conditions in an extreme acid mine drainage environment. Appl. Environ. Microbiol. 1999, 65, 3627–3632. [Google Scholar] [PubMed]
- Gilbert, J.A.; Steele, J.A.; Caporaso, J.G.; Steinbru, L.; Reeder, J.; Temperton, B.; Huse, S.; McHardy, A.C.; Knight, R.; Joint, I.; et al. Defining seasonal marine microbial community dynamics. ISME J. 2012, 6, 298–308. [Google Scholar] [CrossRef] [PubMed]
- Hwang, C.; Wu, W.; Gentry, T.J.; Carley, J.; Corbin, G.A.; Carroll, S.L.; Watson, D.B.; Jardine, P.M.; Zhou, J.; Criddle, C.S.; et al. Bacterial community succession during in situ uranium bioremediation: Spatial similarities along controlled flow paths. ISME J. 2009, 3, 47–64. [Google Scholar] [CrossRef] [PubMed]
- Werker, A.G.; Dougherty, J.M.; McHenry, J.L.; Van Loon, W.A. Treatment variability for wetland wastewater treatment design in cold climates. Ecol. Eng. 2002, 19, 1–11. [Google Scholar] [CrossRef]
- Stein, O.R.; Borden-Stewart, D.J.; Hook, P.B.; Jones, W.L. Seasonal influence on sulfate reduction and zinc sequestration in subsurface treatment wetlands. Water Res. 2007, 41, 3440–3448. [Google Scholar] [CrossRef] [PubMed]
- Purdy, K.J.; Nedwell, D.B.; Embley, T.M. Analysis of the sulfate-reducing bacterial and methanogenic archaeal populations in contrasting Antarctic sediments. Appl. Environ. Microbiol. 2003, 69, 3181–3191. [Google Scholar] [CrossRef] [PubMed]
- Sawicka, J.E.; Jørgensen, B.B.; Brüchert, V. Temperature characteristics of bacterial sulfate reduction in continental shelf and slope sediments. Biogeosciences 2012, 9, 3425–3435. [Google Scholar] [CrossRef]
- Knoblauch, C.; Sahm, K.; Jorgensen, B.B. Psychrophilic sulfate-reducing bacteria isolated from permanently cold Arctic marine sediments: Description of Desulfofrigus oceanense gen. nov., sp. nov., Desulfofrigus fragile sp. nov., Desulfofaba gelida gen. nov., sp. nov., Desulfotalea psychrophila gen. nov., sp. nov. and Desulfotalea arctica sp. nov. Int. J. Syst. Evol. Bacteriol. 1999, 49, 1631–1643. [Google Scholar]
- Knoblauch, C.; Jorgensen, B.B. Effect of temperature on sulphate reduction, growth rate and growth yield in five psychrophilic sulphate-reducing bacteria from Arctic sediments. Environ. Microbiol. 1999, 1, 457–467. [Google Scholar] [CrossRef] [PubMed]
- Abdollahi, H.; Nedwell, D.B. Seasonal temperature as a factor influencing bacterial sulfate reduction in a saltmarsh sediment. Microb. Ecol. 1979, 5, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Robador, A.; Brüchert, V.; Jørgensen, B.B. The impact of temperature change on the activity and community composition of sulfate-reducing bacteria in arctic versus temperate marine sediments. Environ. Microbiol. 2009, 11, 1692–1703. [Google Scholar] [CrossRef] [PubMed]
- Mattes, A.; Evans, L.J.; Gould, D.W.; Duncan, W.F.A.; Glasauer, S. The long term operation of a biologically based treatment system that removes As, S and Zn from industrial (smelter operation) landfill seepage. Appl. Geochem. 2011, 26, 1886–1896. [Google Scholar]
- Baldwin, S.A.; Khoshnoodi, M.; Rezadehbashi, M.; Taupp, M.; Hallam, S.; Mattes, A.; Sanei, H. The microbial community of a passive biochemical reactor treating arsenic, zinc, and sulfate-rich seepage. Front. Bioeng. Biotechnol. 2015, 3. [Google Scholar] [CrossRef] [PubMed]
- Khoshnoodi, M.; Dipple, G.; Baldwin, S. Mineralogical study of a biologically-based treatment system that removes arsenic, zinc and copper from landfill leachate. Minerals 2013, 3, 427–449. [Google Scholar] [CrossRef]
- Carrie, J.; Sanei, H.; Stern, G. Standardisation of Rock–Eval pyrolysis for the analysis of recent sediments and soils. Org. Geochem. 2012, 46, 38–53. [Google Scholar] [CrossRef]
- Chao, A. Nonparametric-estimation of the number of classes in a population. Scand. J. Stat. 1984, 11, 265–270. [Google Scholar]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIMME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Anderson, I.; Ulrich, L.E.; Lupa, B.; Susanti, D.; Porat, I.; Hooper, S.D.; Lykidis, A.; Sieprawska-Lupa, M.; Dharmarajan, L.; Goltsman, E. Genomic characterization of methanomicrobiales reveals three classes of methanogens. PLoS ONE 2009, 4, e5797. [Google Scholar] [CrossRef] [PubMed]
- Corkrey, R.; McMeekin, T.A.; Bowman, J.P.; Ratkowsky, D.A.; Olley, J.; Ross, T. Protein thermodynamics can be predicted directly from biological growth rates. PLoS ONE 2014, 9, e96100. [Google Scholar] [CrossRef]
- Long, R.A.; Azam, F. Antagonistic interactions among marine pelagic bacteria. Appl. Environ. Microbiol. 2001, 67, 4975–4983. [Google Scholar] [CrossRef] [PubMed]
- Leschine, S.B. Cellulose degradation in anaerobic environments. Annu. Rev. Microbiol. 1995, 49, 399–426. [Google Scholar] [CrossRef] [PubMed]
- Wexler, H.M. Bacteroides: The good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 2007, 20, 593–621. [Google Scholar] [CrossRef] [PubMed]
- Kolton, M.; Sela, N.; Elad, Y.; Cytryn, E. Comparative genomic analysis indicates that niche adaptation of terrestrial Flavobacteria is strongly linked to plant glycan metabolism. PLoS ONE 2013, 8, e76704. [Google Scholar] [CrossRef]
- Ventura, M.; Canchaya, C.; Tauch, A.; Chandra, G.; Fitzgerald, G.F.; Chater, K.F.; van Sinderen, D. Genomics of Actinobacteria: Tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 2007, 71, 495–548. [Google Scholar] [CrossRef] [PubMed]
- Suen, G.; Weimer, P.J.; Stevenson, D.M.; Aylward, F.O.; Boyum, J.; Deneke, J.; Drinkwater, C.; Ivanova, N.N.; Mikhailova, N.; Chertkov, O.; et al. The complete genome sequence of Fibrobacter succinogenes S85 reveals a cellulolytic and metabolic specialist. PLoS ONE 2011, 6, e18814. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-R.; Yu, Y.; Luo, W.; Zeng, Y.-X. Marisediminicola antarctica gen. nov., sp. nov., an actinobacterium isolated from the Antarctic. Int. J. Syst. Evol. Microbiol. 2010, 60, 2535–2539. [Google Scholar] [CrossRef] [PubMed]
- Hamada, M.; Ichikawa, N.; Oguchi, A.; Komaki, H.; Tamura, T.; Fujita, N. Draft genome sequences of eight type strains of the genus demequina. Genome Announc. 2015, 3, e00281-15. [Google Scholar] [CrossRef] [PubMed]
- Simpson, A.J.G.; Reinach, F.C.; Arruda, P.; Abreu, F.A.; Acencio, M.; Alvarenga, R.; Alves, L.M.C.; Araya, J.E.; Baia, G.S.; Baptista, C.S.; et al. The genome sequence of the plant pathogen Xylella fastidiosa. The Xylella fastidiosa consortium of the organization for nucleotide sequencing and analysis. Nature 2000, 406, 151–159. [Google Scholar] [PubMed]
- Holmes, D.E.; Nevin, K.P.; Woodard, T.L.; Peacock, A. D.; Lovley, D.R. Prolixibacter bellariivorans gen. nov., sp. nov., a sugar-fermenting, psychrotolerant anaerobe of the phylum Bacteroidetes, isolated from a marine-sediment fuel cell. Int. J. Syst. Evol. Microbiol. 2007, 57, 701–707. [Google Scholar] [CrossRef] [PubMed]
- Meehan, C.J.; Beiko, R.G. A phylogenomic view of ecological specialization in the Lachnospiraceae, a family of digestive tract-associated bacteria. Genome Biol. Evol. 2014, 6, 703–713. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Niu, L.; Zhang, Y. Saccharofermentans acetigenes gen. nov., sp. nov., an anaerobic bacterium isolated from sludge treating brewery wastewater. Int. J. Syst. Evol. Microbiol. 2010, 60, 2735–2738. [Google Scholar] [CrossRef] [PubMed]
- Kudo, H.; Cheng, K-J.; Costerton, J.W. Interactions between Treponema bryantii and cellulolytic bacteria in the in vitro degradation of straw cellulose. Can. J. Microbiol. 1987, 33, 244–248. [Google Scholar] [CrossRef] [PubMed]
- Schmidtova, J.; Baldwin, S.A. Correlation of bacterial communities supported by different organic materials with sulfate reduction in metal-rich landfill leachate. Water Res. 2011, 45, 1115–1128. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Shi, M.; Lu, H.; Wu, D.; Shao, M.-F.; Zhang, T.; Ekama, G.A.; van Loosdrecht, M.C.M.; Chen, G.-H. Microbial community of sulfate-reducing up-flow sludge bed in the SANI® process for saline sewage treatment. Appl. Microbiol. Biotechnol. 2011, 90, 2015–2025. [Google Scholar] [CrossRef] [PubMed]
- Balch, W.E.; Schoberth, S.; Tanner, R.S.; Wolfe, R.S. Acetobacterium, a new genus of hydrogen-oxidizing, carbon dioxide-reducing, anaerobic bacteria. Int. J. Syst. Bacteriol. 1977, 27, 355–361. [Google Scholar] [CrossRef]
- Mattes, T.E.; Alexander, A.K.; Richardson, P.M.; Munk, A.C.; Han, C.S.; Stothard, P.; Coleman, N.V. The genome of Polaromonas sp. strain JS666: Insights into the evolution of a hydrocarbon- and xenobiotic-degrading bacterium, and features of relevance to biotechnology. Appl. Environ. Microbiol. 2008, 74, 6405–6416. [Google Scholar] [CrossRef] [PubMed]
- Masuda, H.; Shiwa, Y.; Yoshikawa, H.; Zylstra, G.J. Draft genome sequence of the versatile alkane-degrading bacterium Aquabacterium sp. strain NJ1. Genome Announc. 2014, 2. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Li, H.; Rensing, C.; Zhao, K.; Johnstone, L.; Wang, G. Genome sequence of the facultative anaerobic arsenite-oxidizing and nitrate-reducing bacterium Acidovorax sp. strain NO1. J. Bacteriol. 2012, 194, 1635–1636. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.; Wang, X.; Li, L.; Li, Z.; Luo, Y. Rhizobia and their bio-partners as novel drivers for functional remediation in contaminated soils. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Nierman, W.C.; Feldblyum, T.V.; Laub, M.T.; Paulsen, I.T.; Nelson, K.E.; Eisen, J.; Heidelberg, J.F.; Alley, M.R.K.; Ohta, N.; Maddock, J.R.; et al. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 2001, 98, 4136–4141. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Cui, H.; Jiang, Z.; Liu, H.; He, H.; Fang, N. Biomineralization processes of calcite induced by bacteria isolated from marine sediments. Braz. J. Microbiol. 2015, 46, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Pecheritsyna, S.A.; Rivkina, E.M.; Akimov, V.N.; Shcherbakova, V.A. Desulfovibrio arcticus sp. nov., a psychrotolerant sulfate-reducing bacterium from a cryopeg. Int. J. Syst. Evol. Microbiol. 2012, 62, 33–37. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaksonen, A.H.; Plumb, J.J.; Robertson, W.J.; Spring, S.; Schumann, P.; Franzmann, P.D.; Puhakka, J.A. Novel thermophilic sulfate-reducing bacteria from a geothermally active underground mine in Japan. Appl. Environ. Microbiol. 2006, 72, 3759–3762. [Google Scholar] [CrossRef] [PubMed]
- Karsten, S.; Jørgensen, B.B. Sulphate Reduction in Marine Sediments. In Marine Geochemistry; Schulz, H.D., Zabel, M., Eds.; Springer: Berlin, Germany, 2000; pp. 263–2812. [Google Scholar]
- Rui, J.; Li, J.; Zhang, S.; Yan, X.; Wang, Y.; Li, X. The core populations and co-occurrence patterns of prokaryotic communities in household biogas digesters. Biotechnol. Biofuels 2015, 8, 158. [Google Scholar] [CrossRef] [PubMed]
- Dollhopf, S.L.; Pariseau, M.L.; Hashsham, S.A.; Tiedje, J.M. Competitive and cooperative interactions affecting a fermentative spirochete in anaerobic chemostats. Microb. Ecol. 2003, 46, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Rohini-Kumar, M.; Saravanan, V.S. Candidate OP phyla: Importance, ecology and cultivation prospects. Indian J. Microbiol. 2010, 50, 474–477. [Google Scholar] [CrossRef] [PubMed]
- Wrighton, K.C.; Thomas, B.C.; Sharon, I.; Miller, C.S.; Castelle, C.J.; VerBerkmoes, N.C.; Wilkins, M.J.; Hettich, R.L.; Lipton, M.S.; Williams, K.H.; et al. Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science 2012, 337, 1661–1665. [Google Scholar] [CrossRef] [PubMed]
- Hibbing, M.E.; Fuqua, C.; Parsek, M.R.; Peterson, S.B. Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 2010, 8, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Mielczarek, A.T.; Kragelund, C.; Eriksen, P.S.; Nielsen, P.H. Population dynamics of filamentous bacteria in Danish wastewater treatment plants with nutrient removal. Water Res. 2012, 46, 3781–3795. [Google Scholar] [CrossRef] [PubMed]
- Eaton, A.D.; Clesceri, L.S.; Rice, E.W.; Greenberg, A.E. Standard Methods for the Examination of Water and Wastewater; American Public Health Association: Washington, D.C., USA, 2005. [Google Scholar]
- Verdouw, H.; van Echteld, C.J.A.; Dekkers, E.M.J. Ammonia determination based on indophenol formation with sodium salicylate. Water Res. 1978, 12, 399–402. [Google Scholar] [CrossRef]
- Behar, F.; Beaumont, V.; Penteado, H.D.B. Rock-Eval 6 technology: Performances and developments. Oil Gas Sci. Technol. 2001, 56, 111–134. [Google Scholar] [CrossRef]
- Pruitt, K.D.; Tatusova, T.; Klimke, W.; Maglott, D.R. NCBI reference sequences: Current status, policy and new initiatives. Nucleic Acids Res. 2009, 37, D32–D36. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Delsuc, F.; Dufayard, J.-F.; Gascuel, O. Estimating maximum likelihood phylogenies with PhyML. Methods Mol. Biol. 2009, 537, 113–137. [Google Scholar] [PubMed]
- Friedman, J.; Alm, E.J. Inferring correlation networks from genomic survey data. PLoS Comput. Biol. 2012, 8, e1002687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, M.T.; Taylor, L.T.; DeLong, E.F. Quantitative analysis of small-subunit rRNA genes in mixed microbial Populations via 5’-nuclease assays. Appl. Environ. Microbiol. 2000, 66, 4605–4614. [Google Scholar] [CrossRef] [PubMed]
- Vigneron, A.; Cruaud, P.; Pignet, P.; Caprais, J.-C.; Cambon-Bonavita, M.-A.; Godfroy, A.; Toffin, L. Archaeal and anaerobic methane oxidizer communities in the Sonora Margin cold seeps, Guaymas Basin (Gulf of California). ISME J. 2013, 7, 1595–1608. [Google Scholar] [CrossRef] [PubMed]





© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baldwin, S.A.; Mattes, A.; Rezadehbashi, M.; Taylor, J. Seasonal Microbial Population Shifts in a Bioremediation System Treating Metal and Sulfate-Rich Seepage. Minerals 2016, 6, 36. https://doi.org/10.3390/min6020036
Baldwin SA, Mattes A, Rezadehbashi M, Taylor J. Seasonal Microbial Population Shifts in a Bioremediation System Treating Metal and Sulfate-Rich Seepage. Minerals. 2016; 6(2):36. https://doi.org/10.3390/min6020036
Chicago/Turabian StyleBaldwin, Susan A., Al Mattes, Maryam Rezadehbashi, and Jon Taylor. 2016. "Seasonal Microbial Population Shifts in a Bioremediation System Treating Metal and Sulfate-Rich Seepage" Minerals 6, no. 2: 36. https://doi.org/10.3390/min6020036
APA StyleBaldwin, S. A., Mattes, A., Rezadehbashi, M., & Taylor, J. (2016). Seasonal Microbial Population Shifts in a Bioremediation System Treating Metal and Sulfate-Rich Seepage. Minerals, 6(2), 36. https://doi.org/10.3390/min6020036