Trends in the Use of Proper Methods for Estimating Mutation Rates in Fluctuation Experiments
Abstract
1. Introduction
2. Material and Methods
2.1. Literature Search Selection Criteria
2.2. Data Curation
3. Results and Discussion
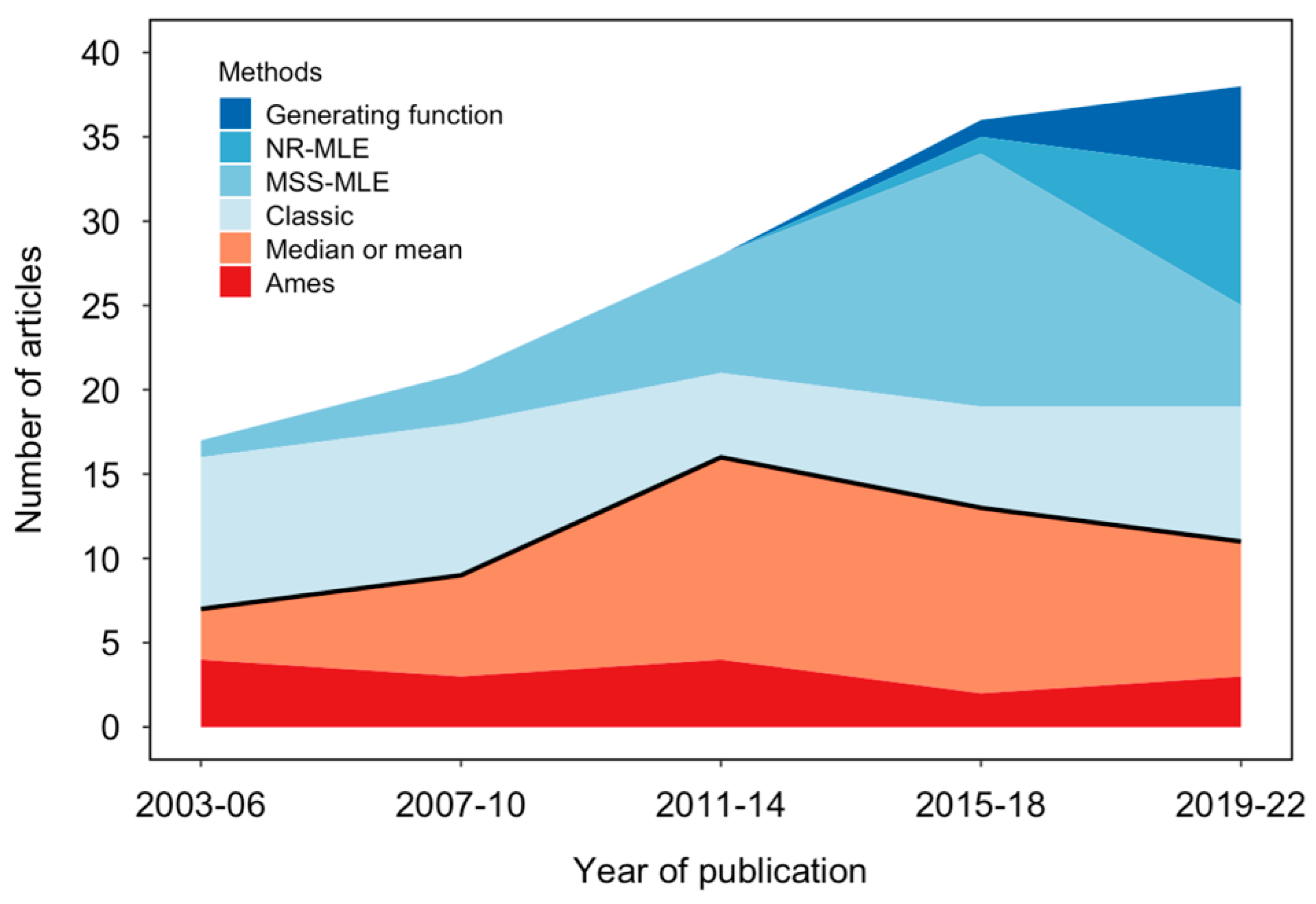
3.1. Proper Methods for the Rise Linked to Increased Computational Availability
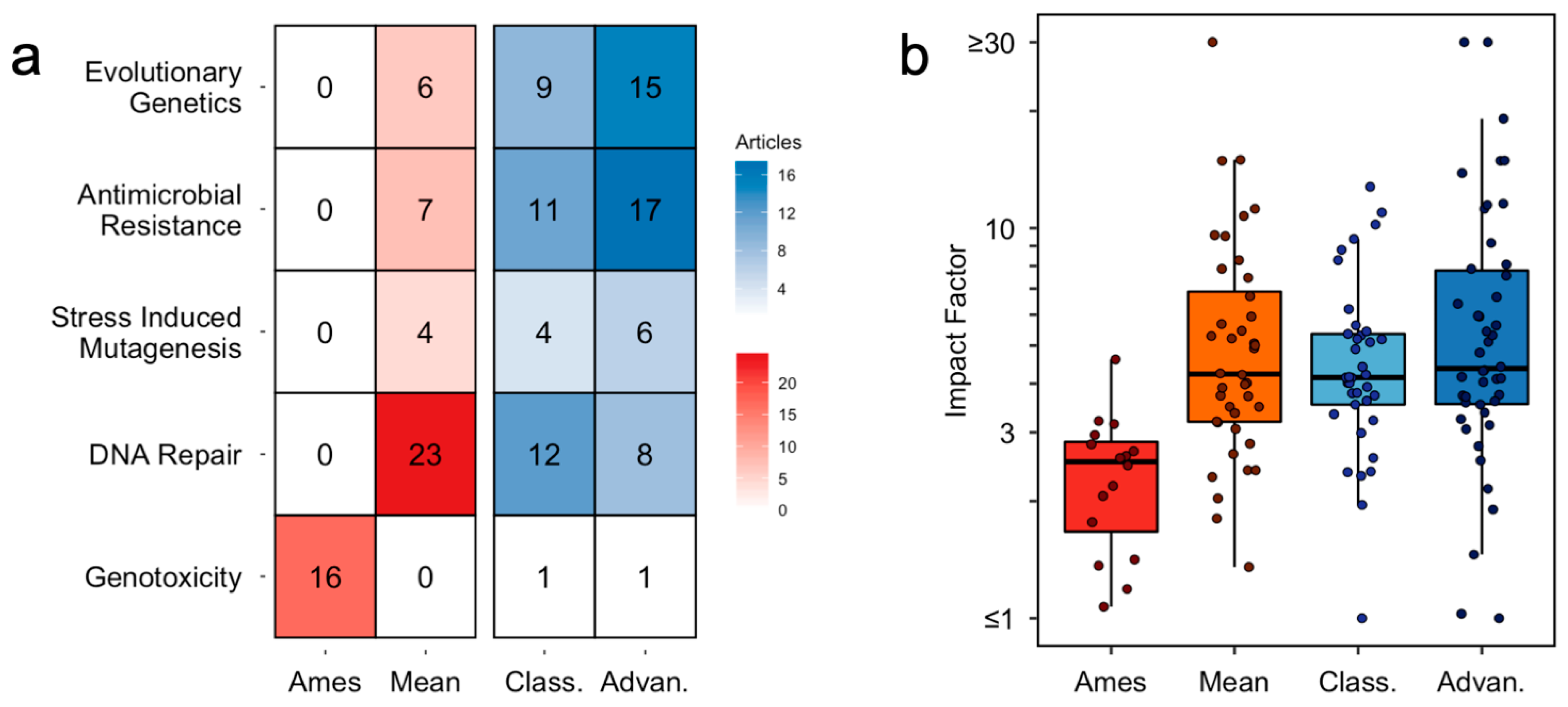
3.2. Inappropriate Methods Are Still Widespread, but Their Prevalence Varies across Sub-Fields
3.3. Impact Factor Fails to Predict Inappropriate Method Usage
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Robert, L.; Ollion, J.; Robert, J.; Song, X.; Matic, I.; Elez, M. Mutation dynamics and fitness effects followed in single cells. Science 2018, 359, 1283–1286. [Google Scholar] [CrossRef] [PubMed]
- Cairns, J. Mutation selection and the natural history of cancer. Nature 1975, 255, 197–200. [Google Scholar] [CrossRef] [PubMed]
- Shanks, G.D.; Edstein, M.D.; Jacobus, D. Evolution from double to triple-antimalarial drug combinations. Trans. R. Soc. Trop. Med. Hyg. 2015, 109, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Lythgoe, K.A.; Gardner, A.; Pybus, O.G.; Grove, J. Short-Sighted Virus Evolution and a Germline Hypothesis for Chronic Viral Infections. Trends Microbiol. 2017, 25, 336–348. [Google Scholar] [CrossRef]
- Baym, M.; Stone, L.K.; Kishony, R. Multidrug evolutionary strategies to reverse antibiotic resistance. Science 2016, 351, aad3292. [Google Scholar] [CrossRef] [PubMed]
- Blazejewski, T.; Ho, H.-I.; Wang, H.H. Synthetic sequence entanglement augments stability and containment of genetic information in cells. Science 2019, 365, 595–598. [Google Scholar] [CrossRef] [PubMed]
- Burt, A. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc. R. Soc. B Biol. Sci. 2003, 270, 921–928. [Google Scholar] [CrossRef]
- Luria, S.E.; Delbrück, M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics 1943, 28, 491–511. [Google Scholar] [CrossRef]
- Wahl, L.M.; Agashe, D. Selection bias in mutation accumulation. Evolution 2022, 76, 528–540. [Google Scholar] [CrossRef]
- Mahilkar, A.; Raj, N.; Kemkar, S.; Saini, S. Selection in a growing colony biases results of mutation accumulation experiments. Sci. Rep. 2022, 12, 15470. [Google Scholar] [CrossRef]
- Bachl, J.; Dessing, M.; Olsson, C.; von Borstel, R.C.; Steinberg, C. An experimental solution for the Luria–Delbrück fluctuation problem in measuring hypermutation rates. Proc. Natl. Acad. Sci. USA 1999, 96, 6847–6849. [Google Scholar] [CrossRef] [PubMed]
- Rosche, W.A.; Foster, P.L. Determining Mutation Rates in Bacterial Populations. Methods 2000, 20, 4–17. [Google Scholar] [CrossRef] [PubMed]
- Lea, D.E.; Coulson, C.A. The distribution of the numbers of mutants in bacterial populations. J. Genet. 1949, 49, 264–285. [Google Scholar] [CrossRef] [PubMed]
- Couce, A.; Blázquez, J. Estimating mutation rates in low-replication experiments. Mutat. Res. Mol. Mech. Mutagen. 2011, 714, 26–32. [Google Scholar] [CrossRef]
- Zheng, Q. Progress of a half century in the study of the Luria–Delbrück distribution. Math. Biosci. 1999, 162, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.E. An improved estimator of spontaneous mutation rates in Luria-Delbrück fluctuation experiments. Mutat. Res. Mutagen. Relat. Subj. 1993, 292, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q. A new practical guide to the Luria–Delbrück protocol. Mutat. Res. Mol. Mech. Mutagen. 2015, 781, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Drake, J.W. A constant rate of spontaneous mutation in DNA-based microbes. Proc. Natl. Acad. Sci. USA 1991, 88, 7160–7164. [Google Scholar] [CrossRef]
- Foster, P.L. Methods for Determining Spontaneous Mutation Rates. Methods Enzymol. 2006, 409, 195–213. [Google Scholar] [CrossRef]
- Sarkar, S.; Ma, W.T.; Sandri, G.v.H. On fluctuation analysis: A new, simple and efficient method for computing the expected number of mutants. Genetica 1992, 85, 173–179. [Google Scholar] [CrossRef]
- Hall, B.M.; Ma, C.-X.; Liang, P.; Singh, K.K. Fluctuation AnaLysis CalculatOR: A web tool for the determination of mutation rate using Luria–Delbrück fluctuation analysis. Bioinformatics 2009, 25, 1564–1565. [Google Scholar] [CrossRef] [PubMed]
- Gillet-Markowska, A.; Louvel, G.; Fischer, G. bz-rates: A Web Tool to Estimate Mutation Rates from Fluctuation Analysis. G3 GenesGenomesGenet. 2015, 5, 2323–2327. [Google Scholar] [CrossRef] [PubMed]
- Mazoyer, A.; Drouilhet, R.; Despréaux, S.; Ycart, B. flan: An R Package for Inference on Mutation Models. R J. 2017, 9, 334–351. [Google Scholar] [CrossRef]
- Zheng, Q. rSalvador: An R Package for the Fluctuation Experiment. G3 GenesGenomesGenet. 2017, 7, 3849–3856. [Google Scholar] [CrossRef]
- Zheng, Q. webSalvador: A Web Tool for the Luria-Delbrük Experiment. Microbiol. Resour. Announc. 2021, 10, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Łazowski, K. Efficient, robust, and versatile fluctuation data analysis using MLE MUtation Rate calculator (mlemur). Mutat. Res. Mol. Mech. Mutagen. 2023, 826, 111816. [Google Scholar] [CrossRef]
- Radchenko, E.A.; McGinty, R.J.; Aksenova, A.Y.; Neil, A.J.; Mirkin, S.M. Quantitative analysis of the rates for repeat-mediated genome instability in a yeast experimental system. Methods Mol. Biol. 2018, 1672, 421–438. [Google Scholar] [CrossRef]
- Ycart, B.; Veziris, N. Unbiased Estimation of Mutation Rates under Fluctuating Final Counts. PLoS ONE 2014, 9, e101434. [Google Scholar] [CrossRef]
- Frenoy, A.; Bonhoeffer, S. Death and population dynamics affect mutation rate estimates and evolvability under stress in bacteria. PLoS Biol. 2018, 16, e2005056. [Google Scholar] [CrossRef]
- Schreck, C.F.; Fusco, D.; Karita, Y.; Martis, S.; Kayser, J.; Duvernoy, M.-C.; Hallatschek, O. Impact of crowding on the diversity of expanding populations. Proc. Natl. Acad. Sci. USA 2023, 120, e2208361120. [Google Scholar] [CrossRef]
- Kindler, O.; Pulkkinen, O.; Cherstvy, A.G.; Metzler, R. Burst statistics in an early biofilm quorum sensing model: The role of spatial colony-growth heterogeneity. Sci. Rep. 2019, 9, 12077. [Google Scholar] [CrossRef] [PubMed]
- Ames, B.N.; Durston, W.E.; Yamasaki, E.; Lee, F.D. Carcinogens are Mutagens: A Simple Test System Combining Liver Homogenates for Activation and Bacteria for Detection. Proc. Natl. Acad. Sci. USA 1973, 70, 2281–2285. [Google Scholar] [CrossRef] [PubMed]
- Ames, B.N.; Lee, F.D.; Durston, W.E. An Improved Bacterial Test System for the Detection and Classification of Mutagens and Carcinogens. Proc. Natl. Acad. Sci. USA 1973, 70, 782–786. [Google Scholar] [CrossRef]
- Dearfield, K.L.; Auletta, A.E.; Cimino, M.C.; Moore, M.M. Considerations in the U.S. Environmental Protection Agency’s testing approach for mutagenicity. Mutat. Res. Genet. Toxicol. 1991, 258, 259–283. [Google Scholar] [CrossRef] [PubMed]
- Center for Food Safety and Applied Nutrition. Guidance for Industry and Other Stakeholders: Redbook. 2000. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/guidance-industry-and-other-stakeholders-redbook-2000 (accessed on 1 August 2023).
- OECD. Test No. 471: Bacterial Reverse Mutation Test; Organisation for Economic Co-operation and Development: Paris, France, 2020. [Google Scholar]
- Zeiger, E.; Anderson, B.; Haworth, S.; Lawlor, T.; Mortelmans, K. Salmonella mutagenicity tests: V. Results from the testing of 311 chemicals. Environ. Mol. Mutagen. 1992, 19, 2–141. [Google Scholar] [CrossRef] [PubMed]
- Al Mamun, A.A.M.; Lombardo, M.-J.; Shee, C.; Lisewski, A.M.; Gonzalez, C.; Lin, D.; Nehring, R.B.; Saint-Ruf, C.; Gibson, J.L.; Frisch, R.L.; et al. Identity and Function of a Large Gene Network Underlying Mutagenic Repair of DNA Breaks. Science 2012, 338, 1344–1348. [Google Scholar] [CrossRef] [PubMed]
- Elez, M.; Radman, M.; Matic, I. The frequency and structure of recombinant products is determined by the cellular level of MutL. Proc. Natl. Acad. Sci. USA 2007, 104, 8935–8940. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, A.; Laureti, L.; Crussard, S.; Abida, H.; Rodríguez-Rojas, A.; Blázquez, J.; Baharoglu, Z.; Mazel, D.; Darfeuille, F.; Vogel, J.; et al. β-lactam antibiotics promote bacterial mutagenesis via an RpoS-mediated reduction in replication fidelity. Nat. Commun. 2013, 4, 1610. [Google Scholar] [CrossRef]
- Thi, T.D.; López, E.; Rodríguez-Rojas, A.; Rodríguez-Beltrán, J.; Couce, A.; Guelfo, J.R.; Castañeda-García, A.; Blázquez, J. Effect of recA inactivation on mutagenesis of Escherichia coli exposed to sublethal concentrations of antimicrobials. J. Antimicrob. Chemother. 2011, 66, 531–538. [Google Scholar] [CrossRef]
- Clarivate Journal Citation Reports. Clarivate Analytics. 2023. Available online: https://jcr.clarivate.com/jcr/home (accessed on 1 August 2023).
- Pinheiro, F.; Warsi, O.; Andersson, D.I.; Lässig, M. Metabolic fitness landscapes predict the evolution of antibiotic resistance. Nat. Ecol. Evol. 2021, 5, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Lee, J.J.; Lee, S.-K.; Kim, S.; Eum, S.-Y.; Eoh, H. Phosphoenolpyruvate depletion mediates both growth arrest and drug tolerance of Mycobacterium tuberculosis in hypoxia. Proc. Natl. Acad. Sci. USA 2021, 118, e2105800118. [Google Scholar] [CrossRef] [PubMed]
- Grimsey, E.M.; Weston, N.; Ricci, V.; Stone, J.W.; Piddock, L.J.V. Overexpression of RamA, Which Regulates Production of the Multidrug Resistance Efflux Pump AcrAB-TolC, Increases Mutation Rate and Influences Drug Resistance Phenotype. Antimicrob. Agents Chemother. 2020, 64, e02460-19. [Google Scholar] [CrossRef]
- Nilsson, A.I.; Berg, O.G.; Aspevall, O.; Kahlmeter, G.; Andersson, D.I. Biological Costs and Mechanisms of Fosfomycin Resistance in Escherichia coli. Antimicrob. Agents Chemother. 2003, 47, 2850–2858. [Google Scholar] [CrossRef]
- Beaufay, F.; Amemiya, H.M.; Guan, J.; Basalla, J.; Meinen, B.A.; Chen, Z.; Mitra, R.; Bardwell, J.C.A.; Biteen, J.S.; Vecchiarelli, A.G.; et al. Polyphosphate drives bacterial heterochromatin formation. Sci. Adv. 2021, 7, eabk0233. [Google Scholar] [CrossRef]
- Katzer, A.; Hockertz, S.; Buchhorn, G.H.; Loehr, J.F. In vitro toxicity and mutagenicity of CoCrMo and Ti6Al wear particles. Toxicology 2003, 190, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Haorah, J.; Chen, S.C.; Wang, X.; Kolar, C.; Lawson, T.A.; Mirvish, S.S. Nitrosation of Glycine Ethyl Ester and Ethyl Diazoacetate To Give the Alkylating Agent and Mutagen Ethyl Chloro(hydroximino)acetate. Chem. Res. Toxicol. 2004, 17, 416–423. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.-K.; Choi, J.-S.; Gil, H.-W.; Yang, J.-O.; Lee, E.-Y.; Jeon, Y.-S.; Lee, Z.-W.; Lee, M.; Hong, M.-Y.; Son, T.-H.; et al. Genotoxicity evaluation of electromagnetic fields generated by 835-MHz mobile phone frequency band. Eur. J. Cancer Prev. 2005, 14, 175–179. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, H.S.; Bicalho, B.; Genari, P.; Santagada, V.; Caliendo, G.; Perissutti, E.; Donato, J.L.; De Nucci, G. In vitro mutagenicity of anti-inflammatory parsalmide analogues PA7, PA10, and PA31 triggered by biotransformation into hydroxy derivatives. Eur. J. Med. Chem. 2006, 41, 408–416. [Google Scholar] [CrossRef]
- Friedrich, R.E. Mutagenicity Testing of Mouthwash Brands. Anticancer Res. 2007, 27, 2091–2098. [Google Scholar] [PubMed]
- Cockell, C.S.; Tsikos, H.; Durante, M.; Parnell, J. Microbe–mineral interactions in naturally radioactive beach sands from Espirito Santo, Brazil: Experiments on mutagenicity. Radiat. Environ. Biophys. 2007, 46, 247–253. [Google Scholar] [CrossRef][Green Version]
- Vargas, V.M.F.; Migliavacca, S.B.; Horn, R.C.; Terra, N.R. Comparative temporal ecotoxicological study in a river basin influenced by petrochemical industries. Sci. Total Environ. 2008, 392, 79–92. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, W.M.; Hussin, W.A.; Al-Faiyz, Y.S.; Ismail, M.A. The position of imidazopyridine and metabolic activation are pivotal factors in the antimutagenic activity of novel imidazo[1,2-a]pyridine derivatives. Eur. J. Pharmacol. 2013, 715, 212–218. [Google Scholar] [CrossRef]
- Thurnham, D.I.; Howard, A.N. Studies on meso-zeaxanthin for potential toxicity and mutagenicity. Food Chem. Toxicol. 2013, 59, 455–463. [Google Scholar] [CrossRef]
- Mölzer, C.; Huber, H.; Steyrer, A.; Ziesel, G.V.; Wallner, M.; Goncharova, I.; Orlov, S.; Urbanová, M.; Ahlfors, C.E.; Vítek, L.; et al. Interaction between TNFone and tetrapyrroles may account for their anti-genotoxic effects—A novel mechanism for DNA-protection. J. Porphyr. Phthalocyanines 2013, 17, 1157–1166. [Google Scholar] [CrossRef]
- Mölzer, C.; Huber, H.; Diem, K.; Wallner, M.; Bulmer, A.C.; Wagner, K.-H. Extracellular and intracellular anti-mutagenic effects of bile pigments in the Salmonella typhimurium reverse mutation assay. Toxicol. In Vitro 2013, 27–360, 433–437. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Liu, X.; Gong, W.; Hou, J. Assessing the Mutagenic Potential of Surface Sediments from Beijing Guanting Reservoir to Salmonella typhimurium. Soil Sediment Contam. Int. J. 2015, 24, 306–324. [Google Scholar] [CrossRef]
- Wang, W.-Q.; Duan, H.-X.; Pei, Z.-T.; Xu, R.-R.; Qin, Z.-T.; Zhu, G.-C.; Sun, L.-W. Evaluation by the Ames Assay of the Mutagenicity of UV Filters Using Benzophenone and Benzophenone-1. Int. J. Environ. Res. Public Health 2018, 15, 1907. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, W.; Pei, Z.; Wu, J.; Yu, R.; Zhang, Y.; Sun, L.; Gao, Y. Mutagenicity Assessment to Pesticide Adjuvants of Toluene, Chloroform, and Trichloroethylene by Ames Test. Int. J. Environ. Res. Public Health 2021, 18, 8095. [Google Scholar] [CrossRef] [PubMed]
- Patenall, B.L.; Hathaway, H.J.; Laabei, M.; Young, A.E.; Thet, N.T.; Jenkins, A.T.A.; Short, R.D.; Allinson, S.L. Assessment of mutations induced by cold atmospheric plasma jet treatment relative to known mutagens in Escherichia coli. Mutagenesis 2021, 36, 380–387. [Google Scholar] [CrossRef]
- Glover, S.A.; Schumacher, R.R. Mutagenicity of N-acyloxy-N-alkoxyamides as an indicator of DNA intercalation: The role of fluorene and fluorenone substituents as DNA intercalators. Mutat. Res. Toxicol. Environ. Mutagen. 2021, 863–864, 503299. [Google Scholar] [CrossRef]
- Ismail, N.; Omar, S.V.; Ismail, N.A.; Peters, R.P.H. In vitro approaches for generation of Mycobacterium tuberculosis mutants resistant to bedaquiline, clofazimine or linezolid and identification of associated genetic variants. J. Microbiol. Methods 2018, 153, 1–9. [Google Scholar] [CrossRef]
- Werngren, J.; Hoffner, S.E. Drug-Susceptible Mycobacterium tuberculosis Beijing Genotype Does Not Develop Mutation-Conferred Resistance to Rifampin at an Elevated Rate. J. Clin. Microbiol. 2003, 41, 1520–1524. [Google Scholar] [CrossRef]
- Bifani, P.; Mathema, B.; Kurepina, N.; Shashkina, E.; Bertout, J.; Blanchis, A.S.; Moghazeh, S.; Driscoll, J.; Gicquel, B.; Frothingham, R.; et al. The Evolution of Drug Resistance in Mycobacterium tuberculosis: From a Mono–Rifampin-Resistant Cluster into Increasingly Multidrug-Resistant Variants in an HIV-Seropositive Population. J. Infect. Dis. 2008, 198, 90–94. [Google Scholar] [CrossRef]
- Levy, D.D.; Sharma, B.; Cebula, T.A. Single-Nucleotide Polymorphism Mutation Spectra and Resistance to Quinolones in Salmonella enterica Serovar Enteritidis with a Mutator Phenotype. Antimicrob. Agents Chemother. 2004, 48, 2355–2363. [Google Scholar] [CrossRef]
- Stumpf, J.D.; Poteete, A.R.; Foster, P.L. Amplification of lac Cannot Account for Adaptive Mutation to Lac+ in Escherichia coli. J. Bacteriol. 2007, 189, 2291. [Google Scholar] [CrossRef]
- Pu, X.-Y.; Zhang, Q.; Pan, J.-C.; Shen, Z.; Zhang, W. Spontaneous mutation frequency and molecular mechanisms of Shigellaflexneri fluoroquinolone resistance under antibiotic selective stress. World J. Microbiol. Biotechnol. 2013, 29, 365–371. [Google Scholar] [CrossRef]
- Li, K.; Wang, X.; Yang, S.; Gu, J.; Deng, J.; Zhang, X.-E. Anti-folates potentiate bactericidal effects of other antimicrobial agents. J. Antibiot. (Tokyo) 2017, 70, 285–291. [Google Scholar] [CrossRef]
- Mead, S.; Vaisman, A.; Valjavec-Gratian, M.; Karata, K.; Vandewiele, D.; Woodgate, R. Characterization of polVR391: A Y-family polymerase encoded by rumA′B from the IncJ conjugative transposon, R391. Mol. Microbiol. 2007, 63, 797–810. [Google Scholar] [CrossRef]
- Criss, A.K.; Bonney, K.M.; Chang, R.A.; Duffin, P.M.; LeCuyer, B.E.; Seifert, H.S. Mismatch Correction Modulates Mutation Frequency and Pilus Phase and Antigenic Variation in Neisseria gonorrhoeae. J. Bacteriol. 2010, 192, 316–325. [Google Scholar] [CrossRef]
- Duvernay, C.; Coulange, L.; Dutilh, B.; Dubois, V.; Quentin, C.; Arpin, C. Duplication of the chromosomal blaSHV-11 gene in a clinical hypermutable strain of Klebsiella pneumoniae. Microbiology 2011, 157, 496–503. [Google Scholar] [CrossRef]
- Cahoon, L.A.; Stohl, E.A.; Seifert, H.S. The Neisseria gonorrhoeae photolyase orthologue phrB is required for proper DNA supercoiling but does not function in photo-reactivation. Mol. Microbiol. 2011, 79, 729–742. [Google Scholar] [CrossRef]
- Robles, A.g.; Reid, K.; Roy, F.; Fletcher, H.m. Porphyromonas gingivalis mutY is involved in the repair of oxidative stress-induced DNA mispairing. Mol. Oral Microbiol. 2011, 26, 175–186. [Google Scholar] [CrossRef]
- Janowska, B.; Kurpios-Piec, D.; Prorok, P.; Szparecki, G.; Komisarski, M.; Kowalczyk, P.; Janion, C.; Tudek, B. Role of damage-specific DNA polymerases in M13 phage mutagenesis induced by a major lipid peroxidation product trans-4-hydroxy-2-nonenal. Mutat. Res. 2012, 729, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Tsai, H.-H.; Shu, H.-W.; Yang, C.-C.; Chen, C.W. Translesion-synthesis DNA polymerases participate in replication of the telomeres in Streptomyces. Nucleic Acids Res. 2012, 40, 1118–1130. [Google Scholar] [CrossRef] [PubMed]
- Keith, B.J.; Jozwiakowski, S.K.; Connolly, B.A. A plasmid-based lacZα gene assay for DNA polymerase fidelity measurement. Anal. Biochem. 2013, 433, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Laureti, L.; Selva, M.; Dairou, J.; Matic, I. Reduction of dNTP levels enhances DNA replication fidelity in vivo. DNA Repair 2013, 12, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Muraoka, W.T.; Wu, Z.; Sahin, O.; Zhang, Q. A single nucleotide change in mutY increases the emergence of antibiotic-resistant Campylobacter jejuni mutants. J. Antimicrob. Chemother. 2015, 70, 2739–2748. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Marroquín, M.; Vidales, L.E.; Debora, B.N.; Santos-Escobar, F.; Obregón-Herrera, A.; Robleto, E.A.; Pedraza-Reyes, M. Role of Bacillus subtilis DNA Glycosylase MutM in Counteracting Oxidatively Induced DNA Damage and in Stationary-Phase-Associated Mutagenesis. J. Bacteriol. 2015, 197, 1963–1971. [Google Scholar] [CrossRef] [PubMed]
- Couce, A.; Alonso-Rodriguez, N.; Costas, C.; Oliver, A.; Blázquez, J. Intrapopulation variability in mutator prevalence among urinary tract infection isolates of Escherichia coli. Clin. Microbiol. Infect. 2016, 22, 566.e1–566.e7. [Google Scholar] [CrossRef]
- Nishimura, I.; Kurokawa, M.; Liu, L.; Ying, B.-W. Coordinated Changes in Mutation and Growth Rates Induced by Genome Reduction. mBio 2017, 8, 10-1128. [Google Scholar] [CrossRef]
- Hu, C.; Zhao, Y.; Sun, H.; Yang, Y. Synergism of Dam, MutH, and MutS in methylation-directed mismatch repair in Escherichia coli. Mutat. Res. Mol. Mech. Mutagen. 2017, 795, 31–33. [Google Scholar] [CrossRef][Green Version]
- Nakano, K.; Yamada, Y.; Takahashi, E.; Arimoto, S.; Okamoto, K.; Negishi, K.; Negishi, T.E. coli mismatch repair enhances AT-to-GC mutagenesis caused by alkylating agents. Mutat. Res. Toxicol. Environ. Mutagen. 2017, 815, 22–27. [Google Scholar] [CrossRef]
- Ukkivi, K.; Kivisaar, M. Involvement of transcription-coupled repair factor Mfd and DNA helicase UvrD in mutational processes in Pseudomonas putida. DNA Repair 2018, 72, 18–27. [Google Scholar] [CrossRef]
- Dupuy, P.; Howlader, M.; Glickman, M.S. A multilayered repair system protects the mycobacterial chromosome from endogenous and antibiotic-induced oxidative damage. Proc. Natl. Acad. Sci. USA 2020, 117, 19517–19527. [Google Scholar] [CrossRef]
- Miyabayashi, H.; Sakai, H.D.; Kurosawa, N. DNA Polymerase B1 Binding Protein 1 Is Important for DNA Repair by Holoenzyme PolB1 in the Extremely Thermophilic Crenarchaeon Sulfolobus acidocaldarius. Microorganisms 2021, 9, 439. [Google Scholar] [CrossRef] [PubMed]
- Vaisman, A.; Łazowski, K.; Reijns, M.A.M.; Walsh, E.; McDonald, J.P.; Moreno, K.C.; Quiros, D.R.; Schmidt, M.; Kranz, H.; Yang, W.; et al. Novel Escherichia coli active site dnaE alleles with altered base and sugar selectivity. Mol. Microbiol. 2021, 116, 909–925. [Google Scholar] [CrossRef] [PubMed]
- Naz, S.; Dabral, S.; Nagarajan, S.N.; Arora, D.; Singh, L.V.; Kumar, P.; Singh, Y.; Kumar, D.; Varshney, U.; Nandicoori, V.K. Compromised base excision repair pathway in Mycobacterium tuberculosis imparts superior adaptability in the host. PLoS Pathog. 2021, 17, e1009452. [Google Scholar] [CrossRef] [PubMed]
- Gifford, D.R.; Krašovec, R.; Aston, E.; Belavkin, R.V.; Channon, A.; Knight, C.G. Environmental pleiotropy and demographic history direct adaptation under antibiotic selection. Heredity 2018, 121, 438–448. [Google Scholar] [CrossRef] [PubMed]
- Kishimoto, T.; Iijima, L.; Tatsumi, M.; Ono, N.; Oyake, A.; Hashimoto, T.; Matsuo, M.; Okubo, M.; Suzuki, S.; Mori, K.; et al. Transition from positive to neutral in mutation fixation along with continuing rising fitness in thermal adaptive evolution. PLoS Genet. 2010, 6, e1001164. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Cai, Z.; Zhang, Y.; Li, Y. Engineering stress tolerance of Escherichia coli by stress-induced mutagenesis (SIM)-based adaptive evolution. Biotechnol. J. 2014, 9, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Giulieri, S.G.; Guérillot, R.; Kwong, J.C.; Monk, I.R.; Hayes, A.S.; Daniel, D.; Baines, S.; Sherry, N.L.; Holmes, N.E.; Ward, P.; et al. Comprehensive Genomic Investigation of Adaptive Mutations Driving the Low-Level Oxacillin Resistance Phenotype in Staphylococcus aureus. mBio 2020, 11, e02882-20. [Google Scholar] [CrossRef]
- Korry, B.J.; Lee, S.Y.E.; Chakrabarti, A.K.; Choi, A.H.; Ganser, C.; Machan, J.T.; Belenky, P. Genotoxic Agents Produce Stressor-Specific Spectra of Spectinomycin Resistance Mutations Based on Mechanism of Action and Selection in Bacillus subtilis. Antimicrob. Agents Chemother. 2021, 65, e0089121. [Google Scholar] [CrossRef]
- Witkowski, T.A.; Grice, A.N.; Stinnett, D.B.; Wells, W.K.; Peterson, M.A.; Hare, J.M. UmuDAb: An Error-Prone Polymerase Accessory Homolog Whose N-Terminal Domain Is Required for Repression of DNA Damage Inducible Gene Expression in Acinetobacter baylyi. PLoS ONE 2016, 11, e0152013. [Google Scholar] [CrossRef]
- Ojha, D.; Jaszczur, M.M.; Sikand, A.; McDonald, J.P.; Robinson, A.; van Oijen, A.M.; Mak, C.H.; Pinaud, F.; Cox, M.M.; Woodgate, R.; et al. Host cell RecA activates a mobile element-encoded mutagenic DNA polymerase. Nucleic Acids Res. 2022, 50, 6854–6869. [Google Scholar] [CrossRef]
- Gillespie, S.H.; Voelker, L.L.; Ambler, J.E.; Traini, C.; Dickens, A. Fluoroquinolone resistance in Streptococcus pneumoniae: Evidence that gyrA mutations arise at a lower rate and that mutation in gyrA or parC predisposes to further mutation. Microb. Drug Resist. Larchmt. N 2003, 9, 17–24. [Google Scholar] [CrossRef]
- Kuntaman, K.; Lestari, E.S.; Severin, J.A.; Kershof, I.M.; Mertaniasih, N.M.; Purwanta, M.; Hadi, U.; Johnson, J.R.; van Belkum, A.; Verbrugh, H.A. Fluoroquinolone-resistant Escherichia coli, Indonesia. Emerg. Infect. Dis. 2005, 11, 1363–1369. [Google Scholar] [CrossRef]
- Glaab, W.E.; Mitchell, L.S.; Miller, J.E.; Vlasakova, K.; Skopek, T.R. 5-fluorouracil forward mutation assay in Salmonella: Determination of mutational target and spontaneous mutational spectra. Mutat. Res. 2005, 578, 238–246. [Google Scholar] [CrossRef]
- Machowski, E.E.; Barichievy, S.; Springer, B.; Durbach, S.I.; Mizrahi, V. In vitro analysis of rates and spectra of mutations in a polymorphic region of the Rv0746 PE_PGRS gene of Mycobacterium tuberculosis. J. Bacteriol. 2007, 189, 2190–2195. [Google Scholar] [CrossRef]
- Arshad, R.; Farooq, S.; Ali, S.S. Effect of mutations induced by N-methyl-N′-nitro-N-nitrosoguanidine on expression of penicillin G acylase and β-lactamase in wild-type Escherichia coli strains. Ann. Microbiol. 2010, 60, 645–652. [Google Scholar] [CrossRef]
- Yang, M.; Gao, C.; Cui, T.; An, J.; He, Z.-G. A TetR-like regulator broadly affects the expressions of diverse genes in Mycobacterium smegmatis. Nucleic Acids Res. 2012, 40, 1009–1020. [Google Scholar] [CrossRef]
- Linkevicius, M.; Sandegren, L.; Andersson, D.I. Mechanisms and fitness costs of tigecycline resistance in Escherichia coli. J. Antimicrob. Chemother. 2013, 68, 2809–2819. [Google Scholar] [CrossRef]
- Leszczynska, D.; Matuszewska, E.; Kuczynska-Wisnik, D.; Furmanek-Blaszk, B.; Laskowska, E. The formation of persister cells in stationary-phase cultures of Escherichia coli is associated with the aggregation of endogenous proteins. PLoS ONE 2013, 8, e54737. [Google Scholar] [CrossRef]
- Wang, D.; Lin, Z.; Ding, X.; Hu, J.; Liu, Y. The Comparison of the Combined Toxicity between Gram-negative and Gram-positive Bacteria: A Case Study of Antibiotics and Quorum-sensing Inhibitors. Mol. Inform. 2016, 35, 54–61. [Google Scholar] [CrossRef]
- Tegova, R.; Tover, A.; Tarassova, K.; Tark, M.; Kivisaar, M. Involvement of error-prone DNA polymerase IV in stationary-phase mutagenesis in Pseudomonas putida. J. Bacteriol. 2004, 186, 2735–2744. [Google Scholar] [CrossRef]
- Mennecier, S.; Coste, G.; Servant, P.; Bailone, A.; Sommer, S. Mismatch repair ensures fidelity of replication and recombination in the radioresistant organism Deinococcus radiodurans. Mol. Genet. Genomics MGG 2004, 272, 460–469. [Google Scholar] [CrossRef]
- Stumpf, J.D.; Foster, P.L. Polyphosphate kinase regulates error-prone replication by DNA polymerase IV in Escherichia coli. Mol. Microbiol. 2005, 57, 751–761. [Google Scholar] [CrossRef]
- Koorits, L.; Tegova, R.; Tark, M.; Tarassova, K.; Tover, A.; Kivisaar, M. Study of involvement of ImuB and DnaE2 in stationary-phase mutagenesis in Pseudomonas putida. DNA Repair 2007, 6, 863–868. [Google Scholar] [CrossRef]
- Lind, P.A.; Andersson, D.I. Whole-genome mutational biases in bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 17878–17883. [Google Scholar] [CrossRef]
- Henderson-Begg, S.K.; Sheppard, C.L.; George, R.C.; Livermore, D.M.; Hall, L.M.C. Mutation frequency in antibiotic-resistant and -susceptible isolates of Streptococcus pneumoniae. Int. J. Antimicrob. Agents 2010, 35, 342–346. [Google Scholar] [CrossRef]
- Martina, P.; Feliziani, S.; Juan, C.; Bettiol, M.; Gatti, B.; Yantorno, O.; Smania, A.M.; Oliver, A.; Bosch, A. Hypermutation in Burkholderia cepacia complex is mediated by DNA mismatch repair inactivation and is highly prevalent in cystic fibrosis chronic respiratory infection. Int. J. Med. Microbiol. IJMM 2014, 304, 1182–1191. [Google Scholar] [CrossRef]
- Hassim, F.; Papadopoulos, A.O.; Kana, B.D.; Gordhan, B.G. A combinatorial role for MutY and Fpg DNA glycosylases in mutation avoidance in Mycobacterium smegmatis. Mutat. Res. 2015, 779, 24–32. [Google Scholar] [CrossRef]
- Castro-Cerritos, K.V.; Yasbin, R.E.; Robleto, E.A.; Pedraza-Reyes, M. Role of Ribonucleotide Reductase in Bacillus subtilis Stress-Associated Mutagenesis. J. Bacteriol. 2017, 199, e00715-16. [Google Scholar] [CrossRef]
- Ivanković, S.; Vujaklija, D.; Đermić, D. Nucleolytic degradation of 3’-ending overhangs is essential for DNA-end resection in RecA-loading deficient recB mutants of Escherichia coli. DNA Repair 2017, 57, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Martin, H.A.; Porter, K.E.; Vallin, C.; Ermi, T.; Contreras, N.; Pedraza-Reyes, M.; Robleto, E.A. Mfd protects against oxidative stress in Bacillus subtilis independently of its canonical function in DNA repair. BMC Microbiol. 2019, 19, 26. [Google Scholar] [CrossRef] [PubMed]
- Grøsvik, K.; Tesfahun, A.N.; Muruzábal-Lecumberri, I.; Haugland, G.T.; Leiros, I.; Ruoff, P.; Kvaløy, J.T.; Knævelsrud, I.; Ånensen, H.; Alexeeva, M.; et al. The Escherichia coli alkA Gene Is Activated to Alleviate Mutagenesis by an Oxidized Deoxynucleoside. Front. Microbiol. 2020, 11, 263. [Google Scholar] [CrossRef] [PubMed]
- Oliver, A.; Levin, B.R.; Juan, C.; Baquero, F.; Blázquez, J. Hypermutation and the preexistence of antibiotic-resistant Pseudomonas aeruginosa mutants: Implications for susceptibility testing and treatment of chronic infections. Antimicrob. Agents Chemother. 2004, 48, 4226–4233. [Google Scholar] [CrossRef] [PubMed]
- Bergval, I.; Kwok, B.; Schuitema, A.; Kremer, K.; van Soolingen, D.; Klatser, P.; Anthony, R. Pre-existing isoniazid resistance, but not the genotype of Mycobacterium tuberculosis drives rifampicin resistance codon preference in vitro. PLoS ONE 2012, 7, e29108. [Google Scholar] [CrossRef] [PubMed]
- Gifford, D.R.; Furió, V.; Papkou, A.; Vogwill, T.; Oliver, A.; MacLean, R.C. Identifying and exploiting genes that potentiate the evolution of antibiotic resistance. Nat. Ecol. Evol. 2018, 2, 1033–1039. [Google Scholar] [CrossRef]
- Huseby, D.L.; Pietsch, F.; Brandis, G.; Garoff, L.; Tegehall, A.; Hughes, D. Mutation Supply and Relative Fitness Shape the Genotypes of Ciprofloxacin-Resistant Escherichia coli. Mol. Biol. Evol. 2017, 34, 1029–1039. [Google Scholar] [CrossRef] [PubMed]
- Kapel, N.; Caballero, J.D.; MacLean, R.C. Localized pmrB hypermutation drives the evolution of colistin heteroresistance. Cell Rep. 2022, 39, 110929. [Google Scholar] [CrossRef] [PubMed]
- Lao, Z.; Matsui, Y.; Ijichi, S.; Ying, B.-W. Global coordination of the mutation and growth rates across the genetic and nutritional variety in Escherichia coli. Front. Microbiol. 2022, 13, 990969. [Google Scholar] [CrossRef]
- Kim, M.; Wolff, E.; Huang, T.; Garibyan, L.; Earl, A.M.; Battista, J.R.; Miller, J.H. Developing a genetic system in Deinococcus radiodurans for analyzing mutations. Genetics 2004, 166, 661–668. [Google Scholar] [CrossRef]
- Pope, C.F.; Gillespie, S.H.; Moore, J.E.; McHugh, T.D. Approaches to measure the fitness of Burkholderia cepacia complex isolates. J. Med. Microbiol. 2010, 59, 679–686. [Google Scholar] [CrossRef]
- Tag ElDein, M.A.; Yassin, A.S.; El-Tayeb, O.; Kashef, M.T. Chlorhexidine leads to the evolution of antibiotic-resistant Pseudomonas aeruginosa. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 2349–2361. [Google Scholar] [CrossRef]
- Lee, G.; Wolff, E.; Miller, J.H. Mutagenicity of the cytidine analog zebularine in Escherichia coli. DNA Repair 2004, 3, 155–161. [Google Scholar] [CrossRef]
- Al Mamun, A.A.M. Elevated expression of DNA polymerase II increases spontaneous mutagenesis in Escherichia coli. Mutat. Res. 2007, 625, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Curti, E.; McDonald, J.P.; Mead, S.; Woodgate, R. DNA polymerase switching: Effects on spontaneous mutagenesis in Escherichia coli. Mol. Microbiol. 2009, 71, 315–331. [Google Scholar] [CrossRef]
- Singh, A.; Karimpour-Fard, A.; Gill, R.T. Increased mutation frequency in redox-impaired Escherichia coli due to RelA- and RpoS-mediated repression of DNA repair. Appl. Environ. Microbiol. 2010, 76, 5463–5470. [Google Scholar] [CrossRef] [PubMed]
- Deventer, A.T.; Bryson, D.; Shortill, M.; Boraston, A.B.; Hobbs, J.K. Molecular Characterization of Clinical Rel Mutations and Consequences for Resistance Expression and Fitness in Staphylococcus aureus. Antimicrob. Agents Chemother. 2022, 66, e0093822. [Google Scholar] [CrossRef] [PubMed]
- Lopatkin, A.J.; Bening, S.C.; Manson, A.L.; Stokes, J.M.; Kohanski, M.A.; Badran, A.H.; Earl, A.M.; Cheney, N.J.; Yang, J.H.; Collins, J.J. Clinically relevant mutations in core metabolic genes confer antibiotic resistance. Science 2021, 371, eaba0862. [Google Scholar] [CrossRef]
- Grinnage-Pulley, T.; Zhang, Q. Genetic Basis and Functional Consequences of Differential Expression of the CmeABC Efflux Pump in Campylobacter jejuni Isolates. PLoS ONE 2015, 10, e0131534. [Google Scholar] [CrossRef]
- Cho, J.; Carr, A.N.; Whitworth, L.; Johnson, B.; Wilson, K.S. MazEF toxin-antitoxin proteins alter Escherichia coli cell morphology and infrastructure during persister formation and regrowth. Microbiol. Read. Engl. 2017, 163, 308–321. [Google Scholar] [CrossRef]
- Wambaugh, M.A.; Shakya, V.P.S.; Lewis, A.J.; Mulvey, M.A.; Brown, J.C.S. High-throughput identification and rational design of synergistic small-molecule pairs for combating and bypassing antibiotic resistance. PLoS Biol. 2017, 15, e2001644. [Google Scholar] [CrossRef]
- Monti, M.R.; Morero, N.R.; Miguel, V.; Argaraña, C.E. nfxB as a novel target for analysis of mutation spectra in Pseudomonas aeruginosa. PLoS ONE 2013, 8, e66236. [Google Scholar] [CrossRef]
- Fehér, T.; Cseh, B.; Umenhoffer, K.; Karcagi, I.; Pósfai, G. Characterization of cycA mutants of Escherichia coli: An assay for measuring in vivo mutation rates. Mutat. Res. Mol. Mech. Mutagen. 2006, 595, 184–190. [Google Scholar] [CrossRef]
- Krašovec, R.; Belavkin, R.V.; Aston, J.A.; Channon, A.; Aston, E.; Rash, B.M.; Kadirvel, M.; Forbes, S.; Knight, C.G. Where antibiotic resistance mutations meet quorum-sensing. Microb. Cell Graz Austria 2014, 1, 250–252. [Google Scholar] [CrossRef] [PubMed]
- Lázár, V.; Nagy, I.; Spohn, R.; Csörgő, B.; Györkei, Á.; Nyerges, Á.; Horváth, B.; Vörös, A.; Busa-Fekete, R.; Hrtyan, M.; et al. Genome-wide analysis captures the determinants of the antibiotic cross-resistance interaction network. Nat. Commun. 2014, 5, 4352. [Google Scholar] [CrossRef] [PubMed]
- Kohlmann, R.; Bähr, T.; Gatermann, S.G. Species-specific mutation rates for ampC derepression in Enterobacterales with chromosomally encoded inducible AmpC β-lactamase. J. Antimicrob. Chemother. 2018, 73, 1530–1536. [Google Scholar] [CrossRef] [PubMed]
- Nyinoh, I.W. Spontaneous mutations conferring antibiotic resistance to antitubercular drugs at a range of concentrations in Mycobacterium smegmatis. Drug Dev. Res. 2019, 80, 147–154. [Google Scholar] [CrossRef]
- Grimsey, E.M.; Fais, C.; Marshall, R.L.; Ricci, V.; Ciusa, M.L.; Stone, J.W.; Ivens, A.; Malloci, G.; Ruggerone, P.; Vargiu, A.V.; et al. Chlorpromazine and Amitriptyline Are Substrates and Inhibitors of the AcrB Multidrug Efflux Pump. mBio 2020, 11, e00465-20. [Google Scholar] [CrossRef] [PubMed]
- Balansky, J.; Pfarr, K.; Szekat, C.; Kehraus, S.; Aden, T.; Grosse, M.; Jansen, R.; Hesterkamp, T.; Schiefer, A.; König, G.M.; et al. The RNA Polymerase Inhibitor Corallopyronin A Has a Lower Frequency of Resistance Than Rifampicin in Staphylococcus aureus. Antibiot. Basel Switz. 2022, 11, 920. [Google Scholar] [CrossRef] [PubMed]
- Deibler, R.W.; Mann, J.K.; Sumners, D.W.L.; Zechiedrich, L. Hin-mediated DNA knotting and recombining promote replicon dysfunction and mutation. BMC Mol. Biol. 2007, 8, 44. [Google Scholar] [CrossRef]
- Li, J.; Yang, Y.; Guevara, J.; Wang, L.; Cao, W. Identification of a prototypical single-stranded uracil DNA glycosylase from Listeria innocua. DNA Repair 2017, 57, 107–115. [Google Scholar] [CrossRef]
- Bodine, T.J.; Evangelista, M.A.; Chang, H.T.; Ayoub, C.A.; Samuel, B.S.; Sucgang, R.; Zechiedrich, L. Escherichia coli DNA ligase B may mitigate damage from oxidative stress. PLoS ONE 2017, 12, e0180800. [Google Scholar] [CrossRef]
- Borsellini, A.; Lebbink, J.H.G.; Lamers, M.H. MutL binds to 3’ resected DNA ends and blocks DNA polymerase access. Nucleic Acids Res. 2022, 50, 6224–6234. [Google Scholar] [CrossRef] [PubMed]
- Cortes, P.R.; Piñas, G.E.; Albarracin Orio, A.G.; Echenique, J.R. Subinhibitory concentrations of penicillin increase the mutation rate to optochin resistance in Streptococcus pneumoniae. J. Antimicrob. Chemother. 2008, 62, 973–977. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Degen, D.; Ebright, Y.W.; Ebright, R.H. Frequency, spectrum, and nonzero fitness costs of resistance to myxopyronin in Staphylococcus aureus. Antimicrob. Agents Chemother. 2012, 56, 6250–6255. [Google Scholar] [CrossRef] [PubMed]
- Otoupal, P.B.; Cordell, W.T.; Bachu, V.; Sitton, M.J.; Chatterjee, A. Multiplexed deactivated CRISPR-Cas9 gene expression perturbations deter bacterial adaptation by inducing negative epistasis. Commun. Biol. 2018, 1, 129. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.-G.; Lu, H.-S.; Buckling, A. Temperature drives diversification in a model adaptive radiation. Proc. Biol. Sci. 2018, 285, 20181515. [Google Scholar] [CrossRef]
- Maharjan, R.P.; Ferenci, T. Escherichia coli mutation rates and spectra with combinations of environmental limitations. Microbiol. Read. Engl. 2018, 164, 1491–1502. [Google Scholar] [CrossRef]
- Snitkin, E.S.; Zelazny, A.M.; Gupta, J.; NISC Comparative Sequencing Program; Palmore, T.N.; Murray, P.R.; Segre, J.A. Genomic insights into the fate of colistin resistance and Acinetobacter baumannii during patient treatment. Genome Res. 2013, 23, 1155–1162. [Google Scholar] [CrossRef]
- Aguilar, C.; Flores, N.; Riveros-McKay, F.; Sahonero-Canavesi, D.; Carmona, S.B.; Geiger, O.; Escalante, A.; Bolívar, F. Deletion of the 2-acyl-glycerophosphoethanolamine cycle improve glucose metabolism in Escherichia coli strains employed for overproduction of aromatic compounds. Microb. Cell Factories 2015, 14, 194. [Google Scholar] [CrossRef]
- Wassermann, T.; Meinike Jørgensen, K.; Ivanyshyn, K.; Bjarnsholt, T.; Khademi, S.M.H.; Jelsbak, L.; Høiby, N.; Ciofu, O. The phenotypic evolution of Pseudomonas aeruginosa populations changes in the presence of subinhibitory concentrations of ciprofloxacin. Microbiol. Read. Engl. 2016, 162, 865–875. [Google Scholar] [CrossRef]
- Dalia, T.N.; Yoon, S.H.; Galli, E.; Barre, F.-X.; Waters, C.M.; Dalia, A.B. Enhancing multiplex genome editing by natural transformation (MuGENT) via inactivation of ssDNA exonucleases. Nucleic Acids Res. 2017, 45, 7527–7537. [Google Scholar] [CrossRef]
- Halperin, S.O.; Tou, C.J.; Wong, E.B.; Modavi, C.; Schaffer, D.V.; Dueber, J.E. CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window. Nature 2018, 560, 248–252. [Google Scholar] [CrossRef] [PubMed]
- Nyerges, Á.; Bálint, B.; Cseklye, J.; Nagy, I.; Pál, C.; Fehér, T. CRISPR-interference-based modulation of mobile genetic elements in bacteria. Synth. Biol. Oxf. Engl. 2019, 4, ysz008. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Rojas, A.; Makarova, O.; Müller, U.; Rolff, J. Cationic Peptides Facilitate Iron-induced Mutagenesis in Bacteria. PLOS Genet. 2015, 11, e1005546. [Google Scholar] [CrossRef] [PubMed]
- Storvik, K.A.M.; Foster, P.L. RpoS, the stress response sigma factor, plays a dual role in the regulation of Escherichia coli’s error-prone DNA polymerase IV. J. Bacteriol. 2010, 192, 3639–3644. [Google Scholar] [CrossRef] [PubMed]
- Maharjan, R.; Ferenci, T. Stress-induced mutation rates show a sigmoidal and saturable increase due to the RpoS sigma factor in Escherichia coli. Genetics 2014, 198, 1231–1235. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Rojas, A.; Makarova, O.; Rolff, J. Antimicrobials, stress and mutagenesis. PLoS Pathog. 2014, 10, e1004445. [Google Scholar] [CrossRef]
- Mo, C.Y.; Manning, S.A.; Roggiani, M.; Culyba, M.J.; Samuels, A.N.; Sniegowski, P.D.; Goulian, M.; Kohli, R.M. Systematically Altering Bacterial SOS Activity under Stress Reveals Therapeutic Strategies for Potentiating Antibiotics. mSphere 2016, 1, e00163-16. [Google Scholar] [CrossRef]
- Arias-Sánchez, F.I.; Allen, R.C.; Hall, A.R. Effects of prior exposure to antibiotics on bacterial adaptation to phages. J. Evol. Biol. 2018, 31, 277–286. [Google Scholar] [CrossRef]
- Zhou, H.; Zhang, L.; Xu, Q.; Zhang, L.; Yu, Y.; Hua, X. The mismatch repair system (mutS and mutL) in Acinetobacter baylyi ADP1. BMC Microbiol. 2020, 20, 40. [Google Scholar] [CrossRef]
- Papkou, A.; Hedge, J.; Kapel, N.; Young, B.; MacLean, R.C. Efflux pump activity potentiates the evolution of antibiotic resistance across S. aureus isolates. Nat. Commun. 2020, 11, 3970. [Google Scholar] [CrossRef]
- Fressatti Cardoso, R.; Martín-Blecua, I.; Pietrowski Baldin, V.; Meneguello, J.E.; Valverde, J.R.; Blázquez, J.; Castañeda-García, A. Noncanonical Mismatch Repair Protein NucS Modulates the Emergence of Antibiotic Resistance in Mycobacterium abscessus. Microbiol. Spectr. 2022, 10, e0222822. [Google Scholar] [CrossRef] [PubMed]
- Parekh, V.J.; Wien, F.; Grange, W.; De Long, T.A.; Arluison, V.; Sinden, R.R. Crucial Role of the C-Terminal Domain of Hfq Protein in Genomic Instability. Microorganisms 2020, 8, 1598. [Google Scholar] [CrossRef] [PubMed]
- Vasse, M.; Bonhoeffer, S.; Frenoy, A. Ecological effects of stress drive bacterial evolvability under sub-inhibitory antibiotic treatments. ISME Commun. 2022, 2, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Behringer, M.G.; Ho, W.-C.; Meraz, J.C.; Miller, S.F.; Boyer, G.F.; Stone, C.J.; Andersen, M.; Lynch, M. Complex Ecotype Dynamics Evolve in Response to Fluctuating Resources. mBio 2022, 13, e0346721. [Google Scholar] [CrossRef] [PubMed]
- Luján, A.M.; Moyano, A.J.; Martino, R.A.; Feliziani, S.; Urretavizcaya, M.; Smania, A.M. ImuB and ImuC contribute to UV-induced mutagenesis as part of the SOS regulon in Pseudomonas aeruginosa. Environ. Mol. Mutagen. 2019, 60, 594–601. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Rosado, A.I.; Valencia, E.Y.; Rodríguez-Rojas, A.; Costas, C.; Galhardo, R.S.; Rodríguez-Beltrán, J.; Blázquez, J. N-acetylcysteine blocks SOS induction and mutagenesis produced by fluoroquinolones in Escherichia coli. J. Antimicrob. Chemother. 2019, 74, 2188–2196. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Ning, Q.; Dong, J.; Brooks, B.W.; You, J. Predicting mixture toxicity and antibiotic resistance of fluoroquinolones and their photodegradation products in Escherichia coli. Environ. Pollut. Barking Essex 1987 2020, 262, 114275. [Google Scholar] [CrossRef]
- Wu, E.Y.; Hilliker, A.K. Identification of Rifampicin Resistance Mutations in Escherichia coli, Including an Unusual Deletion Mutation. J. Mol. Microbiol. Biotechnol. 2017, 27, 356–362. [Google Scholar] [CrossRef]
- Tonoyan, L.; Friel, R.; O’Flaherty, V. Mutation rate and efflux response of bacteria exposed to a novel antimicrobial iodo-thiocyanate complex. J. Glob. Antimicrob. Resist. 2020, 22, 13–17. [Google Scholar] [CrossRef]
- Miyabayashi, H.; Jain, R.; Suzuki, S.; Grogan, D.W.; Kurosawa, N. PolB1 Is Sufficient for DNA Replication and Repair Under Normal Growth Conditions in the Extremely Thermophilic Crenarchaeon Sulfolobus acidocaldarius. Front. Microbiol. 2020, 11, 613375. [Google Scholar] [CrossRef]
- Short, F.L.; Lee, V.; Mamun, R.; Malmberg, R.; Li, L.; Espinosa, M.I.; Venkatesan, K.; Paulsen, I.T. Benzalkonium chloride antagonises aminoglycoside antibiotics and promotes evolution of resistance. EBioMedicine 2021, 73, 103653. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Devin, G.A.; Couce, A. Trends in the Use of Proper Methods for Estimating Mutation Rates in Fluctuation Experiments. Axioms 2023, 12, 1100. https://doi.org/10.3390/axioms12121100
Devin GA, Couce A. Trends in the Use of Proper Methods for Estimating Mutation Rates in Fluctuation Experiments. Axioms. 2023; 12(12):1100. https://doi.org/10.3390/axioms12121100
Chicago/Turabian StyleDevin, Guillem A., and Alejandro Couce. 2023. "Trends in the Use of Proper Methods for Estimating Mutation Rates in Fluctuation Experiments" Axioms 12, no. 12: 1100. https://doi.org/10.3390/axioms12121100
APA StyleDevin, G. A., & Couce, A. (2023). Trends in the Use of Proper Methods for Estimating Mutation Rates in Fluctuation Experiments. Axioms, 12(12), 1100. https://doi.org/10.3390/axioms12121100






