4.1. Simulation Study
In this section, we present a detailed simulation study to evaluate the performance of the proposed smoothed weighted quantile regression method for censored data. We compare our approach with two existing methods, including the weighted quantile regression method and martingale-based quantile regression method [
15,
17]. Portnoy [
15] proposed a recursively reweighted estimation procedure for censored quantile regression, extending the Kaplan–Meier estimator to handle more complex regression settings involving censoring. On the other hand, Peng and Huang [
17] developed a martingale-based estimating equation, which simplifies quantile regression with survival data by minimizing
-type convex functions, ensuring computational efficiency and robustness. For the comparison methods, the
quantreg package in R is used, employing the
crq.fit.por and
crq.fit.pen functions for Portnoy [
15]’s and Peng and Huang [
17]’s methods, respectively. All simulations were executed on a workstation equipped with two 24-core 2.99 GHz CPUs and 128 GB of memory. Each simulation run utilized 8 CPU cores concurrently.
In the simulation study, the covariates
are split into three groups
where the first
covariates
with
follow a multivariate normal distribution
, and the remaining covariates come from a mixture of normal and binomial distributions. Specifically,
with
follow a multivariate normal distribution
, where
is a
covariance matrix with
, for
. The second group of covariates,
with
, follows a multivariate uniform distribution on the interval
with covariance matrix
, and the final group
with
consists of binary variables drawn from a Bernoulli distribution with probability 0.5. The response variable
, where
,
. Let
represent the quantile function of the
t-distribution with 2 degrees of freedom and denote
, the response variable
is generated according to the following model
where
,
with
follows a uniform distribution on interval
and
follows
t-distribution with 2 degrees of freedom. Random censoring
is introduced by sampling censoring times from a mixture distribution:
where
are randomly drawn from the set
. The observed response
is then the minimum of the true response and the censoring time,
. Furthermore, the bandwidth of the Gaussian kernel for our method is selected as
.
For each quantile level , the estimation is conducted using a quantile grid of with values ranging from . The performance of each method is assessed at every quantile level based on the estimation error, average quantile loss, and runtime.The estimation error is quantified using , while the average quantile loss is given by calculating . We consider three main scenarios (1–3) with different sample sizes and numbers of covariates, each replicated 1000 times. Specifically, we have with and , with and , and with and . In addition to these primary settings, we introduce Scenario 4 specifically designed to evaluate the performance of the proposed method under varying censoring rates. This scenario considers a sample size of and , with and , and examines three censoring levels, 20%, 35%, and 50%. This scenario provides insight into how the degree of censoring impacts estimation accuracy and runtime, further testing the robustness of the proposed method. The acronyms used for the methods are: “SWQR” for our proposed Smoothed Weighted Quantile Regression method, “WeightedQR” for Peng and Huang’s method, and “MartingaleQR” for Portnoy’s method. The results are summarized in the following figures and tables, which show the performance of the three methods.
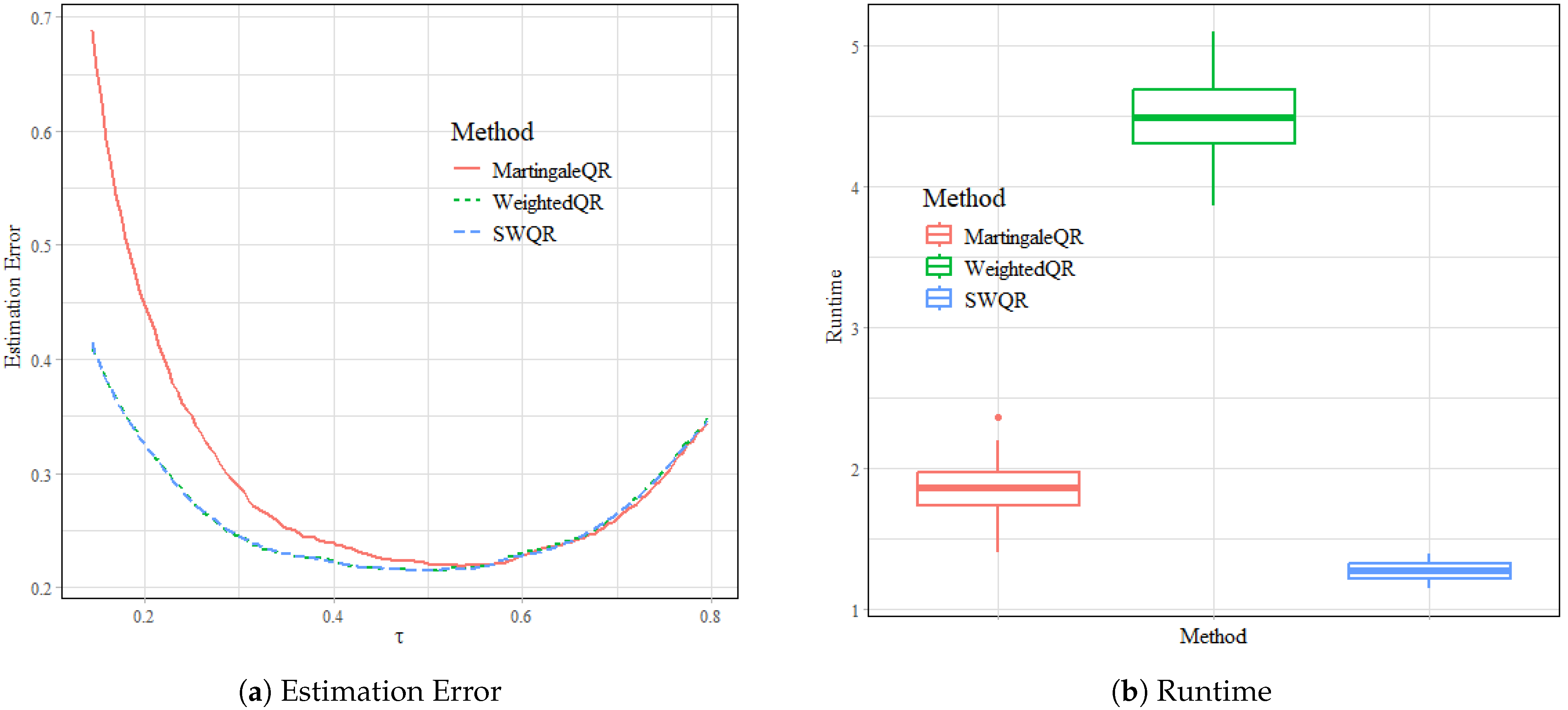
Figure 1,
Figure 2 and
Figure 3 provide a comprehensive comparison of estimation error trends and runtimes across the Scenarios 1–3 with varying sample sizes (
) and covariate dimensions (
). Across all scenarios, SWQR consistently demonstrates competitive performance, achieving estimation errors comparable to or slightly better than WeightedQR and significantly outperforming MartingaleQR, particularly at mid-range quantile levels (
to
). The trends indicate that MartingaleQR suffers from higher errors, especially for lower and upper quantile levels, which could be attributed to its less robust handling of censoring effects. These findings confirm that SWQR balances both robustness and accuracy in quantile estimation across all tested conditions. The runtime analysis, as summarized in the corresponding figures, highlights the computational efficiency of SWQR. SWQR consistently requires the least runtime among the three methods, making it the most efficient in scenarios involving larger datasets. WeightedQR, while competitive in terms of estimation error, incurs substantially higher computational costs, particularly as the sample size and covariate dimensionality grow. MartingaleQR exhibits moderate runtimes but suffers from higher estimation errors, making it less suitable for practical applications.
Table 1,
Table 2 and
Table 3, featuring estimation errors and average quantile losses, provide insights into the comparative performance of the methods across different scenarios. SWQR achieves average quantile loss values closely aligned with, and often slightly lower than, those of WeightedQR, showcasing its robustness and accuracy in quantile estimation. Notably, in Scenario 3 (
), SWQR maintains its effectiveness despite the increased dataset size, demonstrating scalability and suitability for handling large-scale survival data. MartingaleQR, on the other hand, consistently shows higher average quantile losses and estimation errors. The runtime analysis is summarized in
Table 4. SWQR demonstrates a significant computational advantage over WeightedQR, requiring substantially less time to complete estimation tasks across all scenarios. This is particularly evident in Scenarios 2 and 3, where WeightedQR exhibits a steep increase in runtime as the sample size and covariate dimensionality grow. Although MartingaleQR is computationally efficient, its higher average quantile losses and estimation errors make it less favorable for practical use. The overall balance of accuracy and computational efficiency achieved by SWQR underscores its suitability for large-scale survival analysis, offering a reliable alternative to existing methods when both precision and speed are critical considerations.
In Scenario 4, we evaluated the performance of MartingaleQR, WeightedQR, and SWQR under varying censoring rates (20%, 35%, and 50%, respectively) with
and
.
Figure 4,
Figure 5 and
Figure 6 depict the estimation errors and runtimes across quantile levels. The results reveal that SWQR consistently outperforms MartingaleQR in terms of estimation error, particularly at lower quantile levels. Compared to WeightedQR, SWQR demonstrates comparable or slightly better estimation performance across all censoring rates, maintaining its robustness even under higher levels of censoring. Furthermore, the increase in censoring rate slightly elevates the estimation error for all methods; however, the relative performance among the methods remains consistent. Regarding runtime, SWQR achieves the lowest computational time across all scenarios, maintaining efficiency regardless of the censoring rate. WeightedQR, in contrast, exhibits significantly higher runtimes, especially under lower censoring rates.
Table 5,
Table 6,
Table 7 and
Table 8 provide a detailed numerical comparison, highlighting the estimation errors, average quantile losses, and runtimes for each censoring rate. The average quantile losses for SWQR are closely aligned with those of WeightedQR and remain consistently lower than those of MartingaleQR. This indicates SWQR’s ability to handle data with varying censoring rates effectively. Notably, as the censoring rate increases, the performance gap between WeightedQR and SWQR narrows, with SWQR maintaining its advantage in computational efficiency. In terms of recorded runtime in
Table 8, SWQR consistently demonstrates the fastest execution across all censoring levels, highlighting its computational efficiency, while WeightedQR offers competitive accuracy, it suffers from considerably higher computational costs, making it less practical for large-scale applications. MartingaleQR, on the other hand, achieves moderate runtime but struggles with reduced estimation accuracy, particularly under higher censoring rates. This analysis underscores SWQR’s strong balance between speed and accuracy, showcasing its robustness and scalability for datasets with different censoring rates.
In conclusion, the simulation results highlight that the proposed SWQR method consistently surpasses MartingaleQR in estimation accuracy across various quantile levels, particularly as the sample size and number of covariates increase. SWQR balances computational efficiency and estimation accuracy, outperforming WeightedQR, which is limited by its high computational cost in large-scale settings. According to the results, the disparity in runtimes becomes increasingly evident with higher dimensionality, emphasizing SWQR’s capability to deliver reliable performance with minimal computational burden. These results underscore the practicality of SWQR, making it an effective solution for large-scale survival data analysis where both accuracy and resource efficiency are critical considerations.
4.2. Real Data Application
To further evaluate the performance of the proposed smoothed weighted quantile regression method, we applied it to a dataset on primary biliary cirrhosis (PBC), a liver disease dataset extensively used in survival analysis literature [
28]. This dataset consists of information on 418 patients, with their survival times either observed or right-censored. The data include clinical measurements such as age, edema status, and biochemical markers (serum bilirubin, albumin, and prothrombin time). Approximately 40% of the patients in the dataset are censored, making it a suitable candidate for testing the robustness and accuracy of censored quantile regression methods. Our aim is to model the relationship between the logarithm of survival time and the selected covariates at different quantile levels. The response variable of interest is the logarithm of survival time,
, and the covariates include: age of the patient (in years) (
), edema status (
), Logarithm of serum bilirubin concentration (mg/dL) (
), Logarithm of albumin concentration (gm/dL) (
), Logarithm of prothrombin time (in seconds) (
). We model the conditional quantile function of the logarithm of survival time at each quantile level
as:
where the regression coefficients
represent the effect of each covariate on the quantile
of the survival time distribution. To compare the performance of the SWQR method, we apply MartingleQR and WeightedQR [
15,
17]. For the SWQR method, a quantile grid
is selected, ranging from 0.05 to 0.80 in increments of 0.05. The estimation of coefficients is performed for each quantile level, and the performance of all three methods was assessed based on the estimated coefficients.
The estimated regression coefficients for each method are computed across the quantile grid, with the results compared in
Table 9 and
Figure 7. Each covariate’s impact on survival time was examined, highlighting differences in how the methods handle censored observations. For lower quantiles (
to
), the SWQR method closely matches the results from Portnoy [
15]’s method in terms of coefficient magnitude and direction, while showing slightly more stability compared to Peng and Huang [
17]’s method. For example, the effect of serum bilirubin (
) on survival time diminishes as we move toward higher quantiles, reflecting how severe cases impact survival time differently across the population. Similarly, the impact of age on survival (
) shows consistent trends across all methods, with SWQR capturing the effects more consistently across quantiles. At higher quantiles (
and above), the SWQR method continues to provide reliable coefficient estimates, while the performance of Portnoy [
15]’s method starts to deteriorate due to challenges with higher censoring. Peng and Huang [
17]’s method also encounters computational difficulties, particularly with respect to estimating coefficients for
(edema status), as its estimates fluctuate more erratically at these higher quantiles.
In this real-data application, the SWQR method demonstrated its robustness and flexibility in handling censored survival data across a wide range of quantile levels. The method not only provided stable and accurate coefficient estimates, particularly in higher quantiles where censoring becomes more pronounced. The consistent performance of SWQR across different quantile levels, coupled with its computational advantages, makes it a valuable tool for analyzing censored survival data in medical and other applied fields.