Optimising Seniors’ Metabolism of Medications and Avoiding Adverse Drug Events Using Data on How Metabolism by Their P450 Enzymes Varies with Ancestry and Drug–Drug and Drug–Drug–Gene Interactions
Abstract
1. Introduction
2. Materials and Methods
2.1. Systematic Reviews
2.2. Pharmacogenetic Guidelines
2.3. Prescribing Using Curated Summaries of the CPIC and DPWG Pharmacogenomic Guidelines
2.4. Prescribing Using Curated Summaries of Drug–Drug and Drug–Drug–Gene Interactions
3. Results
3.1. Optimising Pharmacogenomic Prescribing for Illnesses Frequently Cared for by Primary Care Physicians
3.1.1. Mental Illness
3.1.2. The Burden of Mental Illness in Primary Care
3.1.3. The Effectiveness of Usual Therapy for Depression
3.1.4. How Pharmacogenomics Affects Response to Antidepressants
3.1.5. Prescribing Using the CPIC and DPWG Guidelines for Depression
3.1.6. Pharmacogenomic Decision Support Tools to Reduce Depressive Symptoms and Relapse Rates and Improve Remission Rates
3.1.7. Cost Effectiveness of Pharmacogenomic-Guided Therapy for Depression
4. Translating Pharmacogenomics into Effective Clinical Practice for Primary Care Patients with Other Than Mental Health Problems
4.1. Studies of Pharmacogenomic-Guided Therapy for Multiple Illnesses
4.2. Pharmacogenomic Studies of Cardiovascular Medication Dosing Requirements
4.2.1. Warfarin and P450 CYP2C9
4.2.2. Warfarin and VKORC1
4.2.3. New Oral Anticoagulants (NOACs)
4.2.4. Direct Oral Anticoagulants (DOACs)
4.2.5. Clopidogrel
4.2.6. Sulfonylureas and Cardiac Arrhythmias
4.3. Tamoxifen and Breast Cancer
4.4. Benzodiazepine Use and Pharmacogenomic-Related Increased Risk of Falls
4.5. Statins and Myopathy
4.6. Genetic Variations in Uric Acid Levels with Allopurinol Treatment, and Adverse Skin Reactions to Allopurinol
5. Becoming Part of a Large Pharmacogenomic Prescribing System
5.1. Improving EMR Medication Recording
5.2. Ensuring DST Systems Provide Comprehensive Advice Valued by Both Patients and Physicians as Improving Care
5.3. Progress in Implementing Pharmacogenetic Guidelines in Large Health Care Systems
5.4. Developing Team Work to Deliver Pharmacogenetic Advice
5.5. Integrating Pharmacogenomic Data into Electronic Health Records
5.6. Decision Support Tools
5.7. Evidence-Based Data on Effectiveness and Costs
6. Discussion
Limitations
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Tobe, S.W.; Stone, J.A.; Walker, K.M.; Anderson, T.; Bhattacharyya, O.; Cheng, A.Y.Y.; Gregoire, J.; Gubitz, G.; L’Abbé, M.; Lau, D.C.W.; et al. Canadian cardiovascular harmonized national guidelines endeavour (C-change). CMAJ 2014, 186, 1299–1305. [Google Scholar] [CrossRef] [PubMed]
- DrugBank. Available online: www.drugbank.ca (accessed on 9 May 2020).
- Flockhart, D.A. Drug Interactions: Cytochrome P450 Drug Interaction Table; Indiana University School of Medicine: Indianapolis, IN, USA, 2020; Available online: https://drug-interactions.medicine.iu.edu (accessed on 5 May 2020).
- Rx Files, 12th ed.; College of Pharmacy and Nutrition, University of Saskatchewan: Saskatoon, SK, Canada, November 2019; Available online: www.RxFiles.ca (accessed on 5 July 2020).
- Dutch Pharmacogenetics Working Group. Available online: https://www.pharmgkb.org/ (accessed on 2 May 2020).
- Clinical Pharmacogenetics Implementation Consortium (CPIC). Available online: https://cpicpgx.org/ (accessed on 2 May 2020).
- Shekhani, R.; Steinacher, L.; Swen, J.J.; Ingelman-Sundberg, M. Evaluation of current regulation and guidelines of pharmacogenomic drug labels: Opportunities for improvements. Clin. Pharmacol. Ther. 2019, 107, 1240–1255. [Google Scholar] [CrossRef] [PubMed]
- Bank, P.C.D.; Swen, J.J.; Guchelaar, H.J. Estimated nationwide impact of implementing a preemptive pharmacogenetic panel approach to guide drug prescribing in primary care in The Netherlands. BMC Med. 2019, 17, 110. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Skierka, J.M.; Blommel, J.H.; Moore, B.E.; VanCuyk, D.L.; Bruflat, J.K.; Peterson, L.M.; Veldhuizen, T.L.; Fadra, N.; Peterson, S.E.; et al. Preemptive pharmacogenomic testing for precision medicine: A comprehensive analysis of five actionable pharmacogenomic genes using next-generation DNA sequencing and a customized CYP2D6 genotyping cascade. J. Mol. Diagn. 2016, 18, 438–445. [Google Scholar] [CrossRef] [PubMed]
- Swen, J.J.; Wilting, I.; de Goede, A.L.; Grandia, L.; Mulder, H.; Touw, D.J.; de Boer, A.; Conemans, J.M.; Egberts, T.C.; Klungel, O.H.; et al. Pharmacogenetics: From bench to byte. Clin. Pharmacol. Ther. 2008, 83, 781–787. [Google Scholar] [CrossRef]
- Swen, J.J.; Nijenhuis, M.; de Boer, A.; Grandia, L.; Maitland-van der Zee, A.H.; Mulder, H.; van der Weide, J.; Wilffert, B.; Deneer, V.H.; Guchelaar, H.J.; et al. Pharmacogenetics: From bench to byte--an update of guidelines. Clin. Pharmacol. Ther. 2011, 89, 662–673. [Google Scholar] [CrossRef]
- Geneesmiddel Informatie Centrum. Informatorium Medicamentorum; KNMP: The Hague, The Netherlands, 2014; Available online: https://www.knmp.nl/producten/boek-informatorium-medicamentorum (accessed on 23 June 2020).
- Johnson, J.A.; Gong, L.; Whirl-Carrillo, M.; Gage, B.F.; Scott, S.A.; Stein, C.M.; Anderson, J.L.; Kimmel, S.E.; Lee, M.T.; Pirmohamed, M. Clinical pharmacogenetics implementation consortium guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing. Clin. Pharmacol. Ther. 2011, 90, 625–629. [Google Scholar] [CrossRef]
- Johnson, J.A.; Caudle, K.E.; Gong, L.; Whirl-Carrillo, M.; Stein, C.M.; Scott, S.A.; Lee, M.T.; Gage, B.F.; Kimmel, S.E.; Perera, M.A.; et al. Clinical pharmacogenetics implementation consortium (cpic) guideline for pharmacogenetics-guided warfarin dosing: 2017 update. Clin. Pharmacol. Ther. 2017, 102, 397–404. [Google Scholar] [CrossRef]
- Hicks, J.K.; Swen, J.J.; Thorn, C.F.; Sangkuhl, K.; Kharasch, E.D.; Ellingrod, V.L.; Skaar, T.C.; Müller, D.J.; Gaedigk, A.; Stingl, J.C. Clinical Pharmacogenetics Implementation Consortium guideline for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants. Clin. Pharmacol. Ther. 2013, 93, 402–408. [Google Scholar] [CrossRef]
- Hicks, J.K.; Sangkuhl, K.; Swen, J.J.; Ellingrod, V.L.; Muller, D.J.; Shimoda, K.; Skaar, T.C.; Müller, D.J.; Gaedigk, A.; Stingl, J.C.; et al. Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin. Pharmacol. Ther. 2017, 102, 37–44. [Google Scholar] [CrossRef]
- Hicks, J.K.; Bishop, J.R.; Sangkuhl, K.; Muller, D.J.; Ji, Y.; Leckband, S.G.; Leeder, J.S.; Graham, R.L.; Chiulli, D.L.; LLerena, A.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and CYP2C19 Genotypes and Dosing of Selective Serotonin Reuptake Inhibitors. Clin. Pharmacol. Ther. Pharmacol. Ther. 2015, 98, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Scott, S.A.; Sangkuhl, K.; Stein, C.M.; Hulot, J.S.; Mega, J.L.; Roden, D.M.; Klein, T.E.; Sabatine, M.S.; Johnson, J.A.; Shuldiner, A.R.; et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin. Pharmacol. Ther. Pharmacol Ther. 2013, 94, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Scott, S.A.; Sangkuhl, K.; Gardner, E.E.; Stein, C.M.; Hulot, J.S.; Johnson, J.A.; Roden, D.M.; Klein, T.E.; Shuldiner, A.R. Clinical pharmacogenetics implementation consortium. clinical pharmacogenetics implementation consortium guidelines for cytochrome P450-2C19 (CYP2C19) genotype and clopidogrel therapy. Clin. Pharmacol. Ther. 2011, 90, 328–332. [Google Scholar] [CrossRef] [PubMed]
- Crews, K.R.; Gaedigk, A.; Dunnenberger, H.M.; Leeder, J.S.; Klein, T.E.; Caudle, K.E.; Haidar, C.E.; Shen, D.D.; Callaghan, J.T.; Sadhasivam, S.; et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin. Pharmacol. Ther. Pharmacol Ther. 2014, 95, 376–382. [Google Scholar] [CrossRef]
- Crews, K.R.; Gaedigk, A.; Dunnenberger, H.M.; Klein, T.E.; Shen, D.D.; Callaghan, J.T.; Kharasch, E.D.; Skaar, T.C. Clinical Pharmacogenetics Implementation Consortium. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for codeine therapy in the context of cytochrome P450 2D6 (CYP2D6) genotype. Clin. Pharmacol. Ther. 2011, 91, 321–326. [Google Scholar] [CrossRef]
- Ramsey, L.B.; Johnson, S.G.; Caudle, K.E.; Haidar, C.E.; Voora, D.; Wilke, R.A.; Maxwell, W.D.; McLeod, H.L.; Krauss, R.M.; Roden, D.M.; et al. The clinical pharmacogenetics implementation consortium guideline for SLCO1B1 and simvastatin-induced myopathy: 2014 update. Clin. Pharmacol. Ther. 2014, 96, 423–428. [Google Scholar] [CrossRef]
- Wilke, R.A.; Ramsey, L.B.; Johnson, S.G.; Maxwell, W.D.; McLeod, H.L.; Voora, D.; Krauss, R.M.; Roden, D.M.; Feng, Q.; Cooper-Dehoff, R.M.; et al. The clinical pharmacogenomics implementation consortium: CPIC guideline for SLCO1B1 and simvastatin induced myopathy. Clin. Pharmacol. Ther. 2012, 92, 112–117. [Google Scholar] [CrossRef]
- Bahar, M.A.; Setiawan, D.; Hak, E.; Wilffert, B. Pharmacogenetics of drug–drug interaction and drug–drug–gene interaction: A systematic review on CYP2C9, CYP2C19 and CYP2D6. Pharmacogenomics 2017, 18, 701–739. [Google Scholar] [CrossRef]
- Beinema, M.J.; de Jong, P.H.; Salden, H.J.; van Wijnen, M.; van der Meer, J.; Brouwers, J.R. The influence of NSAIDs on coumarin sensitivity in patients with CYP2C9 polymorphism after total hip replacement surgery. Mol. Diagn. Ther. 2007, 11, 123–128. [Google Scholar] [CrossRef]
- Visser, L.E.; van Schaik, R.H.; van Vliet, M.; Trienekens, P.H.; De Smet, P.A.; Vulto, A.G.; Hofman, A.; van Duijn, C.M.; Stricker, B.H. Allelic variants of cytochrome P450 2C9 modify the interaction between nonsteroidal anti inflammatory drugs and coumarin anticoagulants. Clin. Pharmacol. Ther. 2005, 77, 479–485. [Google Scholar] [CrossRef]
- Andersson, M.L.; Eliasson, E.; Lindh, J.D. A clinically significant interaction between warfarin and simvastatin is unique to carriers of the CYP2C9*3 allele. Pharmacogenomics 2012, 13, 757–762. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Brundage, R.C.; Oetting, W.S.; Leppik, I.E.; Tracy, T.S. Differential genotype dependent inhibition of CYP2C9 in humans. Drug Metab. Dispos. 2008, 36, 1242–1248. [Google Scholar] [CrossRef] [PubMed]
- Vormfelde, S.V.; Brockmoller, J.; Bauer, S.; Herchenhein, P.; Kuon, J.; Meineke, I.; Roots, I.; Kirchheiner, J. Relative impact of genotype and enzyme induction on the metabolic capacity of CYP2C9 in healthy volunteers. Clin. Pharmacol. Ther. 2009, 86, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Fischer, T.L.; Pieper, J.A.; Graff, D.W.; Rodgers, J.E.; Fischer, J.D.; Parnell, K.J.; Goldstein, J.A.; Greenwood, R.; Patterson, J.H. Evaluation of potential losartan phenytoin drug interactions in healthy volunteers. Clin. Pharmacol. Ther. 2002, 72, 238–246. [Google Scholar] [CrossRef] [PubMed]
- Furuta, T.; Iwaki, T.; Umemura, K. Influences of different proton pump inhibitors on the anti-platelet function of clopidogrel in relation to CYP2C19 genotypes. Br. J. Clin. Pharmacol. 2010, 70, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Depta, J.P.; Lenzini, P.A.; Lanfear, D.E.; Wang, T.Y.; Spertus, J.A.; Bach, R.G.; Cresci, S. Clinical outcomes associated with proton pump inhibitor use among clopidogrel-treated patients within CYP2C19 genotype groups following acute myocardial infarction. Pharmacogenom. J. 2015, 15, 20–25. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hata, M.; Shiono, M.; Akiyama, K.; Sezai, A.; Wakui, S.; Kimura, H.; Sekino, H. Incidence of drug interaction when using proton pump inhibitor and warfarin according to cytochrome P450 2C19 (CYP2C19) genotype in Japanese. Thorac. Cardiovasc. Surg. 2015, 63, 45–50. [Google Scholar]
- Uno, T.; Sugimoto, K.; Sugawara, K.; Tateishi, T. The role of cytochrome P2C19 in R-warfarin pharmacokinetics and its interaction with omeprazole. Ther. Drug Monit. 2008, 30, 276–281. [Google Scholar] [CrossRef]
- Ieiri, I.; Kimura, M.; Irie, S.; Urae, A.; Otsubo, K.; Ishizaki, T. Interaction magnitude, pharmacokinetics and pharmacodynamics of ticlopidine in relation to CYP2C19 genotypic status. Pharmacogenet. Genom. 2005, 15, 851–859. [Google Scholar] [CrossRef]
- Yasui-Furukori, N.; Takahata, T.; Nakagami, T.; Yoshiya, G.; Inoue, Y.; Kaneko, S.; Tateishi, T. Different inhibitory effect of fluvoxamine on omeprazole metabolism between CYP2C19 genotypes. Br. J. Clin. Pharmacol. 2004, 57, 487–494. [Google Scholar] [CrossRef]
- Yasui-Furukori, N.; Saito, M.; Uno, T.; Takahata, T.; Sugawara, K.; Tateishi, T. Effects of fluvoxamine on lansoprazole pharmacokinetics in relation to CYP2C19 genotypes. J. Clin. Pharmacol. 2004, 44, 1223–1229. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, R.S.; Noehr-Jensen, L.; Brosen, K. Inhibitory effect of oral contraceptives on CYP2C19 activity is not significant in carriers of the CYP2C19*17 allele. Clin. Exp. Pharmacol. Physiol. 2013, 40, 683–688. [Google Scholar]
- Uno, T.; Shimizu, M.; Yasui-Furukori, N.; Sugawara, K.; Tateishi, T. Different effects of fluvoxamine on rabeprazole pharmacokinetics in relation to CYP2C19 genotype status. Br. J. Clin. Pharmacol. 2006, 61, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Takahashi, Y.; Imai, K.; Mogami, Y.; Matsuda, K.; Nakai, M.; Kagawa, Y.; Inoue, Y. Interaction between sulthiame and clobazam: Sulthiame inhibits the metabolism of clobazam, possibly via an action on CYP2C19. Epilepsy Behav. 2014, 34, 124–126. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.Y.; Yu, K.S.; Jang, I.J.; Yang, B.H.; Shin, S.G.; Yim, D.S. Omeprazole hydroxylation is inhibited by a single dose of moclobemide in homozygotic EM genotype for CYP2C19. Br. J. Clin. Pharmacol. 2002, 53, 393–397. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.J.; Huang, S.L.; Wang, W.; Zhou, H.H. The induction effect of rifampicin on activity of mephenytoin 4’-hydroxylase related to M1 mutation of CYP2C19 and gene dose. Br. J. Clin. Pharmacol. 1998, 45, 27–29. [Google Scholar] [CrossRef]
- Venkatakrishnan, K.; von Moltke, L.L.; Greenblatt, D.J. Effects of the antifungal agents on oxidative drug metabolism: Clinical relevance. Clin. Pharmacokinet. 2000, 38, 111–180. [Google Scholar] [CrossRef]
- Meyer, U.A. Metabolic interactions of the proton pump inhibitors lansoprazole, omeprazole and pantoprazole with other drugs. Eur. J. Gastroenterol. Hepatol. 1996, 8 (Suppl. S1), S21–S25. [Google Scholar] [CrossRef]
- Ishizaki, T.; Chiba, K.; Manabe, K.; Koyama, E.; Hayashi, M.; Yasuda, S.; Horai, Y.; Tomono, Y.; Yamato, C.; Toyoki, T. Comparison of the interaction potential of a new proton pump inhibitor, E3810, versus omeprazole with diazepam in extensive and poor metabolizers of S mephenytoin 4′ hydroxylation. Clin. Pharmacol. Ther. 1995, 58, 155–164. [Google Scholar] [CrossRef]
- Abdel Rahman, S.M.; Gotschall, R.R.; Kauffman, R.E.; Leeder, J.S.; Kearns, G.L. Investigation of terbinafine as a CYP2D6 inhibitor in vivo. Clin. Pharmacol. Ther. 1999, 65, 465–472. [Google Scholar] [CrossRef]
- Eap, C.B.; Lessard, E.; Baumann, P.; Brawand-Amey, M.; Yessine, M.A.; O’Hara, G.; Turgeon, J. Role of CYP2D6 in the stereoselective disposition of venlafaxine in humans. Pharmacogenetics 2003, 13, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Brosen, K.; Hansen, J.G.; Nielsen, K.K.; Sindrup, S.H.; Gram, L.F. Inhibition by paroxetine of desipramine metabolism in extensive but not in poor metabolizers of sparteine. Eur. J. Clin. Pharmacol. 1993, 44, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Azuma, J.; Hasunuma, T.; Kubo, M.; Miyatake, M.; Koue, T.; Higashi, K.; Fujiwara, T.; Kitahara, S.; Katano, T.; Hara, S. The relationship between clinical pharmacokinetics of aripiprazole and CYP2D6 genetic polymorphism: Effects of CYP enzyme inhibition by coadministration of paroxetine or fluvoxamine. Eur. J. Clin. Pharmacol. 2012, 68, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Pibarot, P.; Pilote, S.; Dumesnil, J.G.; Arsenault, M.; Bélanger, P.M.; Meibohm, B.; Hamelin, B.A. Modulation of metoprolol pharmacokinetics and hemodynamics by diphenhydramine coadministration during exercise testing in healthy premenopausal women. J. Pharmacol. Exp. Ther. 2005, 313, 1172–1181. [Google Scholar] [CrossRef]
- Sharma, A.; Pibarot, P.; Pilote, S.; Dumesnil, J.G.; Arsenault, M.; Bélanger, P.M.; Meibohm, B.; Hamelin, B.A. Toward optimal treatment in women: The effect of sex on metoprolol–diphenhydramine interaction. J. Clin. Pharmacol. 2010, 50, 214–225. [Google Scholar] [CrossRef]
- Hamelin, B.A.; Bouayad, A.; Methot, J.; Jobin, J.; Desgagnés, P.; Poirier, P.; Allaire, J.; Dumesnil, J.G.; Turgeon, J. Significant interaction between the nonprescription antihistamine diphenhydramine and the CYP2D6 substrate metoprolol in healthy men with high or low CYP2D6 activity. Clin. Pharmacol. Ther. 2000, 67, 466–477. [Google Scholar] [CrossRef]
- Werner, U.; Werner, D.; Rau, T.; Fromm, M.F.; Hinz, B.; Brune, K. Celecoxib inhibits metabolism of cytochrome P450 2D6 substrate metoprolol in humans. Clin. Pharmacol. Ther. 2003, 74, 130–137. [Google Scholar] [CrossRef]
- Lessard, E.; Yessine, M.A.; Hamelin, B.A.; Gauvin, C.; Labbé, L.; O’Hara, G.; LeBlanc, J.; Turgeon, J. Diphenhydramine alters the disposition of venlafaxine through inhibition of CYP2D6 activity in humans. J. Clin. Psychopharmacol. 2001, 21, 175–184. [Google Scholar] [CrossRef]
- Stamer, U.M.; Musshoff, F.; Kobilay, M.; Madea, B.; Hoeft, A.; Stuber, F. Concentrations of tramadol and O desmethyltramadol enantiomers in different CYP2D6 genotypes. Clin. Pharmacol. Ther. 2007, 82, 41–47. [Google Scholar] [CrossRef]
- Geber, C.; Ostad Haji, E.; Schlicht, K.; Hiemke, C.; Tadic, A. Severe tremor after cotrimoxazole induced elevation of venlafaxine serum concentrations in a patient with major depressive disorder. Ther. Drug Monit. 2013, 35, 279–282. [Google Scholar] [CrossRef]
- Marusic, S.; Lisicic, A.; Horvatic, I.; Bacic Vrca, V.; Bozina, N. Atorvastatin related rhabdomyolysis and acute renal failurein a genetically predisposed patient with potential drug–drug interaction. Int. J. Clin. Pharm. 2012, 34, 825–827. [Google Scholar] [CrossRef] [PubMed]
- Furuta, T.; Ohashi, K.; Kobayashi, K.; Iida, I.; Yoshida, H.; Shirai, N.; Takashima, M.; Kosuge, K.; Hanai, H.; Chiba, K. Effects of clarithromycin on the metabolism of omeprazole in relation to CYP2C19 genotype status in humans. Clin. Pharmacol. Ther. 1999, 66, 265–274. [Google Scholar] [CrossRef]
- Miura, M.; Tada, H.; Yasui-Furukori, N.; Uno, T.; Sugawara, K.; Tateishi, T.; Suzuki, T. Effect of clarithromycin on the enantioselective disposition of lansoprazole in relation to CYP2C19 genotypes. Chirality 2005, 17, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Niioka, T.; Yasui Furukori, N.; Uno, T.; Sugawara, K.; Kaneko, S.; Tateishi, T. Identification of a single time point for plasma lansoprazole measurement that adequately reflects area under the concentration time curve. Ther. Drug Monit. 2006, 28, 321–325. [Google Scholar] [CrossRef]
- Hassan Alin, M.; Andersson, T.; Niazi, M.; Liljeblad, M.; Persson, B.A.; Rohss, K. Studies on drug interactions between esomeprazole, amoxicillin and clarithromycin in healthy subjects. Int. J. Clin. Pharmacol. Ther. 2006, 44, 119–127. [Google Scholar] [CrossRef]
- Michaud, V.; Kreutz, Y.; Skaar, T.; Ogburn, E.; Thong, N.; Flockhart, D.S.; Desta, Z. Efavirenz mediated induction of omeprazole metabolism is CYP2C19 genotype dependent. Pharmacogenom. J. 2014, 14, 151–159. [Google Scholar] [CrossRef]
- Gasche, Y.; Daali, Y.; Fathi, M.; Chiappe, A.; Cottini, S.; Dayer, P.; Desmeules, J. Codeine intoxication associated with ultrarapid CYP2D6 metabolism. N. Engl. J. Med. 2004, 351, 2827–2831. [Google Scholar] [CrossRef]
- Park, J.Y.; Shon, J.H.; Kim, K.A.; Jung, H.J.; Shim, J.C.; Yoon, Y.R.; Cha, I.J.; Shin, J.G. Combined effects of itraconazole and CYP2D6*10 genetic polymorphism on the pharmacokinetics and pharmacodynamics of haloperidol in healthy subjects. J. Clin. Psychopharmacol. 2006, 26, 135–142. [Google Scholar] [CrossRef]
- Jung, S.M.; Kim, K.A.; Cho, H.K.; Jung, I.G.; Park, P.W.; Byun, W.T.; Park, J.Y. Cytochrome P450 3A inhibitor itraconazole affects plasma concentrations of risperidone and 9 hydroxyrisperidone in schizophrenic patients. Clin. Pharmacol. Ther. 2005, 78, 520–528. [Google Scholar] [CrossRef]
- Malhotra, B.; Sachse, R.; Wood, N. Evaluation of drug–drug interactions with fesoterodine. Eur. J. Clin. Pharmacol. 2009, 65, 551–560. [Google Scholar] [CrossRef]
- Dilger, K.; Greiner, B.; Fromm, M.F.; Hofmann, U.; Kroemer, H.K.; Eichelbaum, M. Consequences of rifampicin treatment on propafenone disposition in extensive and poor metabolizers of CYP2D6. Pharmacogenetics 1999, 9, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Chhun, S.; Verstuyft, C.; Rizzo-Padoin, N.; Simoneau, G.; Becquemont, L.; Peretti, I.; Swaisland, A.; Wortelboer, R. Gefitinibphenytoin interaction is not correlated with the C-erythromycin breath test in healthy male volunteers. Br. J. Clin. Pharmacol. 2009, 68, 226–237. [Google Scholar] [CrossRef] [PubMed]
- Michaud, V.; Kreutz, Y.; Skaar, T. Genotype-based estimation of CYP2C19 contribution to the elimination of omeprazole in healthy subjects. Clin. Pharmacol. Ther. 2012, 91, S64. [Google Scholar]
- Breslow, A.S.; Tran, N.M.; Lu, F.Q.; Alpert, J.E.; Cook, B.L. Depression treatment expenditures for adults in the USA: A systematic review. Curr. Psychiatr. Rep. 2019, 21, 105. [Google Scholar] [CrossRef] [PubMed]
- Johnston, K.J.; Wen, H.; Hockenberry, J.M.; Joynt Maddox, K.E. Association between patient cognitive and functional status and Medicare Total annual cost of care: Implications for value-based Payment Association between patient cognitive and functional status and Medicare cost of Care Association between patient cognitive and functional status and Medicare cost of care. JAMA Intern. Med. 2018, 178, 1489–1497. [Google Scholar]
- Egede, L.E.; Bishu, K.G.; Walker, R.J.; Dismuke, C.E. Impact of diagnosed depression on healthcare costs in adults with and without diabetes: United States, 2004–2011. J. Affect. Dis. 2016, 195, 119–126. [Google Scholar] [CrossRef]
- Undurraga, J.; Baldessarini, R.J. Randomized, placebo-controlled trials of antidepressants for acute major depression: Thirty-year meta-analytic review. Neuropsychopharmacology 2012, 37, 851–864. [Google Scholar] [CrossRef]
- Lee, Y.Y.; Stockings, E.A.; Harris, M.G.; Doi, S.A.R.; Page, I.S.; Davidson, S.K.; Barendregt, J.J. The risk of developing major depression among individuals with subthreshold depression: A systematic review and meta-analysis of longitudinal cohort studies. Psychol. Med. 2019, 49, 92–102. [Google Scholar] [CrossRef]
- Dunlop, B.W. Prediction of treatment outcomes in major depressive disorder. Exp. Rev. Clin. Pharmacol. 2015, 8, 669–672. [Google Scholar] [CrossRef]
- García-González, J.; Tansey, K.E.; Hauser, J.; Henigsberg, N.; Maier, W.; Mors, O.; Placentino, A.; Rietschel, M.; Souery, D.; Žagar, T. Pharmacogenetics of antidepressant response: A polygenic approach. Progr. Neuropsychopharmacol. Biol. Psychiatr. 2017, 75, 128–134. [Google Scholar] [CrossRef]
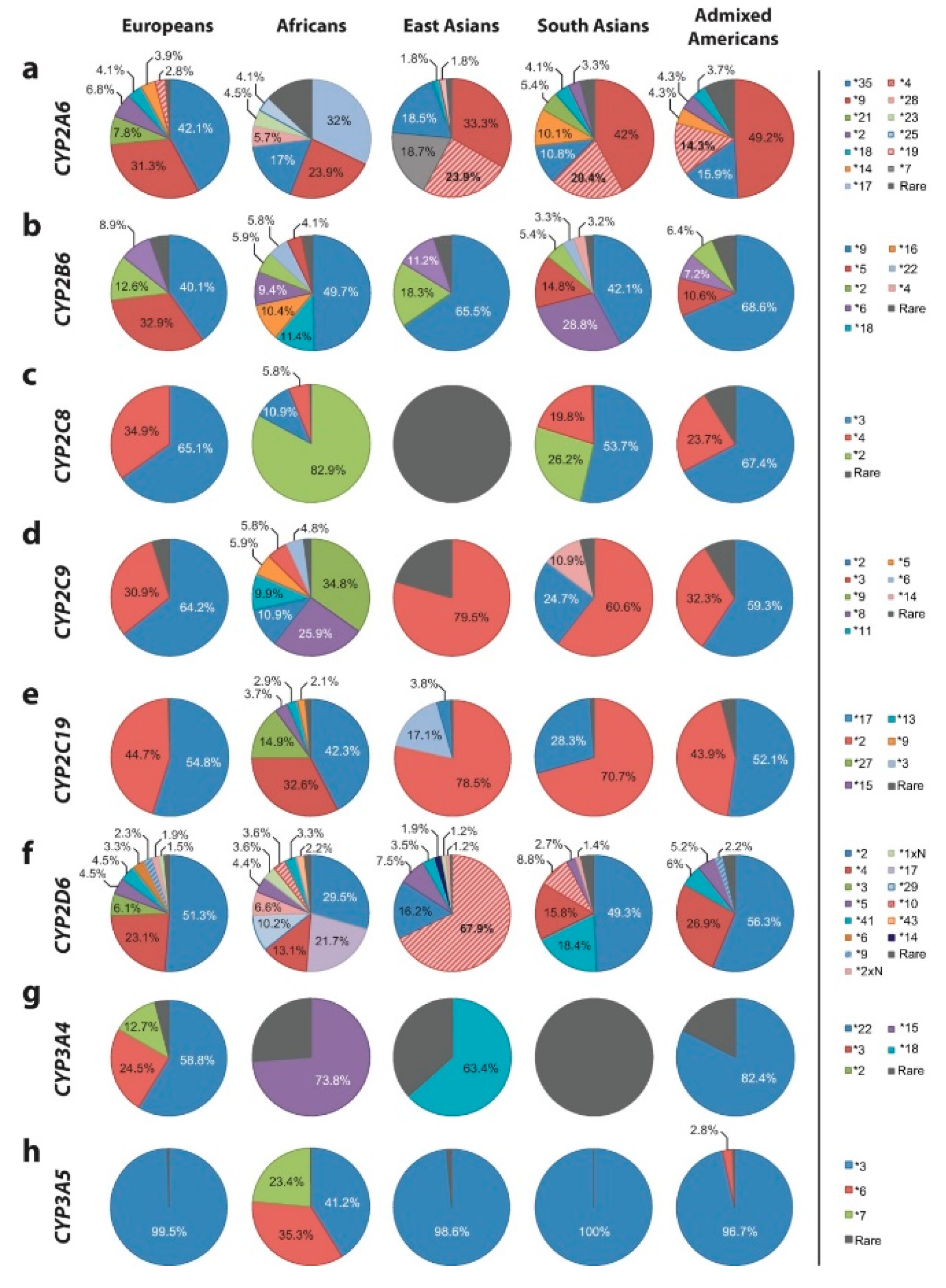
- Zhou, Y.; Ingelman-Sundberg, M.; Lauschke, V.M. Worldwide distribution of cytochrome P450 alleles: A meta-analysis of population-scale sequencing projects. Clin. Pharmacol. Therapeut. 2017, 102, 688–700. [Google Scholar] [CrossRef] [PubMed]
- van Westrhenen, R.; Aitchison, K.J.; Ingelman-Sundberg, M.; Jukić, M.M. Pharmacogenomics of antidepressant and antipsychotic treatment: How far have we got and where are we going? Front. Psychiatr. 2020, 1, 94. [Google Scholar] [CrossRef] [PubMed]
- Jukić, M.M.; Haslemo, T.; Molden, E.; Ingelman-Sundberg, M. Impact of CYP2C19 genotype on escitalopram exposure and therapeutic failure: A retrospective study based on 2087 patients. Am. J. Psychiatr. 2018, 175, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Haslemo, T.; Eliasson, E.; Jukić, M.M.; Ingelman-Sundberg, M.; Molden, E. Significantly lower CYP2D6 metabolism measured as the O/N-desmethylvenlafaxine metabolic ratio in carriers of CYP2D6*41 versus CYP2D6*9 or CYP2D6*10: A study on therapeutic drug monitoring data from 1003 genotyped Scandinavian patients. Br. J. Clin. Pharmacol. 2019, 85, 194–201. [Google Scholar] [CrossRef]
- Fabbri, C.; Tansey, K.E.; Perlis, R.H.; Hauser, J.; Henigsberg, N.; Maier, W.; Mors, O.; Placentino, A.; Rietschel, M.; Souery, D. Effect of cytochrome CYP2C19 metabolizing activity on antidepressant response and side effects: Meta-analysis of data from genome-wide association studies. Eur. Neuropsychopharmacol. 2018, 28, 945–954. [Google Scholar] [CrossRef]
- Maruf, A.A.; Fan, M.; Arnold, P.D.; Müller, D.J.; Aitchison, K.J.; Bousman, C.A. Pharmacogenetic testing options relevant to psychiatry in Canada. Can. J. Psychiatr. 2020, 1–10. [Google Scholar] [CrossRef]
- Bousman, C.A.; Arandjelovic, K.; Mancuso, S.G.; Eyre, H.A.; Dunlop, B.W. Pharmacogenetic tests and depressive symptom remission: A meta-analysis of randomized controlled trials. Pharmacogenomics 2019, 20, 37–47. [Google Scholar] [CrossRef]
- Thase, M.E.; Parikh, S.V.; Rothschild, A.J.; Dunlop, B.W.; DeBattista, C.; Conway, C.R.; Forester, B.P.; Mondimore, F.M.; Li, J.; Brown, K.; et al. Impact of Pharmacogenomics on clinical outcomes for patients taking medications with gene-drug interactions in a randomized controlled trial. J. Clin. Pharmacol. Ther. Psychiatr. 2019, 80. [Google Scholar] [CrossRef]
- Jablonski, M.R.; Lorenz, R.; Li, J.; Dechairo, B.M. Economic outcomes following combinatorial pharmacogenomic testing for elderly psychiatric patients. J. Geriatr. Psychiatr. Neurol. 2019, 17. [Google Scholar] [CrossRef]
- Samwald, M.; Xu, H.; Blagec, K.; Empey, P.E.; Malone, D.C.; Ahmed, S.M.; Ryan, P.; Hofer, S.; Boyce, R.D. Incidence of exposure of patients in the United States to Multiple drugs for which pharmacogenomic guidelines are available. PLoS ONE 2016, 11, e0164972. [Google Scholar] [CrossRef]
- Kim, K.; Magness, J.W.; Nelson, R.; Baron, V.; Brixner, D.I. Clinical utility of pharmacogenetic testing and a clinical decision support tool to enhance the identification of drug therapy problems through medication therapy management in polypharmacy patients. J. Manag. Care Spec. Pharm. 2018, 24, 1250–1259. [Google Scholar] [CrossRef] [PubMed]
- Brixner, D.; Biltaji, E.; Bress, A.; Unni, S.; Ye, X.; Mamiya, T.; Ashcraft, K.; Biskupiak, J. The effect of pharmacogenetic profiling with a clinical decision support tool on healthcare resource utilization and estimated costs in the elderly exposed to polypharmacy. Med. Econ. 2016, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Roden, D.M.; Van Driest, S.L.; Wells, Q.S.; Mosley, J.D.; Denny, J.C.; Peterson, J.F. Opportunities and Challenges in Cardiovascular Pharmacogenomics from Discovery to Implementation. Circ. Res. 2018, 122, 1176–1190. [Google Scholar] [CrossRef] [PubMed]
- Gage, B.F.; Bass, A.R.; Lin, H.; Woller, S.C.; Stevens, S.M.; Al-Hammadi, N.; Li, J.; Rodriguez, T., Jr.; Miller, J.P.; McMillin, G.A.; et al. Effect of genotype-guided warfarin dosing on clinical events and anticoagulation control among patients undergoing hip or knee arthroplasty: The GIFT randomized clinical trial. JAMA 2017, 318, 1115–1124. [Google Scholar] [PubMed]
- Ruff, C.T. Pharmacogenetics of warfarin therapy. Clin. Chem. 2018, 64, 1558–1559. [Google Scholar] [CrossRef] [PubMed]
- Mega, J.L.; Walker, J.R.; Ruff, C.T.; Vandell, A.G.; Nordio, F.; Deenadayalu, N.; Murphy, S.A.; Lee, J.; Mercuri, M.F.; Giugliano, R.P.; et al. Genetics and the clinical response to warfarin and edoxaban: Findings from the randomised, double-blind ENGAGE AF-TIMI 48 trial. Lancet 2015, 385, 2280–2287. [Google Scholar]
- Thrombosis Canada. Clinical Guideline. Apixaban (Eliquis). Available online: thrombosis.canada.ca (accessed on 3 May 2020).
- Thrombosis Canada. Clinical Guideline. NOACs/DOACs: Comparisons and Frequently Asked Questions; PMC: Bethesda, MD, USA, 2014; Available online: thrombosis.canada.ca (accessed on 3 May 2020).
- Thrombosis Canada. Clinical Guideline. NOACs/DOACs: Management of Bleeding; PMC: Betheshda, MD, USA, 2019; Available online: thrombosis.canada.ca (accessed on 3 May 2020).
- Collet, J.P.; Hulot, J.S.; Pena, A.; Villard, E.; Esteve, J.B.; Silvain, J.; Payot, L.; Brugier, D.; Cayla, G.; Beygui, F.; et al. Cytochrome P450 2C19 polymorphism in young patients treated with clopidogrel after myocardial infarction: A cohort study. Lancet 2009, 373, 309–317. [Google Scholar] [CrossRef]
- Simon, T.; Verstuyft, C.; Mary-Krause, M.; Quteineh, L.; Drouet, E.; Méneveau, N.; Steg, P.G.; Ferrières, J.; Danchin, N.; Becquemont, L. French Registry of Acute ST-Elevation and Non-ST-Elevation Myocardial Infarction (FAST-MI) Investigators. Genetic determinants of response to clopidogrel and cardiovascular events. N. Engl. J. Med. 2009, 360, 363–375. [Google Scholar] [CrossRef]
- Mega, J.L.; Close, S.L.; Wiviott, S.D.; Shen, L.; Hockett, R.D.; Brandt, J.T.; Walker, J.R.; Antman, E.M.; Macias, W.; Braunwald, E.; et al. Cytochrome p-450 polymorphisms and response to clopidogrel. N. Engl. J. Med. 2009, 360, 354–362. [Google Scholar] [CrossRef]
- Tornio, A.; Flynn, R.; Morant, S.; Velten, E.; Palmer, C.N.A.; MacDonald, T.M.; Doney, A.S.F. Investigating real-world clopidogrel pharmacogenetics in stroke using a bioresource linked to electronic medical records. Clin. Pharmacol. Ther. 2018, 103, 281–286. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, X.; Lin, J.; Li, H.; Johnston, S.C.; Lin, Y.; Pan, Y.; Liu, L.; Wang, D.; Wang, C.; et al. Association between CYP2C19 loss-of-function allele status and efficacy of clopidogrel for risk reduction among patients with minor stroke or transient ischemic attack. JAMA 2016, 316, 70–78. [Google Scholar] [CrossRef]
- Zheng, L.; Yang, C.; Xiang, L.; Hao, Z. Genotype-guided antiplatelet therapy compared with conventional therapy for patients with acute coronary syndromes: A systematic review and meta-analysis. Biomarkers 2019, 24, 517–523. [Google Scholar] [CrossRef] [PubMed]
- Floyd, J.S.; Sitlani, C.M.; Avery, C.L.; Noordam, R.; Li, X.; Smith, A.V.; Gogarten, S.M.; Li, J.; Broer, L.; Evans, D.S.; et al. Large-scale pharmacogenomic study of sulfonylureas and the QT, JT and QRS intervals: CHARGE Pharmacogenomics Working Group. Pharmacogenom. J. 2018, 18, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Drögemöller, B.I.; Wright, G.E.B.; Shih, J.; Monzon, J.G.; Gelmon, K.A.; Ross, C.J.D.; Amstutz, U.; Carleton, B.C.; CPNDS Clinical Recommendations Group. CYP2D6 as a treatment decision aid for ER-positive non-metastatic breast cancer patients: A systematic review with accompanying clinical practice guidelines. Breast Cancer Res. Treat. 2019, 173, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Province, M.A.; Goetz, M.P.; Brauch, H.; Flockhart, D.A.; Hebert, J.M.; Whaley, R.; Suman, V.J.; Schroth, W.; Winter, S.; Zembutsu, H.; et al. CYP2D6 genotype and adjuvant tamoxifen: Meta-analysis of heterogeneous study populations. Clin. Pharmacol. Ther. 2014, 95, 216–227. [Google Scholar] [CrossRef]
- Province, M.A.; Altman, R.B.; Klein, T.E. Interpreting the CYP2D6 results from the International Tamoxifen Pharmacogenetics Consortium. Clin. Pharmacol. Ther. 2014, 96, 144–146. [Google Scholar] [CrossRef]
- Mao, C.; Yang, Z.Y.; He, B.F.; Liu, S.; Zhou, J.H.; Luo, R.C.; Chen, Q.; Tang, J.L. Toremifene versus tamoxifen for advanced breast cancer. Cochrane Database Syst. Rev. 2012, 7, CD008926. [Google Scholar] [CrossRef]
- Ishiguro, H.; Ohno, S.; Yamamoto, Y.; Takao, S.; Sato, N.; Fujisawa, T.; Kadoya, T.; Kuroi, K.; Bando, H.; Teramura, Y.; et al. Pharmacogenomic-pharmacokinetic study of selective estrogen-receptor modulators with intra-patient dose escalation in breast cancer. Breast Cancer 2019, 26, 535–543. [Google Scholar] [CrossRef]
- Ham, A.C.; Ziere, G.; Broer, L.; Swart, K.M.; Enneman, A.W.; van Dijk, S.C.; van Wijngaarden, J.P.; van der Zwaluw, N.L.; Brouwer-Brolsma, E.M.; Dhonukshe-Rutten, R.A.; et al. CYP2C9 genotypes modify benzodiazepine-related fall risk: original results from three studies with meta-analysis. J. Am. Med. Dir. Assoc. 2017, 18, e1Ce88. [Google Scholar] [CrossRef]
- Xiang, Q.; Chen, S.Q.; Ma, L.Y.; Hu, K.; Zhang, Z.; Mu, G.Y.; Xie, Q.F.; Zhang, X.D.; Cui, Y.M. Association between SLCO1B1 T521C polymorphism and risk of statin-induced myopathy: A meta-analysis. Pharmacogenom. J. 2018, 18, 721–729. [Google Scholar] [CrossRef]
- Vassy, J.L.; Chun, S.; Advani, S.; Ludin, S.A.; Smith, J.G.; Alligood, E.C. Impact of SLCO1B1 pharmacogenetic testing on patient and healthcare outcomes: A systematic review. Clin. Pharmacol. Ther. 2019, 106, 360–373. [Google Scholar] [CrossRef]
- Whirl-Carillo, M.; McDonagh, E.M.; Hebert, J.M.; Sangkuhl, K.; Thorn, C.F.; Altman, R.B.; Klein, T.E. Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 2012, 92, 414–417. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.C.; Yee, S.W.; Liang, X.; Hoffmann, T.J.; Kvale, M.N.; Banda, Y.; Jorgenson, E.; Schaefer, C.; Risch, N.; Giacomini, K.M. Genome-widea association study identifies ABCG2 (BCRP) as an allopurinol transporter and a determinant of drug response. Clin. Pharmacol. Ther. 2015, 97, 518–525. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Cheng, Y.J.; Zhu, L.L.; Yu, L.; Zhao, X.K.; Jia, M.; Wen, C.H.; Long, X.Z.; Tang, T.; He, A.J.; et al. Impact of HLA-B*58:01 allele and allopurinol-induced cutaneous adverse drug reactions: Evidence from 21 pharmacogenetic studies. Oncotarget 2016, 7, 81870–81879. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Available online: www.genome.gov (accessed on 23 July 2020).
- Available online: https://www.nimh.nih.gov/about/strategic-planning-reports/index.shtml (accessed on 23 July 2020).
- Bousman, C. Genes, Neurocognition, and HIV Risk Behaviors in the Context of Methamphetamine and HIV. Ph.D. Thesis, University of Calgary, Calgary, AB, Canada, 2020. [Google Scholar]
- Kulenovic, A.; Lagumdzija-Kulenovic, A. PM-TOM: A method for finding personalized polypharmacy therapies with minimal adverse drug-drug, drug-gene and drug-condition interactions. Stud. Health Technol. Inform. 2020, 270, 648–652. [Google Scholar]
- van der Wouden, C.H.; Bank, P.C.D.; Özokcu, K.; Swen, J.J.; Guchelaar, H.-J. Pharmacist-initiated pre-emptive pharmacogenetic panel testing with clinical decision support in primary care: record of pgx results and real-world impact. Genes 2019, 10, 416. [Google Scholar] [CrossRef] [PubMed]
- Seidling, H.M.; Mahler, C.; Strasner, C.; Straus, B.; Bernhard, G.; Szecsenyi, J.; Haefeli, W.W.; Wehrmann, U. Use of medication lists: A population-based approach to increase the prevalence of medication lists within a region in Germany: A pe-post study. Int. J. Clin. Pharmacol. Ther. 2019, 57, 375–383. [Google Scholar] [CrossRef]
- The Office of the US National Coordinator for Health Information Technology (ONC). Available online: https://www.healthit.gov/topic/innovation/state-innovation-model-resource-center (accessed on 3 May 2020).
- Arndt, B.G.; Beasley, J.W.; Watkinson, M.D.; Temte, J.L.; Tuan, W.-J.; Sinsky, C.A.; Gilchrist, V.I. Tethered to the EHR: primary care physician workload assessment using EHR Event log data and time-motion observations. Ann. Fam. Med. 2017, 15, 419–426. [Google Scholar] [CrossRef]
- Montague, E.; Asan, O. Dynamic modeling of patient and physician eye gaze to understand the effects of electronic health records on doctor-patient communication and attention. Int. J. Med. Inform. 2014, 83, 225–234. [Google Scholar] [CrossRef]
- National Human Genome Research Institute. US National Institutes of Health: Bethesda, MA, USA. Available online: www.genome.gov (accessed on 22 April 2020).
- Levy, K.D.; Blake, K.; Fletcher-Hoppe, C.; Franciosi, J.; Goto, D.; Hicks, J.K.; Holmes, A.M.; Kanuri, S.H.; Madden, E.B.; Musty, M.D.; et al. Opportunities to implement a sustainable genomic medicine program: Lessons learned from the IGNITE Network. Genet. Med. 2019, 21, 743–747. [Google Scholar] [CrossRef]
- Rosenman, M.B.; Decker, B.; Levy, K.D.; Holmes, A.M.; Pratt, V.M.; Eadon, M.T. Lessons learned when introducing pharmacogenomic panel testing into clinical practice. Value Health 2017, 20, 54–59. [Google Scholar] [CrossRef]
- Sperber, N.R.; Carpenter, J.S.; Cavallari, L.H.; Damschroder, L.J.; Cooper-DeHoff, R.M.; Denny, J.C.; Ginsburg, G.S.; Guan, Y.; Horowitz, C.R.; Levy, K.D.; et al. Challenges and strategies for implementing genomic services in diverse settings: Experiences from the Implementing GeNomics In pracTicE (IGNITE) network. BMC Med. Genom. 2017, 10, 35. [Google Scholar]
- Clinical Decision Support Knowledgebase Educational Materials (CDS KnowledgeBase). Available online: https://cdskb.org (accessed on 28 April 2020).
- Rohrer Vitek, C.R.; Abul-Husn, N.S.; Connolly, J.J.; Hartzler, A.L.; Kitchner, T.; Peterson, J.F.; Rasmussen, L.V.; Smith, M.E.; Stallings, S.; Williams, M.S. Healthcare provider education to support integration of pharmacogenomics in practice: The eMERGE Network experience. Pharmacogenomics 2017, 18, 1013–1025. [Google Scholar] [CrossRef] [PubMed]
- Caraballo, P.J.; Bielinski, S.J.; St. Sauver, J.L.; Weinshilboum, R.M. Electronic medical record-integrated pharmacogenomics and related clinical decision support concepts. Clin. Pharmacol. Ther. 2017, 102, 254–264. [Google Scholar] [CrossRef] [PubMed]
- SPARK Toolbox. Available online: https://ignite-genomics.org/spark-toolbox/clinicians/ (accessed on 10 April 2020).
- Implementing Genomics in Practice (Ignite). Implementation Guidelines. Available online: https://www.genome.gov/Funded-Programs-Projects/Implementing-Genomics-in-Practice-IGNITE (accessed on 24 April 2020).
- Hicks, J.K.; Dunnenberger, H.M.; Gumpper, H.M.; Haidar, C.E.; Hoffman, J.M. Integrating pharmacogenomics into electronic health records with clinical decision support. Am. J. Health-Syst. Pharm. 2016, 73, 1967–1976. [Google Scholar] [CrossRef] [PubMed]
- Ignite. Available online: https://gmkb.org/ (accessed on 10 May 2020).
- Zhou, Y.; Fujikura, K.; Mkrtchian, S.; Lauschke, V.M. Computational methods for the pharmacogenetic interpretation of next generation sequencing data. Front. Pharmacol. 2018, 9. [Google Scholar] [CrossRef]
| A. Genotypes and Enzymatic Activity of CYP2C19 and CYP2D6 | |||
|---|---|---|---|
| Genotype Functional | Diplotype | Categorisation 1 | Enzymatic Capacity 1 |
| CYP2C19 | |||
| CYP2C19 Null/Null | PM/PM | Poor | 0% |
| CYP2C19Null/Wt | PM/NM | Intermediate | 50% |
| CYP2C19Null/*17 | PM/UM | Intermediate | 60% |
| CYP2C19Wt/Wt | NM/NM | Normal | 100% |
| CYP2C19Wt/*17 | NM/UM | Ultrarapid | 110% |
| CYP2C19*17/*17 | UM/UM | Ultrarapid | 120% |
| CYP2D6 | |||
| CYP2D6Null/Null | PM/PM | Poor | 0% |
| CYP2D6Null/*41 | PM/IM | Intermediate | 5% |
| CYP2D6Null/*9-10 | PM/IM | Intermediate | 15% |
| CYP2D6*41/*9-10 | IM/IM | Intermediate OR Normal | 20% |
| CYP2D6*9-10/*9-10 | IM/IM | Intermediate OR Normal | 30% |
| CYP2D6Wt/Null | NM/PM | Intermediate OR Normal | 50% |
| CYP2D6Wt/*41 | NM/IM | Normal | 55% |
| CYP2D6Wt/*9-10 | NM/IM | Normal | 65% |
| CYP2D6Wt/Wt | NM/NM | Normal | 100% |
| CYP2D6WtX3 | UM/UM | Ultrarapid | 150% |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thomas, R.E. Optimising Seniors’ Metabolism of Medications and Avoiding Adverse Drug Events Using Data on How Metabolism by Their P450 Enzymes Varies with Ancestry and Drug–Drug and Drug–Drug–Gene Interactions. J. Pers. Med. 2020, 10, 84. https://doi.org/10.3390/jpm10030084
Thomas RE. Optimising Seniors’ Metabolism of Medications and Avoiding Adverse Drug Events Using Data on How Metabolism by Their P450 Enzymes Varies with Ancestry and Drug–Drug and Drug–Drug–Gene Interactions. Journal of Personalized Medicine. 2020; 10(3):84. https://doi.org/10.3390/jpm10030084
Chicago/Turabian StyleThomas, Roger E. 2020. "Optimising Seniors’ Metabolism of Medications and Avoiding Adverse Drug Events Using Data on How Metabolism by Their P450 Enzymes Varies with Ancestry and Drug–Drug and Drug–Drug–Gene Interactions" Journal of Personalized Medicine 10, no. 3: 84. https://doi.org/10.3390/jpm10030084
APA StyleThomas, R. E. (2020). Optimising Seniors’ Metabolism of Medications and Avoiding Adverse Drug Events Using Data on How Metabolism by Their P450 Enzymes Varies with Ancestry and Drug–Drug and Drug–Drug–Gene Interactions. Journal of Personalized Medicine, 10(3), 84. https://doi.org/10.3390/jpm10030084





