Predictive Role of Biopsy Based Biomarkers for Radiotherapy Treatment in Rectal Cancer
Abstract
:1. Translational Relevance
2. Introduction
3. Materials and Methods
3.1. Patients and Samples
3.2. Immunohistochemistry
3.3. Evaluation of RT Impact on Biomarkers and Potential Value of Biomarkers in BS in Different Groups
3.4. Statistical Analysis
4. Results
4.1. RT Impact on Biomarkers in PT
4.2. The Differences between BS and PT in the Unmatched or Matched Cases (RT Group or Non-RT Group, Respectively)
4.3. Diagnostic Value of Biomarkers in BS
4.4. The Prognostic and Predictive Value of Biomarkers in BS
5. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Majano, S.B.; Di Girolamo, C.; Rachet, B.; Maringe, C.; Guren, M.G.; Glimelius, B.; Iversen, L.H.; Schnell, E.A.; Lundqvist, K.; Christensen, J.; et al. Surgical treatment and survival from colorectal cancer in Denmark, England, Norway, and Sweden: A population-based study. Lancet Oncol. 2019, 20, 74–87. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Zheng, R.; Baade, P.D.; Zhang, S.; Zeng, H.; Bray, F.; Jemal, A.; Yu, X.Q.; He, J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016, 66, 115–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salazar, R.; Ciardiello, F. Optimizing Anti-EGFR Therapy in Colorectal Cancer. Clin. Cancer Res. 2015, 21, 5415–5416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Aguilar, J.; Chow, O.S.; Smith, D.D.; Marcet, J.E.; Cataldo, P.A.; Varma, M.G.; Kumar, A.S.; Oommen, S.; Coutsoftides, T.; Hunt, S.R.; et al. Effect of adding mFOLFOX6 after neoadjuvant chemoradiation in locally advanced rectal cancer: A multicentre, phase 2 trial. Lancet Oncol. 2015, 16, 957–966. [Google Scholar] [CrossRef] [Green Version]
- Kiriyama, S.; Saito, Y.; Yamamoto, S.; Soetikno, R.; Matsuda, T.; Nakajima, T.; Kuwano, H. Comparison of endoscopic submucosal dissection with laparoscopic-assisted colorectal surgery for early-stage colorectal cancer: A retrospective analysis. Endoscopy 2012, 44, 1024–1030. [Google Scholar] [CrossRef]
- Dupré, A.; Melodelima, D.; Pérol, D.; Chen, Y.; Vincenot, J.; Chapelon, J.-Y.; Rivoire, M. First Clinical Experience of Intra-Operative High Intensity Focused Ultrasound in Patients with Colorectal Liver Metastases: A Phase I-IIa Study. PLoS ONE 2015, 19, e0118212. [Google Scholar] [CrossRef] [Green Version]
- Nordholm-Carstensen, A.; Wille-Jørgensen, P.A.; Jorgensen, L.N.; Harling, H. Indeterminate Pulmonary Nodules at Colorectal Cancer Staging: A Systematic Review of Predictive Parameters for Malignancy. Ann. Surg. Oncol. 2013, 20, 4022–4030. [Google Scholar] [CrossRef] [PubMed]
- Pettersson, D.; Cedermark, B.; Holm, T.; Radu, C.; Påhlman, L.; Glimelius, B.; Martling, A. Interim analysis of the Stockholm III trial of preoperative radiotherapy regimens for rectal cancer. Br. J. Surg. 2010, 97, 580–587. [Google Scholar] [CrossRef]
- Lea, D.; Håland, S.; Hagland, H.R.; Søreide, K. Accuracy of TNM staging in colorectal cancer: A review of current culprits, the modern role of morphology and stepping-stones for improvements in the molecular era. Scand. J. Gastroenterol. 2014, 49, 1153–1163. [Google Scholar] [CrossRef]
- Hathout, L.; Maloney-Patel, N.; Malhotra, U.; Wang, S.-J.; Chokhavatia, S.; Dalal, I.; Poplin, E.; Jabbour, S.K. Management of locally advanced rectal cancer in the elderly: A critical review and algorithm. J. Gastrointest. Oncol. 2018, 9, 363–376. [Google Scholar] [CrossRef]
- Takahashi, H.; Ishikawa, T.; Ishiguro, M.; Okazaki, S.; Mogushi, K.; Kobayashi, H.; Iida, S.; Mizushima, H.; Tanaka, H.; Uetake, H.; et al. Prognostic significance of Traf2- and Nck-interacting kinase (TNIK) in colorectal cancer. BMC Cancer 2015, 15, 794. [Google Scholar] [CrossRef] [Green Version]
- Morton, D.L.; Thompson, J.F.; Cochran, A.J.; Mozzillo, N.; Nieweg, O.E.; Roses, D.F.; Hoekstra, H.J.; Karakousis, C.P.; Puleo, C.A.; Coventry, B.J.; et al. Final Trial Report of Sentinel-Node Biopsy versus Nodal Observation in Melanoma. N. Engl. J. Med. 2014, 370, 599–609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blechacz, B.; Mishra, L. Biopsy for liver cancer: How to balance research needs with evidence-based clinical practice. Hepatology 2015, 62, 1645. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.J.; Garcia-Aguilar, J. Advances and Challenges in Treatment of Locally Advanced Rectal Cancer. J. Clin. Oncol. 2015, 33, 1797–1808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koelzer, V.H.; Zlobec, I.; Lugli, A. Tumor budding in colorectal cancer—Ready for diagnostic practice? Hum. Pathol. 2016, 47, 4–19. [Google Scholar] [CrossRef]
- Varghese, S.; Newton, R.; Ross-Innes, C.S.; Lao-Sirieix, P.; Krishnadath, K.K.; O’Donovan, M.; Novelli, M.; Wernisch, L.; Bergman, J.; Fitzgerald, R.C. Analysis of dysplasia in patients with barrett’s esophagus based on expression pattern of 90 genes. Gastroenterology 2015, 149, 1511–1518.e5. [Google Scholar] [CrossRef]
- Amatu, A.; Barault, L.; Moutinho, C.; Cassingena, A.; Bencardino, K.; Ghezzi, S.; Palmeri, L.; Bonazzina, E.; Tosi, F.; Ricotta, R.; et al. Tumor MGMT promoter hypermethylation changes over time limit temozolomide efficacy in a phase II trial for metastatic colorectal cancer. Ann. Oncol. 2016, 27, 1062–1067. [Google Scholar] [CrossRef]
- Wei, Q.; Huang, X.; Fu, B.; Liu, J.; Zhong, L.; Yang, Q.; Zhao, T. IMP3 expression in biopsy specimens of colorectal cancer predicts lymph node metastasis and TNM stage. Int. J. Clin. Exp. Pathol. 2015, 8, 11024–11032. [Google Scholar]
- Meng, W.-J.; Pathak, S.; Ding, Z.-Y.; Zhang, H.; Adell, G.; Holmlund, B.; Li, Y.; Zhou, Z.-G.; Sun, X.-F. Special AT-rich sequence binding protein 1 expression correlates with response to preoperative radiotherapy and clinical outcome in rectal cancer. Cancer Biol. Ther. 2015, 16, 1738–1745. [Google Scholar] [CrossRef] [Green Version]
- Wallin, Å.R.; Svanvik, J.; Adell, G.; Sun, X.-F. Expression of PRL proteins at invasive margin of rectal cancers in relation to preoperative radiotherapy. Int. J. Radiat. Oncol. Biol. Phys. 2006, 65, 452–458. [Google Scholar] [CrossRef]
- Shabo, I.; Olsson, H.; Sun, X.-F.; Svanvik, J. Expression of the macrophage antigen CD163 in rectal cancer cells is associated with early local recurrence and reduced survival time. Int. J. Cancer 2009, 125, 1826–1831. [Google Scholar] [CrossRef] [PubMed]
- Pachkoria, K.; Zhang, H.; Adell, G.; Jarlsfelt, I.; Sun, X.-F. Significance of Cox-2 expression in rectal cancers with or without preoperative radiotherapy. Int. J. Radiat. Oncol. Biol. Phys. 2005, 63, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Moparthi, S.B.; Bergman, V.; Adell, G.; Thorstensson, S.; Sun, X.F. pRb2/p130 protein in relation to clinicopathological and biological variables in rectal cancers with a clinical trial of preoperative radiotherapy. Int. J. Colorectal Dis. 2009, 24, 1303–1310. [Google Scholar] [CrossRef] [PubMed]
- Schlenker, E.H. Tips and Tricks for Successful Application of Statistical Methods to Biological Data. Methods Mol. Biol. 2016, 1366, 271–285. [Google Scholar] [CrossRef]
- Fadhil, W.; Ibrahem, S.; Seth, R.; AbuAli, G.; Ragunath, K.; Kaye, P.; Ilyas, M. The utility of diagnostic biopsy specimens for predictive molecular testing in colorectal cancer. Histopathology 2012, 61, 1117–1124. [Google Scholar] [CrossRef] [PubMed]
- Ceylan, Y.; Lekili, M.; Müezzinoğlu, T.; Neşe, N.; Işisağ, A. Predictive value of Cyclooxygenase-2 over expression for identifying prostate cancer from benign prostatic hyperplasia in prostate biopsy specimens. Minerva Urol. Nefrol. 2015, 68, 255–262. [Google Scholar]
- Martling, A.; Holm, T.; Johansson, H.; Rutqvist, L.E.; Cedermark, B. Stockholm Colorectal Cancer Study Group. The Stockholm II trial on preoperative radiotherapy in rectal carcinoma: Long-term follow-up of a population-based study. Cancer 2001, 92, 896–902. [Google Scholar] [CrossRef]
- Shia, J.; Stadler, Z.; Weiser, M.R.; Rentz, M.; Gonen, M.; Tang, L.H.; Vakiani, E.; Katabi, N.; Xiong, X.; Markowitz, A.J.; et al. Immunohistochemical Staining for DNA Mismatch Repair Proteins in Intestinal Tract Carcinoma. How reliable are biopsy samples? Am. J. Surg. Pathol. 2011, 35, 447–454. [Google Scholar] [CrossRef]
- Waaijer, L.; Willems, S.M.; Verkooijen, H.M.; Buck, D.B.; Van Der Pol, C.C.; Van Diest, P.J.; Witkamp, A.J. Impact of preoperative evaluation of tumour grade by core needle biopsy on clinical risk assessment and patient selection for adjuvant systemic treatment in breast cancer. Br. J. Surg. 2015, 102, 1048–1055. [Google Scholar] [CrossRef]
- Blanco-Calvo, M.; Concha, A.; Figueroa, A.; Garrido, F.; Valladares-Ayerbes, M. Colorectal Cancer Classification and Cell Heterogeneity: A Systems Oncology Approach. Int. J. Mol. Sci. 2015, 16, 13610–13632. [Google Scholar] [CrossRef] [Green Version]
- Ohashi, R.; Matsubara, M.; Watarai, Y.; Yanagihara, K.; Yamashita, K.; Tsuchiya, S.-I.; Takei, H.; Naito, Z. Diagnostic value of fine needle aspiration and core needle biopsy in special types of breast cancer. Breast Cancer 2016, 23, 675–683. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.-H.; An, X.; Lin, X.; Gao, Y.-H.; Liu, G.-C.; Kong, L.-H.; Pan, Z.-Z.; Ding, P.-R. The value of forceps biopsy and core needle biopsy in prediction of pathologic complete remission in locally advanced rectal cancer treated with neoadjuvant chemoradiotherapy. Oncotarget 2015, 6, 33919–33925. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.-C.; Ng, K.-F.; Lee, S.-E.; Chen, K.-H.; Yeh, T.-S.; Chen, T.-C. HER2 testing in paired biopsy and excision specimens of gastric cancer: The reliability of the scoring system and the clinicopathological factors relevant to discordance. Gastric Cancer 2016, 19, 176–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Rossum, P.S.; Goense, L.; Meziani, J.; Reitsma, J.B.; Siersema, P.D.; Vleggaar, F.P.; Van Vulpen, M.; Meijer, G.J.; Ruurda, J.P.; Van Hillegersberg, R. Endoscopic biopsy and EUS for the detection of pathologic complete response after neoadjuvant chemoradiotherapy in esophageal cancer: A systematic review and meta-analysis. Gastrointest. Endosc. 2016, 83, 866–879. [Google Scholar] [CrossRef]
- Sato, T.; Yoneyama, T.; Tobisawa, Y.; Hatakeyama, S.; Yamamoto, H.; Kojima, Y.; Mikami, J.; Mori, K.; Hashimoto, Y.; Koie, T.; et al. Core 2 beta-1, 6-N-acetylglucosaminyltransferase-1 expression in prostate biopsy specimen is an indicator of prostate cancer aggressiveness. Biochem. Biophys. Res. Commun. 2016, 470, 150–156. [Google Scholar] [CrossRef] [Green Version]
- Feng, G.; Wang, X.; Cao, X.; Shen, L.; Zhu, J. ZEB1 Expression in Endometrial Biopsy Predicts Lymph Node Metastases in Patient with Endometrial Cancer. Dis. Markers 2014, 2014, 680361. [Google Scholar] [CrossRef]
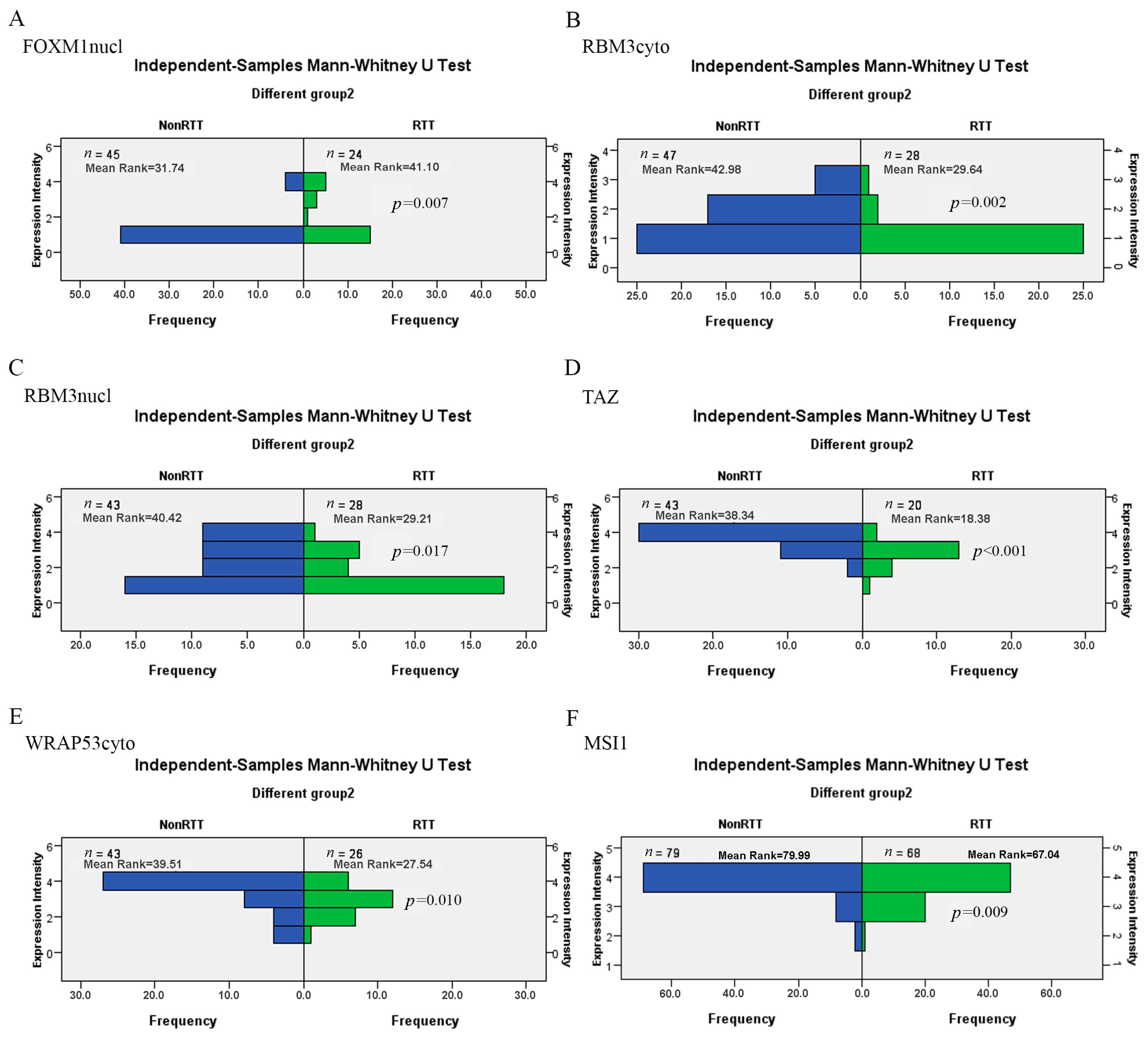

| Marker | BS | DN | p-Value | ||
|---|---|---|---|---|---|
| N | Mean Rank | N | Mean Rank | ||
| AEG1-NonRT | 58 | 52.46 | 46 | 40.54 | 0.026 |
| AEG1-RT | 42 | 32.02 | 37 | 29.98 | 0.210 |
| CD163-NonRT | 58 | 53.00 | 46 | 49.05 | 0.080 |
| CD163-RT | 43 | 38.50 | 36 | 28.50 | 0.039 |
| COX2-NonRT | 44 | 30.81 | 38 | 42.19 | 0.001 |
| COX2-RT | 41 | 34.25 | 35 | 40.10 | 0.044 |
| FOXM1cyto-NonRT | 56 | 52.00 | 46 | 41.00 | 0.032 |
| FOXM1nucl-NonRT | 54 | 54.18 | 46 | 38.00 | <0.001 |
| FOXM1nucl-RT | 41 | 31.24 | 34 | 18.50 | <0.001 |
| FOXO3Acyto-NonRT | 54 | 56.49 | 44 | 32.51 | <0.001 |
| XO3Acyto-RT | 43 | 34.05 | 27 | 21.72 | 0.002 |
| Ki67-NonRT | 45 | 55.21 | 41 | 52.34 | 0.346 |
| Ki67-RT | 28 | 32.25 | 26 | 30.87 | 0.259 |
| LIVIN-NonRT | 59 | 57.85 | 46 | 35.91 | <0.001 |
| LIVIN-RT | 42 | 34.14 | 30 | 26.00 | 0.053 |
| LOXcyto-NonRT | 57 | 45.92 | 46 | 47.08 | 0.811 |
| LOXcyto-RT | 40 | 28.48 | 22 | 36.52 | 0.019 |
| LOXnucl-NonRT | 57 | 44.80 | 45 | 47.22 | 0.613 |
| LOXnucl-RT | 42 | 29.96 | 23 | 38.22 | 0.024 |
| MSI1-NonRT | 57 | 83.55 | 63 | 39.64 | <0.001 |
| MSI1-RT | 43 | 65.63 | 55 | 36.89 | <0.001 |
| NFKBP65cyto-NonRT | 56 | 51.54 | 46 | 41.46 | 0.050 |
| NFKBP65cyto-RT | 43 | 35.44 | 25 | 35.58 | 0.975 |
| NFKBP65nucl-NonRT | 56 | 44.43 | 47 | 49.51 | 0.047 |
| NFKBP65nucl-RT | 43 | 34.56 | 24 | 35.46 | 0.675 |
| P73cyto-NonRT | 58 | 28.50 | 20 | 37.30 | 0.012 |
| P73cyto-RT | 48 | 24.32 | 21 | 36.10 | 0.001 |
| P130cyto-NonRT | 52 | 40.95 | 20 | 40.05 | 0.820 |
| P130cyto-RT | 42 | 30.91 | 21 | 32.09 | 0.699 |
| PINCH-NonRT | 56 | 51.29 | 27 | 38.71 | 0.026 |
| PINCH-RT | 48 | 46.25 | 21 | 27.55 | 0.016 |
| PPARcyto-NonRT | 54 | 53.08 | 43 | 33.15 | <0.001 |
| PPARcyto-RT | 42 | 32.57 | 27 | 23.26 | 0.022 |
| RBM3cyto-NonRT | 58 | 57.81 | 47 | 37.98 | <0.001 |
| RBM3cyto-RT | 41 | 33.22 | 28 | 22.96 | 0.001 |
| RBM3nucl-RT | 42 | 31.94 | 28 | 24.20 | 0.060 |
| SATB1-NonRT | 51 | 45.50 | 41 | 37.50 | 0.068 |
| SATB1-RT | 40 | 31.85 | 30 | 30.12 | 0.616 |
| SIRT6cyto-NonRT | 55 | 56.95 | 47 | 37.27 | <0.001 |
| SIRT6cyto-RT | 44 | 39.84 | 24 | 29.96 | 0.008 |
| SIRT6nucl-NonRT | 53 | 47.39 | 46 | 45.61 | 0.737 |
| SIRT6nucl-RT | 45 | 37.19 | 26 | 36.81 | 0.923 |
| TAZ-NonRT | 57 | 55.12 | 43 | 31.88 | <0.001 |
| TAZ-RT | 44 | 33.18 | 30 | 27.82 | 0.188 |
| WRAP53cyto-NonRT | 54 | 52.87 | 40 | 30.31 | <0.001 |
| WRAP53cyto-RT | 44 | 31.96 | 26 | 21.04 | 0.006 |
| WRAP53nucl-NonRT | 55 | 47.15 | 44 | 40.92 | 0.231 |
| RT Group | Non-RT Group | |||||||
|---|---|---|---|---|---|---|---|---|
| Univariate Analysis | Multivariate Analysis | Univariate Analysis | Multivariate Analysis | |||||
| HR (95% CI) | p | HR (95% CI) | p | HR (95% CI) | p | HR (95% CI) | p | |
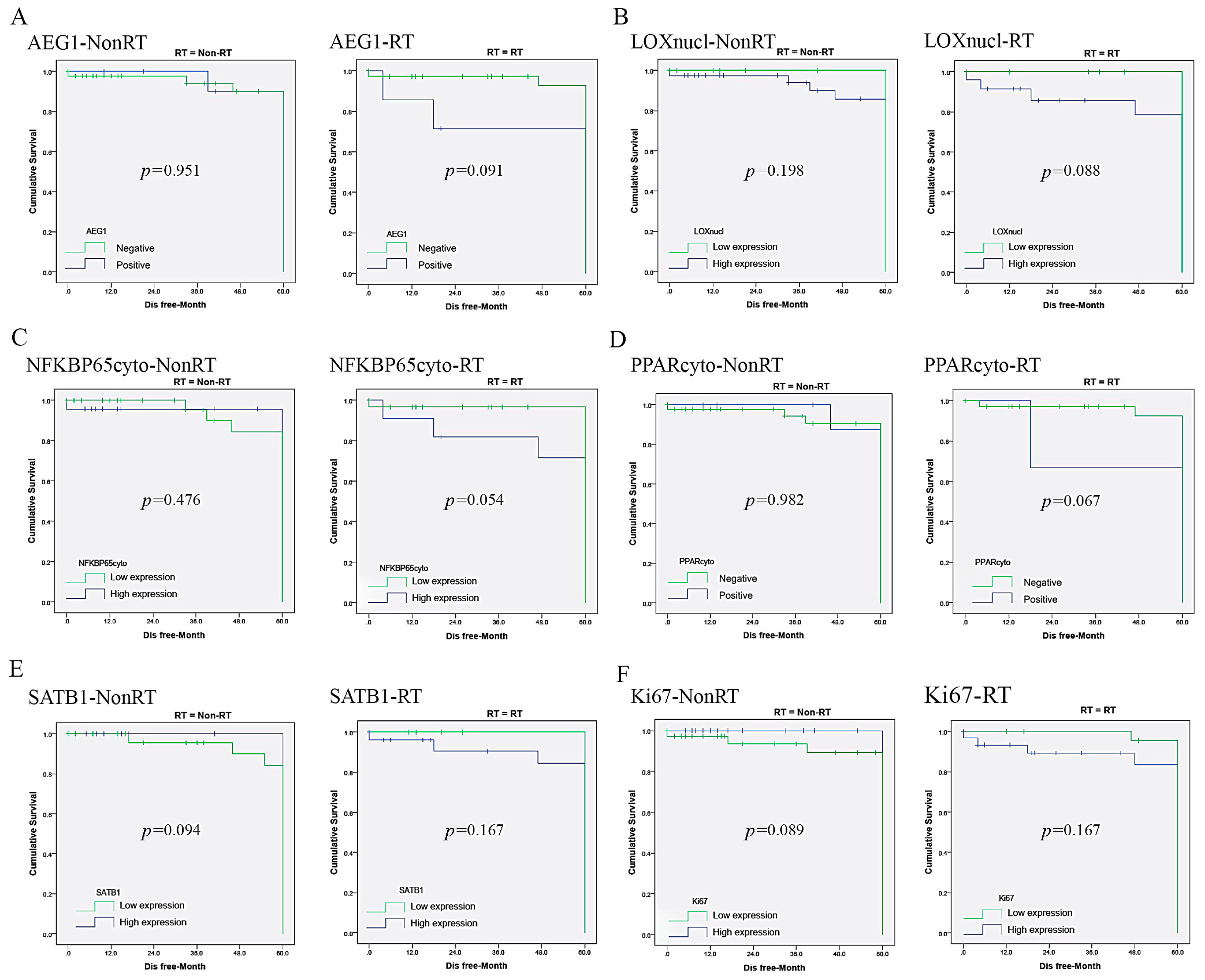
| AEG1 | 1.820 (1.213–3.124) | 0.045 | 1.581 (1.108–2.915) | 0.074 | 1.005 (0.521–1.252) | 0.681 | − | − |
| CD163 | 0.960 (0.576–1.345) | 0.465 | − | − | 0.880 (0.462–1.099) | 0.442 | − | − |
| COX2 | 1.233 (0.951–2.218) | 0.044 | 1.088 (0.710–1.632) | 0.178 | 0.861 (0.509–1.417) | 0.498 | − | − |
| FOXM1cyto | 0.875 (0.665–1.127) | 0.546 | − | − | 0.719 (0.636–1.308) | 0.421 | − | − |
| FOXM1nucl | 1.265 (0.896−2.127) | 0.043 | 0.768 (0.551–1.301) | 0.432 | 1.287 (0.855–1.973) | 0.039 | 0.815 (0.768–1.545) | 0.616 |
| FOXO3Acyto | 1.001 (0.601–1.236) | 0.687 | − | − | 1.026 (0.705–1.617) | 0.449 | − | − |
| Ki67 | 0.756 (0.433–1.224) | 0.437 | − | − | 0.712 (0.412–1.156) | 0.385 | − | − |
| LIVIN | 1.129 (0.711–1.237) | 0.700 | − | − | 0.995 (0.532–1.211) | 0.575 | − | − |
| LOXcyto | 0.923 (0.494–1.268) | 0.543 | − | − | 0.897 (0.561–1.265) | 0.458 | − | − |
| LOXnucl | 1.356 (0.987–2.826) | 0.037 | 1.253 (0.914–2.435) | 0.096 | 0.625 (0.419–1.087) | 0.444 | − | − |
| MSI1 | 0.910 (0.523–1.336) | 0.249 | − | − | 0.858 (0.469–1.217) | 0.311 | − | − |
| NFKBP65cyto | 2.401 (1.832–4.365) | 0.011 | 2.225 (1.787–4.064) | 0.042 | 1.354 (1.017–2.620) | 0.027 | 0.989 (0.659–1.578) | 0.590 |
| NFKBP65nucl | 0.562 (0.387–0.993) | 0.505 | − | − | 0.611 (0.365–1.120) | 0.242 | − | − |
| P53 | 0.741 (0.411–1.103) | 0.314 | − | − | 0.722 (0.405–1.008) | 0.206 | − | − |
| P73cyto | 0.891 (0.637–1.124) | 0.784 | − | − | 0.712 (0.440–1.432) | 0.483 | − | − |
| P130cyto | 1.057 (0.501–1.401) | 0.350 | − | − | 2.656 (1.913–4.016) | 0.010 | 2.532 (1.811–3.989) | 0.048 |
| PINCH | 0.921 (0.585–1.356) | 0.454 | − | − | 3.541 (1.942–5.308) | <0.001 | 3.239 (1.901–5.003) | <0.001 |
| PPARcyto | 2.587 (1.684–3.889) | 0.023 | 2.565 (1.591–3.776) | 0.049 | 0.543 (0.336–1.028) | 0.959 | − | − |
| PPARstrom | 0.879 (0.535–1.433) | 0.125 | − | − | 0.885 (0.638–1.584) | 0.299 | − | − |
| PRL | 0.586 (0.371–1.113) | 0.450 | − | − | 0.595 (0.389–1.215) | 0.371 | − | − |
| RBM3cyto | 0.506 (0.393–0.904) | 0.891 | − | − | 0.612 (0.387–0.947) | 0.832 | − | − |
| RBM3nucl | 0.715 (0.421–1.019) | 0.966 | − | − | 0.857 (0.569–1.272) | 0.837 | − | − |
| SATB1 | 0.812 (0.564–1.541) | 0.178 | − | − | 1.350 (1.118–2.469) | 0.049 | 1.251 (0.973–2.061) | 0.062 |
| SIRT6cyto | 0.602 (0.418–0.902) | 0.381 | − | − | 0.525 (0.381–0.865) | 0.451 | − | − |
| SIRT6nucl | 1.310 (1.058–2.379) | 0.034 | 1.265 (1.024–1.960) | 0.052 | 0.649 (0.417–1.059) | 0.279 | − | − |
| TAZ | 0.791 (0.607–1.019) | 0.555 | − | − | 0.541 (0.349–0.945) | 0.319 | − | − |
| WRAP53cyto | 1.267 (0.826–2.358) | 0.042 | 0.896 (0.356–1.321) | 0.662 | 1.216 (0.959–1.837) | 0.048 | 0.861 (0.510–1.530) | 0.510 |
| WRAP53nucl | 0.859 (0.538–1.252) | 0.678 | − | − | 0.716 (0.433–1.201) | 0.273 | − | − |
| WRAP53stromcyto | 0.708 (0.423–1.071) | 0.312 | − | − | 0.641 (0.356–0.952) | 0.361 | − | − |
| WRAP53stromnucl | 0.856 (0.579–1.134) | 0.872 | − | − | 0.764 (0.496–1.082) | 0.160 | − | − |
| RT Group | Non-RT Group | |||||||
|---|---|---|---|---|---|---|---|---|
| Univariate Analysis | Multivariate Analysis | Univariate Analysis | Multivariate Analysis | |||||
| HR (95% CI) | p | HR (95% CI) | p | HR (95% CI) | p | HR (95% CI) | p | |
| AEG1 | 1.520 (1.217–2.648) | 0.041 | 1.418 (1.103–2.306) | 0.091 | 0.738 (0.623–1.237) | 0.951 | − | − |
| CD163 | 0.718 (0.465–1.021) | 0.320 | − | − | 0.695 (0.433–1.125) | 0.608 | − | − |
| COX2 | 3.467 (1.725–4.763) | 0.008 | 3.345 (1.643–4.568) | 0.022 | 1.523 (0.987–3.126) | 0.044 | 1.386 (0.921–1.953) | 0.084 |
| FOXM1cyto | 0.763 (0.458–1.034) | 0.770 | − | − | 0.637 (0.357–0.998) | 0.624 | − | − |
| FOXM1nucl | 1.347 (0.778–1.995) | 0.035 | 0.876 (0.580–1.426) | 0.359 | 1.268 (0.795–2.101) | 0.038 | 0.741 (0.411–1.132) | 0.608 |
| FOXO3Acyto | 0.595 (0.310–0.872) | 0.914 | − | − | 0.624 (0.398–1.013) | 0.386 | − | − |
| Ki67 | 1.298 (0.756–1.741) | 0.045 | 1.021 (0.616–1.442) | 0.167 | 1.545 (0.977–2.978) | 0.034 | 1.247 (0.915–1.868) | 0.089 |
| LIVIN | 0.692 (0.482–1.145) | 0.781 | - | - | 0.793 (0.586–1.335) | 0.375 | - | - |
| LOXcyto | 0.781 (0.433–1.325) | 0.541 | − | − | 0.763 (0.464–1.250) | 0.944 | − | − |
| LOXnucl | 1.529 (1.186–3.243) | 0.037 | 1.453 (1.151–2.247) | 0.088 | 1.067 (0.834–1.996) | 0.198 | − | − |
| MSI1 | 0.769 (0.443–1.156) | 0.128 | − | − | 0.849 (0.515–1.131) | 0.210 | − | − |
| NFKBP65cyto | 1.530 (1.126–2.898) | 0.015 | 1.312 (0.949–2.356) | 0.054 | 1.253 (0.881–2.440) | 0.045 | 0.623 (0.418–1.037) | 0.476 |
| NFKBP65nucl | 0.629 (0.318–0.925) | 0.498 | - | - | 0.722 (0.421–1.214) | 0.445 | - | - |
| P53 | 0.884 (0.569–1.217) | 0.115 | - | - | 0.865 (0.510–1.237) | 0.315 | - | - |
| P73cyto | 0.887 (0.536–1.213) | 0.452 | - | - | 0.689 (0.480–1.121) | 0.766 | - | - |
| P130cyto | 0.659 (0.347–1.224) | 0.420 | − | − | 1.035 (0.626–1.435) | 0.418 | − | − |
| PINCH | 0.720 (0.451–1.279) | 0.527 | − | − | 4.153 (1.672–6.783) | 0.001 | 3.879 (1.525–5.963) | 0.005 |
| PPARcyto | 1.586 (0.926–3.310) | 0.037 | 1.325 (0.915–2.256) | 0.067 | 0.645 (0.416–1.275) | 0.982 | − | − |
| PPARstrom | 0.854 (0.571–1.401) | 0.429 | − | − | 0.717 (0.426–1.319) | 0.972 | − | − |
| PRL | 0.530 (0.352–1.011) | 0.519 | − | − | 0.601 (0.421–1.009) | 0.449 | − | − |
| RBM3cyto | 0.648 (0.379–1.096) | 0.433 | − | − | 1.326 (0.919–2.148) | 0.046 | 0.788 (0.513–1.345) | 0.218 |
| RBM3nucl | 0.516 (0.405–1.107) | 0.734 | − | − | 0.753 (0.452–1.130) | 0.625 | − | − |
| SATB1 | 0.858 (0.532–1.610) | 0.167 | − | − | 1.561 (1.022–2.962) | 0.048 | 1.453 (0.986–2.207) | 0.094 |
| SIRT6cyto | 0.653 (0.420–0.933) | 0.433 | − | − | 0.661 (0.392–0.946) | 0.714 | − | − |
| SIRT6nucl | 0.715 (0.514–1.211) | 0.840 | − | − | 0.803 (0.568–1.315) | 0.419 | − | − |
| TAZ | 1.335 (0.898–2.312) | 0.039 | 0.737 (0.512–1.210) | 0.628 | 0.783 (0.448–1.213) | 0.424 | − | − |
| WRAP53cyto | 1.211 (0.865–2.074) | 0.049 | 0.696 (0.347–1.101) | 0.645 | 3.762 (1.564–5.568) | 0.003 | 3.556 (1.512–5.210) | 0.007 |
| WRAP53nucl | 0.641 (0.368–1.105) | 0.698 | − | − | 1.527 (1.189–2.991) | 0.040 | 1.463 (1.032–2.313) | 0.077 |
| WRAP53stromcyto | 0.550 (0.312–1.069) | 0.526 | − | − | 0.613 (0.338–1.195) | 0.471 | − | − |
| WRAP53stromnucl | 0.677 (0.481–1.156) | 0.873 | − | − | 0.862 (0.501–1.357) | 0.394 | − | − |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, Y.; Zhao, S.; Holmqvist, A.; Hahn-Stromberg, V.; Adell, G.; Holmlund, B.; Pathak, S.; Peng, Z.; Sun, X.-F. Predictive Role of Biopsy Based Biomarkers for Radiotherapy Treatment in Rectal Cancer. J. Pers. Med. 2020, 10, 168. https://doi.org/10.3390/jpm10040168
Wen Y, Zhao S, Holmqvist A, Hahn-Stromberg V, Adell G, Holmlund B, Pathak S, Peng Z, Sun X-F. Predictive Role of Biopsy Based Biomarkers for Radiotherapy Treatment in Rectal Cancer. Journal of Personalized Medicine. 2020; 10(4):168. https://doi.org/10.3390/jpm10040168
Chicago/Turabian StyleWen, Yugang, Senlin Zhao, Annica Holmqvist, Victoria Hahn-Stromberg, Gunnar Adell, Birgitta Holmlund, Surajit Pathak, Zhihai Peng, and Xiao-Feng Sun. 2020. "Predictive Role of Biopsy Based Biomarkers for Radiotherapy Treatment in Rectal Cancer" Journal of Personalized Medicine 10, no. 4: 168. https://doi.org/10.3390/jpm10040168





