Systems Biology and Experimental Model Systems of Cancer
Abstract
:1. Introduction to Cancer Systems Biology
2. Cancer Systems Biology for Precision Medicine
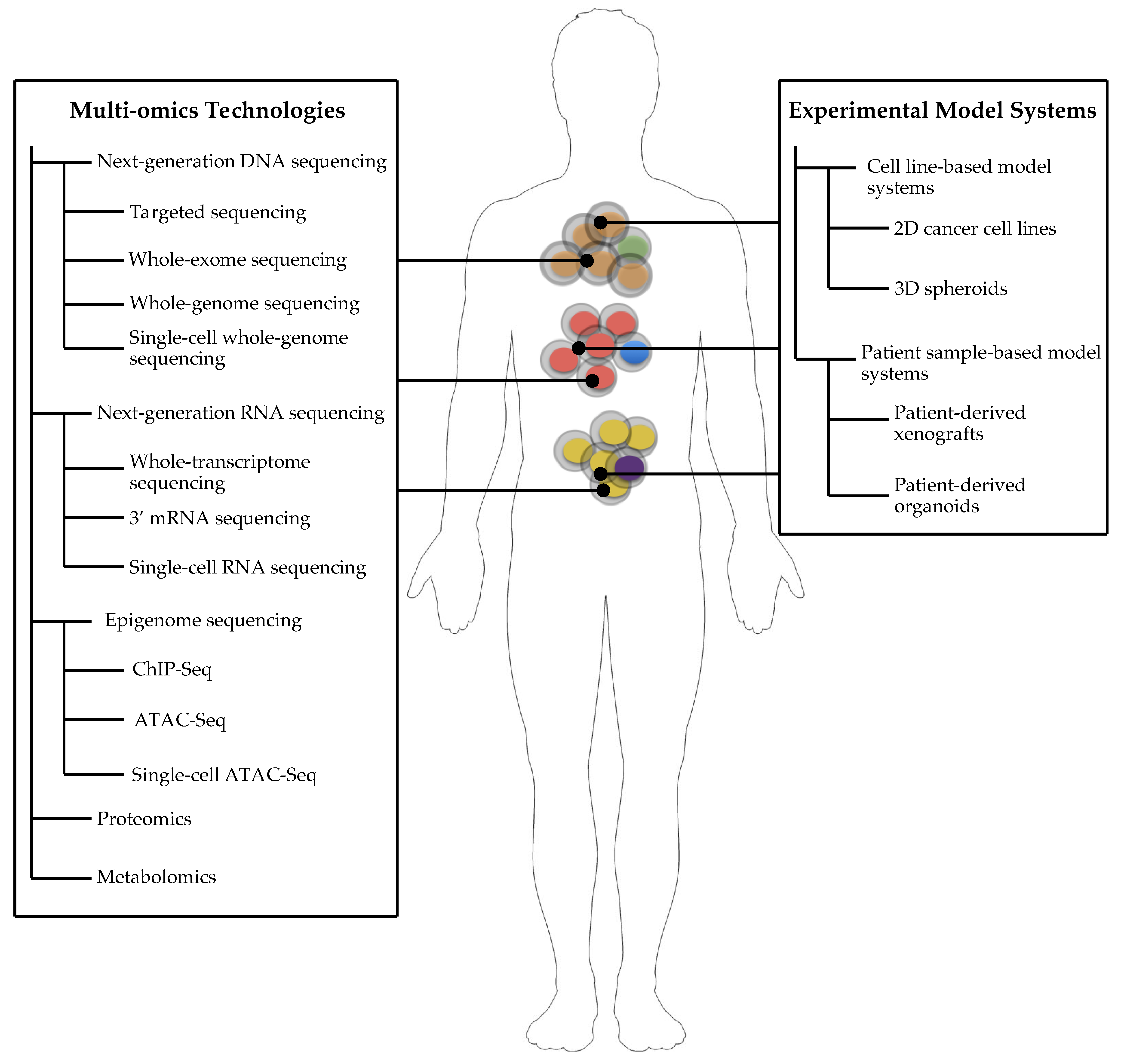
3. Experimental Model Systems of Cancer
4. Cell Line-Based Model Systems
5. Patient Sample-Based Model Systems
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pietras, K.; Östman, A. Hallmarks of cancer: Interactions with the tumor stroma. Exp. Cell Res. 2010, 316, 1324–1331. [Google Scholar] [CrossRef]
- Han, X.; Zhou, Z.; Fei, L.; Sun, H.; Wang, R.; Chen, Y.; Chen, H.; Wang, J.; Tang, H.; Ge, W.; et al. Construction of a human cell landscape at single-cell level. Nature 2020, 581, 303–309. [Google Scholar] [CrossRef]
- Campbell, P.J.; Getz, G.; Korbel, J.O.; Stuart, J.M.; Jennings, J.L.; Stein, L.D.; Perry, M.D.; Nahal-Bose, H.K.; Ouellette, B.F.F.; Li, C.H.; et al. Pan-cancer analysis of whole genomes. Nature 2020, 578, 82–93. [Google Scholar] [CrossRef] [Green Version]
- Zehir, A.; Benayed, R.; Shah, R.H.; Syed, A.; Middha, S.; Kim, H.R.; Srinivasan, P.; Gao, J.; Chakravarty, D.; Devlin, S.M.; et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 2017, 23, 703–713. [Google Scholar] [CrossRef] [PubMed]
- Jamal-Hanjani, M.; Wilson, G.A.; McGranahan, N.; Birkbak, N.J.; Watkins, T.B.K.; Veeriah, S.; Shafi, S.; Johnson, D.H.; Mitter, R.; Rosenthal, R.; et al. Tracking the evolution of non–small-cell lung cancer. N. Eng. J. Med. 2017, 376, 2109–2121. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, S.; Hosen, M.I.; Ahmed, M.; Shekhar, H.U. Onco-Multi-OMICS Approach: A New Frontier in Cancer Research. BioMed Res. Int. 2018, 2018. [Google Scholar] [CrossRef] [Green Version]
- Filipp, F.V. Precision medicine driven by cancer systems biology. Cancer Metastasis Rev. 2017, 36, 91–108. [Google Scholar] [CrossRef] [Green Version]
- McGranahan, N.; Swanton, C. Clonal heterogeneity and tumor evolution: Past, present, and the future. Cell 2017, 168, 613–628. [Google Scholar] [CrossRef]
- Hinohara, K.; Polyak, K. Intratumoral Heterogeneity: More Than Just Mutations. Trends Cell Biol. 2019, 29, 569–579. [Google Scholar] [CrossRef]
- Guo, M.; Peng, Y.; Gao, A.; Du, C.; Herman, J.G. Epigenetic heterogeneity in cancer. Biomark. Res. 2019, 7, 23. [Google Scholar] [CrossRef] [PubMed]
- Turajlic, S.; Xu, H.; Litchfield, K.; Rowan, A.; Chambers, T.; Lopez, J.I.; Nicol, D.; O’Brien, T.; Larkin, J.; Horswell, S.; et al. Tracking cancer evolution reveals constrained routes to metastases: TRACERx renal. Cell 2018, 173, 581–594.e12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acar, A.; Hidalgo-Sastre, A.; Leverentz, M.K.; Mills, C.G.; Woodcock, S.; Baron, M.; Collu, G.M.; Brennan, K. Inhibition of Wnt signalling by Notch via two distinct mechanisms. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Collu, G.M.; Hidalgo-Sastre, A.; Brennan, K. Wnt-Notch signalling crosstalk in development and disease. Cell. Mol. Life Sci. 2014, 71, 3553–3567. [Google Scholar] [CrossRef]
- Stylianou, S.; Clarke, R.B.; Brennan, K. Aberrant activation of Notch signaling in human breast cancer. Cancer Res. 2006, 66, 1517–1525. [Google Scholar] [CrossRef] [Green Version]
- Marusyk, A.; Janiszewska, M.; Polyak, K. Intratumor heterogeneity: The Rosetta Stone of therapy resistance. Cancer Cell 2020, 37, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Werner, H.M.J.; Mills, G.B.; Ram, P.T. Cancer systems biology: A peek into the future of patient care? Nat. Rev. Clin. Oncol. 2014, 11, 167–176. [Google Scholar] [CrossRef] [Green Version]
- Turajlic, S.; Sottoriva, A.; Graham, T.; Swanton, C. Resolving genetic heterogeneity in cancer. Nat. Rev. Genet. 2019, 20, 406–416. [Google Scholar] [CrossRef]
- Greaves, M.; Maley, C.C. Clonal evolution in cancer. Nature 2012, 481, 306–313. [Google Scholar] [CrossRef]
- Caravagna, G.; Heide, T.; Williams, M.J.; Zapata, L.; Nichol, D.; Chkhaidze, K.; Cross, W.; Cresswell, G.D.; Werner, B.; Acar, A.; et al. Subclonal reconstruction of tumors by using machine learning and population genetics. Nat. Genet. 2020, 52, 898–907. [Google Scholar] [CrossRef]
- Levy, S.E.; Myers, R.M. Advancements in next-generation sequencing. Annu. Rev. Genom. Hum. Genet. 2016, 17, 95–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antman, E.; Weiss, S.; Loscalzo, J. Systems pharmacology, pharmacogenetics, and clinical trial design in network medicine. Wiley Interdiscip. Rev. Syst. Biol. Med. 2012, 4, 367–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barry, P.; Vatsiou, A.; Spiteri, I.; Nichol, D.; Cresswell, G.D.; Acar, A.; Trahearn, N.; Hrebien, S.; Garcia-Murillas, I.; Chkhaidze, K.; et al. The spatiotemporal evolution of lymph node spread in early breast cancer. Clin. Cancer Res. 2018, 24, 4763–4770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spiteri, I.; Caravagna, G.; Cresswell, G.D.; Vatsiou, A.; Nichol, D.; Acar, A.; Ermini, L.; Chkhaidze, K.; Werner, B.; Mair, R.; et al. Evolutionary dynamics of residual disease in human glioblastoma. Ann. Oncol. 2019, 30, 456–463. [Google Scholar] [CrossRef] [Green Version]
- Cross, W.; Kovac, M.; Mustonen, V.; Temko, D.; Davis, H.; Baker, A.M.; Biswas, S.; Arnold, R.; Chegwidden, L.; Gatenbee, C.; et al. The evolutionary landscape of colorectal tumorigenesis. Nat. Ecol. Evol. 2018, 2, 1661–1672. [Google Scholar] [CrossRef] [Green Version]
- Kelso, T.W.R.; Porter, D.K.; Amaral, M.L.; Shokhirev, M.N.; Benner, C.; Hargreaves, D.C. Chromatin accessibility underlies synthetic lethality of SWI/SNF subunits in ARID1A-mutant cancers. eLife 2017, 6. [Google Scholar] [CrossRef]
- Kim, H.; Zheng, S.; Amini, S.S.; Virk, S.M.; Mikkelsen, T.; Brat, D.J.; Grimsby, J.; Sougnez, C.; Muller, F.; Hu, J.; et al. Whole-genome and multisector exome sequencing of primary and post-treatment glioblastoma reveals patterns of tumor evolution. Genome Res. 2015, 25, 316–327. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Choi, P.S.; Francis, J.M.; Imielinski, M.; Watanabe, H.; Cherniack, A.D.; Meyerson, M. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat. Genet. 2016, 48. [Google Scholar] [CrossRef]
- Calabrese, C.; Davidson, N.R.; Demircioğlu, D.; Fonseca, N.A.; He, Y.; Kahles, A.; van Lehmann, K.; Liu, F.; Shiraishi, Y.; Soulette, C.M.; et al. Genomic basis for RNA alterations in cancer. Nature 2020, 578. [Google Scholar] [CrossRef] [Green Version]
- Reyna, M.A.; Haan, D.; Paczkowska, M.; Verbeke, L.P.C.; Vazquez, M.; Kahraman, A.; Pulido-Tamayo, S.; Barenboim, J.; Wadi, L.; Dhingra, P.; et al. Pathway and network analysis of more than 2500 whole cancer genomes. Nat. Commun. 2020, 11. [Google Scholar] [CrossRef]
- Rheinbay, E.; Nielsen, M.M.; Abascal, F.; Wala, J.A.; Shapira, O.; Tiao, G.; Hornshøj, H.; Hess, J.M.; Juul, R.I.; Lin, Z.; et al. Analyses of non-coding somatic drivers in 2658 cancer whole genomes. Nature 2020, 578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhang, H.E.C.; Ruddy, D.A.; Radhakrishna, V.K.; Caushi, J.X.; Zhao, R.; Hims, M.M.; Singh, A.P.; Kao, I.; Rakiec, D.; Shaw, P.; et al. Studying clonal dynamics in response to cancer therapy using high-complexity barcoding. Nat. Med. 2015, 21, 440–448. [Google Scholar] [CrossRef] [PubMed]
- Acar, A.; Nichol, D.; Fernandez-Mateos, J.; Cresswell, G.D.; Barozzi, I.; Hong, S.P.; Trahearn, N.; Spiteri, I.; Stubbs, M.; Burke, R.; et al. Exploiting evolutionary steering to induce collateral drug sensitivity in cancer. Nat. Commun. 2020, 11, 1923. [Google Scholar] [CrossRef] [Green Version]
- Tang, F.; Barbacioru, C.; Wang, Y.; Nordman, E.; Lee, C.; Xu, N.; Wang, X.; Bodeau, J.; Tuch, B.B.; Siddiqui, A.; et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 2009, 6, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Macosko, E.Z.; Basu, A.; Satija, R.; Nemesh, J.; Shekhar, K.; Goldman, M.; Tirosh, I.; Bialas, A.R.; Kamitaki, N.; Martersteck, E.M.; et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 2015, 161, 1202–1214. [Google Scholar] [CrossRef] [Green Version]
- Klein, A.M.; Mazutis, L.; Akartuna, I.; Tallapragada, N.; Veres, A.; Li, V.; Peshkin, L.; Weitz, D.A.; Kirschner, M.W. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 2015, 161, 1187–1201. [Google Scholar] [CrossRef] [Green Version]
- Baron, C.S.; van Oudenaarden, A. Unravelling cellular relationships during development and regeneration using genetic lineage tracing. Nat. Rev. Mol. Cell Biol. 2019, 20, 753–765. [Google Scholar] [CrossRef]
- González-Silva, L.; Quevedo, L.; Varela, I. Tumor Functional Heterogeneity Unraveled by scRNA-seq Technologies. Trends Cancer 2020, 6, 13–19. [Google Scholar] [CrossRef] [Green Version]
- Navin, N.; Kendall, J.; Troge, J.; Andrews, P.; Rodgers, L.; McIndoo, J.; Cook, K.; Stepansky, A.; Levy, D.; Esposito, D.; et al. Tumour evolution inferred by single-cell sequencing. Nature 2011, 472, 90–94. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.; Gao, R.; Sei, E.; Brandt, R.; Hartman, J.; Hatschek, T.; Crosetto, N.; Foukakis, T.; Navin, N.E. Chemoresistance evolution in triple-negative breast cancer delineated by single-cell sequencing. Cell 2018, 173, 879–893.e13. [Google Scholar] [CrossRef] [Green Version]
- Laks, E.; McPherson, A.; Zahn, H.; Lai, D.; Steif, A.; Brimhall, J.; Biele, J.; Wang, B.; Masud, T.; Ting, J.; et al. Clonal decomposition and DNA replication states defined by scaled single-cell genome sequencing. Cell 2019, 179, 1207–1207.e22. [Google Scholar] [CrossRef] [PubMed]
- Cusanovich, D.A.; Daza, R.; Adey, A.; Pliner, H.A.; Christiansen, L.; Gunderson, K.L.; Steemers, F.J.; Trapnell, C.; Shendure, J. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 2015, 348, 910–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Granja, J.M.; Klemm, S.; McGinnis, L.M.; Kathiria, A.S.; Mezger, A.; Corces, M.R.; Parks, B.; Gars, E.; Liedtke, M.; Zheng, G.X.Y.; et al. Single-cell multiomic analysis identifies regulatory programs in mixed-phenotype acute leukemia. Nat. Biotechnol. 2019, 37, 1458–1465. [Google Scholar] [CrossRef] [PubMed]
- Macaulay, I.C.; Haerty, W.; Kumar, P.; Li, Y.I.; Hu, T.X.; Teng, M.J.; Goolam, M.; Saurat, N.; Coupland, P.; Shirley, L.M.; et al. G&T-seq: Parallel sequencing of single-cell genomes and transcriptomes. Nat. Methods 2015, 12, 519–522. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Cusanovich, D.A.; Ramani, V.; Aghamirzaie, D.; Pliner, H.A.; Hill, A.J.; Daza, R.M.; McFaline-Figueroa, J.L.; Packer, J.S.; Christiansen, L.; et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science 2018, 361, 1380–1385. [Google Scholar] [CrossRef] [Green Version]
- Cao, Q.; Zhou, M.; Wang, X.; Meyer, C.A.; Zhang, Y.; Chen, Z.; Li, C.; Liu, X.S. CaSNP: A database for interrogating copy number alterations of cancer genome from SNP array data. Nucleic Acids Res. 2011, 39, D968–D974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Anda-Jáuregui, G.; Hernández-Lemus, E. Computational oncology in the multi-omics era: State of the Art. Front. Oncol. 2020, 10, 423. [Google Scholar]
- Huret, J.L.; Ahmad, M.; Arsaban, M.; Bernheim, A.; Cigna, J.; Desangles, F.; Guignard, J.C.; Jacquemot-Perbal, M.C.; Labarussias, M.; Leberre, V.; et al. Atlas of genetics and cytogenetics in oncology and haematology in 2013. Nucleic Acids Res. 2013, 41, D920–D924. [Google Scholar] [CrossRef]
- Gorohovski, A.; Tagore, S.; Palande, V.; Malka, A.; Raviv-Shay, D.; Frenkel-Morgenstern, M. ChiTaRS-3.1-the enhanced chimeric transcripts and RNA-seq database matched with protein-protein interactions. Nucleic Acids Res. 2017, 45, D790–D795. [Google Scholar] [CrossRef]
- Halling-Brown, M.D.; Bulusu, K.C.; Patel, M.; Tym, J.E.; Al-Lazikani, B. canSAR: An integrated cancer public translational research and drug discovery resource. Nucleic Acids Res. 2012, 40, D947–D956. [Google Scholar] [CrossRef] [Green Version]
- Su, W.H.; Chao, C.C.; Yeh, S.H.; Chen, D.S.; Chen, P.J.; Jou, Y.S. OncoDB.HCC: An integrated oncogenomic database of hepatocellular carcinoma revealed aberrant cancer target genes and loci. Nucleic Acids Res. 2007, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forbes, S.A.; Beare, D.; Boutselakis, H.; Bamford, S.; Bindal, N.; Tate, J.; Cole, C.G.; Ward, S.; Dawson, E.; Ponting, L.; et al. COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017, 45, D777–D783. [Google Scholar] [CrossRef]
- Samur, M.K.; Yan, Z.; Wang, X.; Cao, Q.; Munshi, N.C.; Li, C.; Shah, P.K. canEvolve: A web portal for integrative oncogenomics. PLoS ONE 2013, 8, e56228. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, A.; Tuknait, A.; Anand, P.; Gupta, S.; Sharma, M.; Mathur, D.; Joshi, A.; Singh, S.; Gautam, A.; Raghava, G.P.S. CancerPPD: A database of anticancer peptides and proteins. Nucleic Acids Res. 2015, 43, D837–D843. [Google Scholar] [CrossRef] [Green Version]
- Cutts, R.J.; Gadaleta, E.; Hahn, S.A.; Crnogorac-Jurcevic, T.; Lemoine, N.R.; Chelala, C. The pancreatic expression database: 2011 update. Nucleic Acids Res. 2011, 39, D1023–D1028. [Google Scholar] [CrossRef] [Green Version]
- Hudson, T.J.; Anderson, W.; Aretz, A.; Barker, A.D.; Bell, C.; Bernabé, R.R.; Bhan, M.K.; Calvo, F.; Eerola, I.; Gerhard, D.S.; et al. International network of cancer genome projects. Nature 2010, 464, 993–998. [Google Scholar]
- He, X.; Chang, S.; Zhang, J.; Zhao, Q.; Xiang, H.; Kusonmano, K.; Yang, L.; Sun, Z.S.; Yang, H.; Wang, J. MethyCancer: The database of human DNA methylation and cancer. Nucleic Acids Res. 2008, 36, D836–D841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whiteaker, J.R.; Halusa, G.N.; Hoofnagle, A.N.; Sharma, V.; MacLean, B.; Yan, P.; Wrobel, J.A.; Kennedy, J.; Mani, D.R.; Zimmerman, L.J.; et al. CPTAC Assay Portal: A repository of targeted proteomic assays. Nat. Methods 2014, 11, 703–704. [Google Scholar] [CrossRef] [Green Version]
- Perez-Llamas, C.; Gundem, G.; Lopez-Bigas, N. Integrative Cancer Genomics (IntOGen) in Biomart. Database 2011, 2011. [Google Scholar] [CrossRef] [Green Version]
- Parkinson, H.; Kapushesky, M.; Shojatalab, M.; Abeygunawardena, N.; Coulson, R.; Farne, A.; Holloway, E.; Kolesnykov, N.; Lilja, P.; Lukk, M.; et al. ArrayExpress—A public database of microarray experiments and gene expression profiles. Nucleic Acids Res. 2007, 35. [Google Scholar] [CrossRef]
- Liu, S.H.; Shen, P.C.; Chen, C.Y.; Hsu, A.N.; Cho, Y.C.; Lai, Y.L.; Chen, F.H.; Li, C.Y.; Wang, S.C.; Chen, M.; et al. DriverDBv3: A multi-omics database for cancer driver gene research. Nucleic Acids Res. 2020, 48. [Google Scholar] [CrossRef] [Green Version]
- Thomas, J.K.; Kim, M.S.; Balakrishnan, L.; Nanjappa, V.; Raju, R.; Marimuthu, A.; Radhakrishnan, A.; Muthusamy, B.; Khan, A.A.; Sakamuri, S.; et al. Pancreatic Cancer Database: An integrative resource for pancreatic cancer. Cancer Biol. Ther. 2014, 15. [Google Scholar] [CrossRef] [Green Version]
- Kumar, R.; Chaudhary, K.; Gupta, S.; Singh, H.; Kumar, S.; Gautam, A.; Kapoor, P.; Raghava, G.P.S. CancerDR: Cancer drug resistance database. Sci. Rep. 2013, 3. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.E.V.; Blundell, T.L.; Ascher, D.B. Platinum: A database of experimentally measured effects of mutations on structurally defined protein-ligand complexes. Nucleic Acids Res. 2015, 43. [Google Scholar] [CrossRef]
- Jemal, A.; Siegel, R.; Ward, E.; Hao, Y.; Xu, J.; Thun, M.J. Cancer Statistics, 2009. CA Cancer J. Clin. 2009, 59. [Google Scholar] [CrossRef]
- Hidalgo, M.; Amant, F.; Biankin, A.V.; Budinská, E.; Byrne, A.T.; Caldas, C.; Clarke, R.B.; de Jong, S.; Jonkers, J.; Mælandsmo, G.M.; et al. Patient-derived Xenograft models: An emerging platform for translational cancer research. Cancer Discov. 2014, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sachs, N.; Clevers, H. Organoid cultures for the analysis of cancer phenotypes. Curr. Opin. Genet. Dev. 2014, 24, 68–73. [Google Scholar] [CrossRef]
- Ben-David, U.; Beroukhim, R.; Golub, T.R. Genomic evolution of cancer models: Perils and opportunities. Nat. Rev. Cancer 2019, 19, 97–109. [Google Scholar] [CrossRef]
- Dugger, S.A.; Platt, A.; Goldstein, D.B. Drug development in the era of precision medicine. Nat. Rev. Drug Discov. 2018, 17, 183–196. [Google Scholar] [CrossRef] [PubMed]
- Dhandapani, M.; Goldman, A. Preclinical Cancer Models and Biomarkers for Drug Development: New Technologies and Emerging Tools. J. Mol. Biomark. Diagn. 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gey, G.O.; Coffmann, W.D.; Kubicek, M.T. Tissue culture studies of the proliferative capacity of cervical carcinoma and normal epithelium. Cancer Res. 1952, 12, 264–265. [Google Scholar]
- Masters, J.R.W. Human cancer cell lines: Fact and fantasy. Nat. Rev. Mol. Cell Biol. 2000, 1, 233–236. [Google Scholar] [CrossRef]
- Kapałczyńska, M.; Kolenda, T.; Przybyła, W.; Zajączkowska, M.; Teresiak, A.; Filas, V.; Ibbs, M.; Bliźniak, R.; Łuczewski, Ł.; Lamperska, K. 2D and 3D cell cultures—A comparison of different types of cancer cell cultures. Arch. Med. Sci. 2018, 14. [Google Scholar] [CrossRef] [PubMed]
- Pampaloni, F.; Reynaud, E.G.; Stelzer, E.H.K. The third dimension bridges the gap between cell culture and live tissue. Nat. Rev. Mol. Cell Biol. 2007, 8, 839–845. [Google Scholar] [CrossRef] [PubMed]
- Hamburger, A.W.; Salmon, S.E. Primary bioassay of human tumor stem cells. Science 1977, 197. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, J.; Nakazawa, K. Orderly arrangement of hepatocyte spheroids on a microfabricated chip. Tissue Eng. 2005, 11, 1254–1262. [Google Scholar] [CrossRef]
- Desroches, B.R.; Zhang, P.; Choi, B.R.; King, M.E.; Maldonado, A.E.; Li, W.; Rago, A.; Liu, G.; Nath, N.; Hartmann, K.M.; et al. Functional scaffold-free 3-D cardiac microtissues: A novel model for the investigation of heart cells. Am. J. Physiol. Heart Circ. Physiol. 2012, 302. [Google Scholar] [CrossRef] [Green Version]
- Achilli, T.M.; Meyer, J.; Morgan, J.R. Advances in the formation, use and understanding of multi-cellular spheroids. Exp. Opin. Biol. Ther. 2012, 12, 1347–1360. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Cuddihy, M.J.; Kotov, N.A. Three-dimensional cell culture matrices: State of the art. Tissue Eng. Part B Rev. 2008, 14, 61–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schilsky, R.L. Personalized medicine in oncology: The future is now. Nat. Rev. Drug Discov. 2010, 9, 363–366. [Google Scholar] [CrossRef]
- Karlsson, H.; Fryknäs, M.; Larsson, R.; Nygren, P. Loss of cancer drug activity in colon cancer HCT-116 cells during spheroid formation in a new 3-D spheroid cell culture system. Exp. Cell Res. 2012, 318. [Google Scholar] [CrossRef]
- Lai, Y.; Wei, X.; Lin, S.; Qin, L.; Cheng, L.; Li, P. Current status and perspectives of patient-derived xenograft models in cancer research. J. Hematol. Oncol. 2017, 10, 106. [Google Scholar] [CrossRef]
- Chdiwa, T.; Kawai, K.; Noguchi, A.; Sato, H.; Hayashi, A.; Cho, H.; Shiozawa, M.; Kishida, T.; Morinaga, S.; Yokose, T.; et al. Establishment of patient-derived cancer xenografts in immunodeficient NOG mice. Int. J. Oncol. 2015, 47. [Google Scholar] [CrossRef] [Green Version]
- Jhan, J.R.; Andrechek, E.R. Effective personalized therapy for breast cancer based on predictions of cell signaling pathway activation from gene expression analysis. Oncogene 2017, 36. [Google Scholar] [CrossRef] [PubMed]
- Byrne, A.T.; Alférez, D.G.; Amant, F.; Annibali, D.; Arribas, J.; Biankin, A.V.; Bruna, A.; Budinská, E.; Caldas, C.; Chang, D.K.; et al. Interrogating open issues in cancer precision medicine with patient-derived xenografts. Nat. Rev. Cancer 2017, 17, 254–268. [Google Scholar] [CrossRef]
- Lupo, B.; Sassi, F.; Pinnelli, M.; Galimi, F.; Zanella, E.R.; Vurchio, V.; Migliardi, G.; Gagliardi, P.A.; Puliafito, A.; Manganaro, D.; et al. Colorectal cancer residual disease at maximal response to EGFR blockade displays a druggable Paneth cell–like phenotype. Sci. Transl. Med. 2020, 12, eaax8313. [Google Scholar] [CrossRef]
- Bertotti, A.; Papp, E.; Jones, S.; Adleff, V.; Anagnostou, V.; Lupo, B.; Sausen, M.; Phallen, J.; Hruban, C.A.; Tokheim, C.; et al. The genomic landscape of response to EGFR blockade in colorectal cancer. Nature 2015, 526. [Google Scholar] [CrossRef]
- Bertotti, A.; Migliardi, G.; Galimi, F.; Sassi, F.; Torti, D.; Isella, C.; Corà, D.; di Nicolantonio, F.; Buscarino, M.; Petti, C.; et al. A molecularly annotated platform of patient- derived xenografts (“xenopatients”) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov. 2011, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lazzari, L.; Corti, G.; Picco, G.; Isella, C.; Montone, M.; Arcela, P.; Durinikova, E.; Zanella, E.R.; Novara, L.; Barbosa, F.; et al. Patient-derived xenografts and matched cell lines identify pharmacogenomic vulnerabilities in colorectal cancer. Clin. Cancer Res. 2019, 25. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Sun, L.; Liu, M.; Mao, Y. Patient-derived organoids: A promising model for personalized cancer treatment. Gastroenterol. Rep. 2018, 6, 243–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vlachogiannis, G.; Hedayat, S.; Vatsiou, A.; Jamin, Y.; Fernández-Mateos, J.; Khan, K.; Lampis, A.; Eason, K.; Huntingford, I.; Burke, R.; et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 2018, 359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weeber, F.; van de Wetering, M.; Hoogstraat, M.; Dijkstra, K.K.; Krijgsman, O.; Kuilman, T.; Gadellaa-Van Hooijdonk, C.G.M.; van der Velden, D.L.; Peeper, D.S.; Cuppen, E.P.J.G.; et al. Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. Proc. Natl. Acad. Sci. USA 2015, 112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van de Wetering, M.; Francies, H.E.; Francis, J.M.; Bounova, G.; Iorio, F.; Pronk, A.; van Houdt, W.; van Gorp, J.; Taylor-Weiner, A.; Kester, L.; et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015, 161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boj, S.F.; Hwang, C.I.; Baker, L.A.; Chio, I.I.C.; Engle, D.D.; Corbo, V.; Jager, M.; Ponz-Sarvise, M.; Tiriac, H.; Spector, M.S.; et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 2015, 160. [Google Scholar] [CrossRef] [Green Version]
- Gao, D.; Vela, I.; Sboner, A.; Iaquinta, P.J.; Karthaus, W.R.; Gopalan, A.; Dowling, C.; Wanjala, J.N.; Undvall, E.A.; Arora, V.K.; et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 2014, 159. [Google Scholar] [CrossRef]
- Lee, S.H.; Hu, W.; Matulay, J.T.; Silva, M.V.; Owczarek, T.B.; Kim, K.; Chua, C.W.; Barlow, L.M.J.; Kandoth, C.; Williams, A.B.; et al. Tumor evolution and drug response in patient-derived organoid models of bladder cancer. Cell 2018, 173. [Google Scholar] [CrossRef] [Green Version]
- Sachs, N.; de Ligt, J.; Kopper, O.; Gogola, E.; Bounova, G.; Weeber, F.; Balgobind, A.V.; Wind, K.; Gracanin, A.; Begthel, H.; et al. A Living Biobank of Breast Cancer Organoids Captures Disease Heterogeneity. Cell 2018, 172. [Google Scholar] [CrossRef] [Green Version]
- Hubert, C.G.; Rivera, M.; Spangler, L.C.; Wu, Q.; Mack, S.C.; Prager, B.C.; Couce, M.; McLendon, R.E.; Sloan, A.E.; Rich, J.N. A three-dimensional organoid culture system derived from human glioblastomas recapitulates the hypoxic gradients and cancer stem cell heterogeneity of tumors found in vivo. Cancer Res. 2016, 76. [Google Scholar] [CrossRef] [Green Version]
- Nelson, L.; Tighe, A.; Golder, A.; Littler, S.; Bakker, B.; Moralli, D.; Murtuza Baker, S.; Donaldson, I.J.; Spierings, D.C.J.; Wardenaar, R.; et al. A living biobank of ovarian cancer ex vivo models reveals profound mitotic heterogeneity. Nat. Commun. 2020, 11. [Google Scholar] [CrossRef]
- Aboulkheyr Es, H.; Montazeri, L.; Aref, A.R.; Vosough, M.; Baharvand, H. Personalized Cancer Medicine: An Organoid Approach. Trends Biotechnol. 2018, 36, 358–371. [Google Scholar] [CrossRef]
| Name | Description | Website | Reference |
|---|---|---|---|
| CaSNP | CaSNP performs quantitative analysis of copy number variation from SNP arrays in multiple cancer types | https://bioinformaticshome.com/tools/cnv/descriptions/CaSNP.html | [46] |
| OncoLand | OncoLand provides oncology data access in sample and gene directions. | https://omicsoftdocs.github.io/ArraySuiteDoc/tutorials/OncoLand/Introduction/ | [47] |
| AGCOH | The Atlas of Genetics, Cytogenetics in Oncology and Hematology perform comprehensive genomic characterization and analysis of multiple cancer types | http://atlasgeneticsoncology.org/BackpageAbout.html | [48] |
| PCWAG | PCWAG—Pan-cancer Analysis of Whole Genomes provides common patterns of mutations from more than 2600 cancer whole genomes | http://dcc.icgc.org/pcawg | [4] |
| ChiTaRS | ChiTaRS contains chimeric transcripts and RNA-Seq data | http://chitars.bioinfo.cnio.es/ | [49] |
| CanSAR | CanSAR provides information about translational research and drug discovery knowledgebase | https://cansarblack.icr.ac.uk/ | [50] |
| OncoDB.HCC | Oncogenomics Database of Hepatocellular Carcinoma provides genomic, transcriptomic, and proteomic data | http://oncodb.hcc.ibms.sinica.edu.tw/index.htm | [51] |
| COSMIC | COSMIC performs a comprehensive database of somatic mutation in multiple cancer types | https://cancer.sanger.ac.uk/cosmic | [52] |
| canEvolve | canEvolve is a comprehensive database including genes, miRNA, and protein expression profiles; copy number changes for a variety of cancer types and protein–protein interactions | http://www.canevolve.org/AnalysisResults/AnalysisResults.html | [53] |
| CancerPPD | CancerPPD provides information about anticancer peptides and proteins in multiple cancer types | http://crdd.osdd.net/raghava/cancerppd/ | [54] |
| PED | The Pancreatic Expression Database performs a comprehensive meta-analysis of pancreatic cancer | http://www.pancreasexpression.org/ | [55] |
| CGP | Cancer Genome Project provides genotype and copy number changes information in tumors | https://www.sanger.ac.uk/group/cancer-genome-project | [56] |
| MethyCancer | MethyCancer provides information about DNA methylation and gene expression in a variety of cancer types | http://methycancer.psych.ac.cn/ | [57] |
| CPTAC | Clinical Proteomic Tumor Analysis Consortium is a database containing an integration of genomic and proteomic data | https://proteomics.cancer.gov/ | [58] |
| intOGen | Integrative Onco Genomics performs comprehensive genomic data of multiple cancer types | https://www.intogen.org/search | [59] |
| ArrayExpress | ArrayExpress focuses on microarray gene expression data | https://www.ebi.ac.uk/arrayexpress/ | [60] |
| DriverDBv3 | DriverDBv3 is a database of cancer omics | http://driverdb.tms.cmu.edu.tw/ | [61] |
| PCDB | The Pancreatic Cancer Database provides genetic information in pancreatic cancer | http://www.pancreaticcancerdatabase.org | [62] |
| CancerDR | CancerDR contains anticancer drugs and their effectiveness against a variety of cell lines | http://crdd.osdd.net/raghava/cancerdr/ | [63] |
| Platinum | Platinum provides knowledge about missense mutations on ligand–proteome interactions | http://biosig.unimelb.edu.au/platinum/ | [64] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yalcin, G.D.; Danisik, N.; Baygin, R.C.; Acar, A. Systems Biology and Experimental Model Systems of Cancer. J. Pers. Med. 2020, 10, 180. https://doi.org/10.3390/jpm10040180
Yalcin GD, Danisik N, Baygin RC, Acar A. Systems Biology and Experimental Model Systems of Cancer. Journal of Personalized Medicine. 2020; 10(4):180. https://doi.org/10.3390/jpm10040180
Chicago/Turabian StyleYalcin, Gizem Damla, Nurseda Danisik, Rana Can Baygin, and Ahmet Acar. 2020. "Systems Biology and Experimental Model Systems of Cancer" Journal of Personalized Medicine 10, no. 4: 180. https://doi.org/10.3390/jpm10040180
APA StyleYalcin, G. D., Danisik, N., Baygin, R. C., & Acar, A. (2020). Systems Biology and Experimental Model Systems of Cancer. Journal of Personalized Medicine, 10(4), 180. https://doi.org/10.3390/jpm10040180





