Impact of Single-Nucleotide Polymorphisms of CTLA-4, CD80 and CD86 on the Effectiveness of Abatacept in Patients with Rheumatoid Arthritis
Abstract
1. Introduction
2. Material and Methods
2.1. Study Design
2.2. Ethics Statements
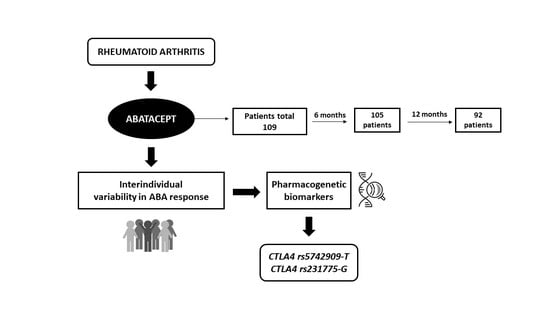
2.3. Study Population
2.4. Sociodemographic and Clinical Variables
2.5. Genetic Variables
2.5.1. DNA Isolation
2.5.2. Detection of Gene Polymorphisms
2.6. Response Variables
2.7. Statistical Analysis
3. Results
3.1. Patient Characteristics
3.2. Clinical Effectiveness of ABA
3.3. Genotype Distribution
3.4. ABA Response Predictors at 6 Months
3.4.1. EULAR Response
3.4.2. Low Disease Activity (LDA)
3.4.3. Remission
3.5. ABA Response Predictors at 12 Months
3.5.1. EULAR Response
3.5.2. Low Disease Activity (LDA)
3.5.3. Remission
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ABA | Abatacept |
| ACPA | anti-cyclic citrullinated peptide antibodies |
| ACR | American College of Rheumatology |
| bDMARDs | Biologic disease-modifying antirheumatic drugs |
| BT | biological therapy |
| CRP | C-reactive protein |
| csDMARDs | conventional synthetic disease-modifying antirheumatic drugs |
| CTLA-4 | cytotoxic T-lymphocyte-associated antigen 4 |
| DAS28 | 28-joints Disease Activity Score |
| DMARDs | disease-modifying antirheumatic drugs |
| ESR | erythrocyte sedimentation rate |
| EULAR | European League Against Rheumatism |
| GC | glucocorticoid |
| HAQ | Health Assessment Questionnaire score |
| IV | intravenous |
| LDA | Low-activity disease |
| LFN | leflunomide |
| MTX | methotrexate |
| NIJ | number of inflamed joints |
| NPJ | number of painful joints |
| PVAS | patient’s visual analogue scale |
| RA | rheumatoid arthritis |
| RF | rheumatoid factor |
| SC | subcutaneous |
| TNFi | tumor necrosis factor inhibitor |
| tsDMARDs | targeted synthetic disease-modifying antirheumatic drugs |
References
- Soubrier, M.; Lahaye, C.; Tatar, Z. Abatacept for treatment of rheumatoid arthritis: Special focus on the elderly. Drugs Aging 2018, 35, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Carmona, L.; Villaverde, V.; Hernández-García, C.; Ballina, J.; Gabriel, R.; Laffon, A. The prevalence of rheumatoid arthritis in the general population of Spain. Rheumatology 2002, 41, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Croft, P.; Silman, A.; Hochberg, M. Epidemiology of the Rheumatic Diseases; Oxford Medical Publications: Oxford, UK, 1993. [Google Scholar]
- Carbonell, J.; Cobo, T.; Balsa, A.; Descalzo, M.A.; Carmona, L.; SERAP Study Group. The incidence of rheumatoid arthritis in Spain: Results from a nationwide primary care registry. Rheumatology 2008, 47, 1088–1092. [Google Scholar] [CrossRef] [PubMed]
- Alhambra-Expósito, M.R.; Arjonilla-Sampedro, M.E.; Molina-Puerta, M.J.; Tenorio-Jiménez, C.; Manzano-García, G.; Moreno-Moreno, P.; Benito-López, P. Recomendaciones dietéticas en la artritis reumatoide. Rev. Española De Nutr. Hum. Y Dietética 2013, 17, 165–171. [Google Scholar]
- Smolen, J.S.; Aletaha, D.; McInnes, I.B. Rheumatoid arthritis. Lancet 2016, 388, 2023–2038. [Google Scholar] [CrossRef]
- Smolen, J.S.; Landewe, R.B.M.; Bijlsma, J.W.J.; Burmester, G.R.; Dougados, M.; Kerschbaumer, A.; McInnes, I.B.; Sepriano, A.; van Vollenhoven, R.F.; de Wit, M.; et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2019 update. Ann. Rheum. Dis. 2020, 79, 685–699. [Google Scholar] [CrossRef] [PubMed]
- Blair, H.A.; Deeks, E.D. Abatacept: A review in rheumatoid arthritis. Drugs 2017, 77, 1221–1233. [Google Scholar] [CrossRef]
- Fu, J.; van Wietmarschen, H.A.; van der Kooij, A.; Cuppen, B.V.J.; Schroën, Y.; Marijnissen, A.K.; Meulman, J.J.; Lafeber, F.P.J.G.; van der Greef, J. Systems approach for classifying the response to biological therapies in patients with rheumatoid arthritis in clinical practice. Eur. J. Integr. Med. 2018, 19, 65–71. [Google Scholar] [CrossRef]
- Morales, A.J.; Maldonado-Montoro, M.; de la Plata, J.E.M.; Ramírez, C.P.; Daddaoua, A.; Payer, C.A.; Ruiz, M.E.; Collado, C.G. FCGR2A/FCGR3A gene polymorphisms and clinical variables as predictors of response to tocilizumab and rituximab in patients with rheumatoid arthritis. J. Clin. Pharm. 2019, 59, 517–531. [Google Scholar] [CrossRef] [PubMed]
- Maldonado-Montoro, M.; Cañadas-Garre, M.; González-Utrilla, A.; Plaza-Plaza, J.C.; Calleja-Hernández, M.Ÿ. Genetic and clinical biomarkers of tocilizumab response in patients with rheumatoid arthritis. Pharm. Res. 2016, 111, 264–271. [Google Scholar] [CrossRef]
- Cañete, J.D.; Suárez, B.; Hernández, M.V.; Sanmartí, R.; Rego, I.; Celis, R.; Moll, C.; Pinto, J.A.; Blanco, F.J.; Lozano, F. Influence of variants of Fc gamma receptors IIA and IIIA on the American College of Rheumatology and European League Against Rheumatism responses to anti-tumour necrosis factor alpha therapy in rheumatoid arthritis. Ann. Rheum. Dis. 2009, 68, 1547–1552. [Google Scholar] [CrossRef] [PubMed]
- Bek, S.; Bojesen, A.; Nielsen, J.; Sode, J.; Bank, S.; Vogel, U.; Andersen, V. Systematic review and meta-analysis: Pharmacogenetics of anti-TNF treatment response in rheumatoid arthritis. Pharm. J. 2017, 17, 403–411. [Google Scholar] [CrossRef]
- Maldonado-Montoro, M.; Cañadas-Garre, M.; González-Utrilla, A.; Calleja-Hernández, M.Á. Influence of IL6R gene polymorphisms in the effectiveness to treatment with tocilizumab in rheumatoid arthritis. Pharm. J. 2018, 18, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Kavvoura, F.K.; Akamizu, T.; Awata, T.; Ban, Y.; Chistiakov, D.A.; Frydecka, I.; Ghaderi, A.; Gough, S.C.; Hiromatsu, Y.; Ploski, R.; et al. Cytotoxic T-lymphocyte associated antigen 4 gene polymorphisms and autoimmune thyroid disease: A meta-analysis. J. Clin. Endocrinol. Metab. 2007, 92, 3162–3170. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, A.M.; Renzoni, E.A.; Anevlavis, S.; Lagan, A.L.; Munkonge, F.M.; Fonseca, C.; Black, C.M.; Briggs, D.; Wells, A.U.; Marshall, S.E.; et al. A polymorphism in the promoter region of the CD86 (B7.2) gene is associated with systemic sclerosis. Int. J. Immunogenet. 2006, 33, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.-L.; Chen, R.-H.; Lin, H.-J.; Liu, Y.-H.; Chen, W.-C.; Tsai, Y.; Wan, L.; Tsai, F.-J. The association between polymorphisms of B7 molecules (CD80 and CD86) and Graves’ ophthalmopathy in a Taiwanese population. Ophthalmology 2011, 118, 553–557. [Google Scholar] [CrossRef]
- Talotta, R.; Bagnato, G.L.; Atzeni, F.; Ditto, M.C.; Bitto, A.; Squadrito, F.; Lo Gullo, A.; Sarzi-Puttini, P.; Bagnato, G.F. Polymorphic alleles in exon 1 of the CTLA4 gene do not predict the response to abatacept. Clin. Exp. Rheumatol. 2013, 31, 813. [Google Scholar]
- Wu, P.; Wang, Z.; Lu, S.; Zhao, X. CD86 +1057G/A polymorphism and risk of chronic immune thrombocytopenia. Autoimmunity 2014, 47, 482–485. [Google Scholar] [CrossRef]
- Jellis, C.L.; Wang, S.S.; Rennert, P.; Borriello, F.; Sharpe, A.H.; Green, N.R.; Gray, G.S. Genomic organization of the gene coding for the costimulatory human B-lymphocyte antigen B7-2 (CD86). Immunogenetics 1995, 42, 85–89. [Google Scholar] [CrossRef]
- Pawlak, E.; Karabon, L.; Wlodarska-Polinska, I.; Jedynak, A.; Jonkisz, A.; Tomkiewicz, A.; Kornafel, J.; Stepien, M.; Ignatowicz, A.; Lebioda, A. Influence of CTLA-4/CD28/ICOS gene polymorphisms on the susceptibility to cervical squamous cell carcinoma and stage of differentiation in the Polish population. Hum. Immunol. 2010, 71, 195–200. [Google Scholar] [CrossRef]
- Kailashiya, V.; Barun Sharma, H.; Kailashiya, J. Role of CTLA4 A49G polymorphism in systemic lupus erythematosus and its geographical distribution. J. Clin. Pathol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Villalobos, E.F.; Carrillo-Ballesteros, F.J.; Munoz-Valle, J.F.; Palafox-Sanchez, C.A.; Valle, Y.; Orozco-Barocio, G.; Oregon-Romero, E. Association of CD28 and CTLA4 haplotypes with susceptibility to primary Sjogren’s syndrome in Mexican population. J. Clin. Lab. Anal. 2019, 33, e22620. [Google Scholar] [CrossRef] [PubMed]
- Schildberg, F.A.; Klein, S.R.; Freeman, G.J.; Sharpe, A.H. Coinhibitory pathways in the B7-CD28 ligand-receptor family. Immunity 2016, 44, 955–972. [Google Scholar] [CrossRef] [PubMed]
- Valk, E.; Rudd, C.E.; Schneider, H. CTLA-4 trafficking and surface expression. Trends Immunol. 2008, 29, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Choi, S.J.; Ji, J.D.; Song, G.G. CTLA-4 and TNF-α promoter-308 A/G polymorphisms and ANCA-associated vasculitis susceptibility: A meta-analysis. Mol. Biol. Rep. 2012, 39, 319–326. [Google Scholar] [CrossRef]
- Daikh, K.J.; Daikh, D.I. CTLA-4: A key regulatory point in the control of autoimmune disease. Immunol. Rev. 2008, 223, 143–155. [Google Scholar]
- Li, F.; Ma, X.; Du, L.; Shi, L.; Cao, Q.; Li, N.; Pang, T.; Liu, Y.; Kijlstra, A.; Yang, P. Identification of susceptibility SNPs in CTLA-4 and PTPN22 for scleritis in Han Chinese. Clin. Exp. Immunol. 2019, 197, 230–236. [Google Scholar] [CrossRef]
- Felson, D.T.; Anderson, J.J. Methodological and statistical approaches to criteria development in rheumatic diseases. Bailliere’s Clin. Rheumatol. 1995, 9, 253–266. [Google Scholar] [CrossRef]
- Wells, G.; Becker, J.; Teng, J.; Dougados, M.; Schiff, M.; Smolen, J.; Aletaha, D.; van Riel, P. Validation of the 28-joint Disease Activity Score (DAS28) and European League Against Rheumatism response criteria based on C-reactive protein against disease progression in patients with rheumatoid arthritis, and comparison with the DAS28 based on erythrocyte sedimentation rate. Ann. Rheum. Dis. 2009, 68, 954–960. [Google Scholar]
- Prevoo, M.; Hof, M.A.V.; Kuper, H.; van Leeuwen, M.; van de Putte, L.; van Riel, P. Modified disease activity scores that include twenty-eight-joint counts development and validation in a prospective longitudinal study of patients with rheumatoid arthritis. Arthritis Rheum. Off. J. Am. Coll. Rheumatol. 1995, 38, 44–48. [Google Scholar] [CrossRef]
- Van Gestel, A.M.; Anderson, J.J.; van Riel, P.L.; Boers, M.; Haagsma, C.J.; Rich, B.; Wells, G.; Lange, M.L.; Felson, D.T. ACR and EULAR improvement criteria have comparable validity in rheumatoid arthritis trials. American College of Rheumatology European League of Associations for Rheumatology. J. Rheumatol. 1999, 26, 705–711. [Google Scholar] [PubMed]
- Singh, J.A.; Saag, K.G.; Bridges, S.L., Jr.; Akl, E.A.; Bannuru, R.R.; Sullivan, M.C.; Vaysbrot, E.; McNaughton, C.; Osani, M.; Shmerling, R.H.; et al. 2015 American College of Rheumatology Guideline for the Treatment of Rheumatoid Arthritis. Arthritis Rheumatol. 2016, 68, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Felson, D.T.; Smolen, J.S.; Wells, G.; Zhang, B.; van Tuyl, L.H.D.; Funovits, J.; Aletaha, D.; Allaart, C.F.; Bathon, J.; Bombardieri, S.; et al. American College of Rheumatology/European League Against Rheumatism provisional definition of remission in rheumatoid arthritis for clinical trials. Arthritis Rheum. 2011, 63, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Faul, F.; Erdfelder, E.; Lang, A.-G.; Buchner, A. G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behavior Res. Methods. 2007, 39, 175–191. [Google Scholar] [CrossRef] [PubMed]
- RC Team. R: A Language and Environment for Statistical Computing; RC Team: Vienna, Austria, 2013. [Google Scholar]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Zhai, C.; Li, S.; Feng, W.; Shi, W.; Wang, J.; Wang, Q.; Chai, L.; Zhang, Q.; Yan, X.; Li, M. Association of interleukin-17a rs2275913 gene polymorphism and asthma risk: A meta-analysis. Arch. Med. Sci. AMS 2018, 14, 1204. [Google Scholar] [CrossRef]
- Rentería, M.E.; Cortes, A.; Medland, S.E. Using PLINK for genome-wide association studies (GWAS) and data analysis. In Genome-Wide Association Studies and Genomic Prediction; Humana Press: Totowa, NJ, USA, 2013; pp. 193–213. [Google Scholar] [CrossRef]
- Solé, X.; Guinó, E.; Valls, J.; Iniesta, R.; Moreno, V. SNPStats: A web tool for the analysis of association studies. Bioinformatics 2006, 22, 1928–1929. [Google Scholar] [CrossRef]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef]
- Alyousef, Y.M.; Borgio, J.F.; AbdulAzeez, S.; Al-Masoud, N.; Al-Ali, A.A.; Al-Shwaimi, E.; Al-Ali, A.K. Association of MBL2 gene polymorphism with dental caries in Saudi children. Caries Res. 2017, 51, 12–16. [Google Scholar] [CrossRef]
- AbdulAzeez, S.; Al-Nafie, A.N.; Al-Shehri, A.; Borgio, J.F.; Baranova, E.V.; Al-Madan, M.S.; Al-Ali, R.A.; Al-Muhanna, F.; Al-Ali, A.; Al-Mansori, M. Intronic polymorphisms in the CDKN2B-AS1 gene are strongly associated with the risk of myocardial infarction and coronary artery disease in the Saudi population. Int. J. Mol. Sci. 2016, 17, 395. [Google Scholar] [CrossRef] [PubMed]
- Al Asoom, L.I.; Alsuwat, H.S.; Rafique, N.; al Makhaita, M.; Alamoudi, W.; AbdulAzeez, S.; Borgio, J.F. Functional DNA variations associated with Saudi female with low VO2max: A pilot microarray study. Am. J. Transl. Res. 2019, 11, 3659. [Google Scholar] [PubMed]
- Cagnotto, G.; Willim, M.; Nilsson, J.A.; Compagno, M.; Jacobsson, L.T.H.; Saevarsdottir, S.; Turesson, C. Abatacept in rheumatoid arthritis: Survival on drug, clinical outcomes, and their predictors-data from a large national quality register. Arthritis Res. 2020, 22, 15. [Google Scholar] [CrossRef] [PubMed]
- Alten, R.; Mariette, X.; Lorenz, H.M.; Galeazzi, M.; Cantagrel, A.; Nüßlein, H.G.; Chartier, M.; Elbez, Y.; Rauch, C.; le Bars, M. Real-world predictors of 12-month intravenous abatacept retention in patients with rheumatoid arthritis in the ACTION observational study. RMD Open 2017, 3, e000538. [Google Scholar] [CrossRef] [PubMed]
- Kremer, J.M.; Genant, H.K.; Moreland, L.W.; Russell, A.S.; Emery, P.; Abud-Mendoza, C.; Szechinski, J.; Li, T.; Ge, Z.; Becker, J.C.; et al. Effects of abatacept in patients with methotrexate-resistant active rheumatoid arthritis: A randomized trial. Ann. Intern. Med. 2006, 144, 865–876. [Google Scholar] [CrossRef]
- Schiff, M.; Keiserman, M.; Codding, C.; Songcharoen, S.; Berman, A.; Nayiager, S.; Saldate, C.; Aranda, R.; Becker, J.C.; Nys, M.; et al. Clinical response and tolerability to abatacept in patients with rheumatoid arthritis previously treated with infliximab or abatacept: Open-label extension of the ATTEST study. Ann. Rheum. Dis. 2011, 70, 2003–2007. [Google Scholar] [CrossRef]
- Takahashi, N.; Kojima, T.; Kaneko, A.; Kida, D.; Hirano, Y.; Fujibayashi, T.; Yabe, Y.; Takagi, H.; Oguchi, T.; Miyake, H.; et al. Longterm efficacy and safety of abatacept in patients with rheumatoid arthritis treated in routine clinical practice: Effect of concomitant methotrexate after 24 weeks. J. Rheumatol. 2015, 42, 786–793. [Google Scholar] [CrossRef]
- Kubo, S.; Nakayamada, S.; Nakano, K.; Sawamukai, N.; Hirata, S.; Hanami, K.; Saito, K.; Tanaka, Y. Comparison of efficacy of TNF inhibitors and abatacept in patients with rheumatoid arthritis; Adjusted with propensity score matching. Clin. Immunol. 2018, 191, 67–74. [Google Scholar] [CrossRef]
- Gottenberg, J.E.; Ravaud, P.; Cantagrel, A.; Combe, B.; Flipo, R.M.; Schaeverbeke, T.; Houvenagel, E.; Gaudin, P.; Loeuille, D.; Rist, S.; et al. Positivity for anti-cyclic citrullinated peptide is associated with a better response to abatacept: Data from the ‘Orencia and Rheumatoid Arthritis’ registry. Ann. Rheum. Dis. 2012, 71, 1815–1819. [Google Scholar] [CrossRef]
- Gazeau, P.; Alegria, G.C.; Devauchelle-Pensec, V.; Jamin, C.; Lemerle, J.; Bendaoud, B.; Brooks, W.H.; Saraux, A.; Cornec, D.; Renaudineau, Y. Memory B cells and response to abatacept in rheumatoid arthritis. Clin. Rev. Allergy Immunol. 2017, 53, 166–176. [Google Scholar] [CrossRef]
- AlFadhli, S. Overexpression and secretion of the soluble CTLA-4 splice variant in various autoimmune diseases and in cases with overlapping autoimmunity. Genet. Test. Mol. Biomark. 2013, 17, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Giscombe, R.; Wang, X.; Huang, D.; Lefvert, A.K. Coding sequence 1 and promoter single nucleotide polymorphisms in the CTLA-4 gene in Wegener’s granulomatosis. J. Rheumatol. 2002, 29, 950–953. [Google Scholar] [PubMed]
- Balbi, G.; Ferrera, F.; Rizzi, M.; Piccioli, P.; Morabito, A.; Cardamone, L.; Ghio, M.; Palmisano, G.; Carrara, P.; Pedemonte, S. Association of− 318 C/T and+ 49 A/G cytotoxic T lymphocyte antigen-4 (CTLA-4) gene polymorphisms with a clinical subset of Italian patients with systemic sclerosis. Clin. Exp. Immunol. 2007, 149, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhao, X.; Giscombe, R.; Lefvert, A. A CTLA-4 gene polymorphism at position− 318 in the promoter region affects the expression of protein. Genes Immun. 2002, 3, 233–234. [Google Scholar] [CrossRef]
- Abdulqader, A.R.; Mohammed, A.I.; Rachid, S. Polymorphisms in the cytotoxic T lymphocyte-associated protein-4 immune regulatory gene and their impact on inhibitor development in patients with hemophilia A. J. Int. Med. Res. 2019, 47, 4981–4992. [Google Scholar] [CrossRef]
- Liu, M.F.; Wang, C.R.; Chen, P.C.; Fung, L.L. Increased expression of soluble cytotoxic T-lymphocyte-associated antigen-4 molecule in patients with systemic lupus erythematosus. Scand. J. Immunol. 2003, 57, 568–572. [Google Scholar] [CrossRef]
- Braun, J.; Donner, H.; Siegmund, T.; Walfish, P.; Usadel, K.; Badenhoop, K. CTLA-4 promoter variants in patients with Graves’ disease and Hashimoto’s thyroiditis. Tissue Antigens 1998, 51, 563–566. [Google Scholar] [CrossRef]
- Torres-Carrillo, N.; Ontiveros-Mercado, H.; Torres-Carrillo, N.M.; Parra-Rojas, I.; Rangel-Villalobos, H.; Ramírez-Dueñas, M.G.; Gutiérrez-Ureña, S.R.; Valle, Y.; Muñoz-Valle, J.F. The− 319C/+ 49G/CT60G haplotype of CTLA-4 gene confers susceptibility to rheumatoid arthritis in Mexican population. Cell Biochem. Biophys. 2013, 67, 1217–1228. [Google Scholar] [CrossRef]
- Harper, K.; Balzano, C.; Rouvier, E.; Mattéi, M.-G.; Luciani, M.; Golstein, P. CTLA-4 and CD28 activated lymphocyte molecules are closely related in both mouse and human as to sequence, message expression, gene structure, and chromosomal location. J. Immunol. 1991, 147, 1037–1044. [Google Scholar]
- Ueda, H.; Howson, J.M.; Esposito, L.; Heward, J.; Chamberlain, G.; Rainbow, D.B.; Hunter, K.M.; Smith, A.N.; di Genova, G.; Herr, M.H. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature 2003, 423, 506–511. [Google Scholar] [CrossRef]
- Repnik, K.; Potočnik, U. CTLA4 CT60 single-nucleotide polymorphism is associated with Slovenian inflammatory bowel disease patients and regulates expression of CTLA4 isoforms. DNA Cell Biol. 2010, 29, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Downie-Doyle, S.; Bayat, N.; Rischmueller, M.; Lester, S. Influence of CTLA4 haplotypes on susceptibility and some extraglandular manifestations in primary Sjögren’s syndrome. Arthritis Rheum. Off. J. Am. Coll. Rheumatol. 2006, 54, 2434–2440. [Google Scholar] [CrossRef] [PubMed]
- Nisticò, L.; Buzzetti, R.; Pritchard, L.E.; van der Auwera, B.; Giovannini, C.; Bosi, E.; Larrad, M.T.M.; Rios, M.S.; Chow, C.; Cockram, C.S. The CTLA-4 gene region of chromosome 2q33 is linked to, and associated with, type 1 diabetes. Hum. Mol. Genet. 1996, 5, 1075–1080. [Google Scholar] [CrossRef] [PubMed]
- Anjos, S.; Nguyen, A.; Ounissi-Benkalha, H.; Tessier, M.-C.; Polychronakos, C. A common autoimmunity predisposing signal peptide variant of the cytotoxic T-lymphocyte antigen 4 results in inefficient glycosylation of the susceptibility allele. J. Biol. Chem. 2002, 277, 46478–46486. [Google Scholar] [CrossRef] [PubMed]
- Mäurer, M.; Loserth, S.; Kolb-Mäurer, A.; Ponath, A.; Wiese, S.; Kruse, N.; Rieckmann, P. A polymorphism in the human cytotoxic T-lymphocyte antigen 4 (CTLA4) gene (exon 1+ 49) alters T-cell activation. Immunogenetics 2002, 54, 1–8. [Google Scholar] [PubMed]
- Jonson, C.O.; Hedman, M.; Faresjo, M.K.; Casas, R.; Ilonen, J.; Ludvigsson, J.; Vaarala, O.; ABIS Study Group. The association of CTLA-4 and HLA class II autoimmune risk genotype with regulatory T cell marker expression in 5-year-old children. Clin. Exp. Immunol. 2006, 145, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Masterman, T.; Ligersa, A.; Zhangb, Z.; Hellgrenb, D.; Salterb, H.; Anvretb, M.; Hillerta, J. CTLA4 dimorphisms and the multiple sclerosis phenotype. J. Neuroimmunol. 2002, 131, 208–212. [Google Scholar] [CrossRef]
- Kouki, T.; Sawai, Y.; Gardine, C.A.; Fisfalen, M.-E.; Alegre, M.-L.; DeGroot, L.J. CTLA-4 gene polymorphism at position 49 in exon 1 reduces the inhibitory function of CTLA-4 and contributes to the pathogenesis of Graves’ disease. J. Immunol. 2000, 165, 6606–6611. [Google Scholar] [CrossRef]
- Lee, W.Y.; Chang, Y.H.; Lo, M.K.; Chang, C.P.; Yang, S.C.; Yang, T.P.; Ho, K.T.; Juan, C.W.; Shiau, M.Y. Polymorphisms of cytotoxic T lymphocyte-associated antigen-4 and cytokine genes in Taiwanese patients with ankylosing spondylitis. Tissue Antigens 2010, 75, 119–126. [Google Scholar] [CrossRef]
- Shojaa, M.; Aghaie, M.; Amoli, M.; Javid, N.; Shakeri, F.; Khashayar, P.; Tabarraei, A.; Keshtkar, A.A.; Joshaghani, H.R.; Kouroshnia, A. Association between 318 C/T polymorphism of the CTLA-4 gene and systemic lupus erythematosus in Iranian patients. Int. J. Rheum. Dis. 2017, 20, 2040–2044. [Google Scholar] [CrossRef]
- Muñoz-Valle, J.F.; Valle, Y.; Padilla-Gutiérrez, J.R.; Parra-Rojas, I.; Rangel-Villalobos, H.; del Mercado, M.V.; Ledezma-Lozano, I.Y.; Villafan-Bernal, J.R.; Armendáriz-Borunda, J.; Pereira-Suárez, A.L. The+ 49A> G CTLA-4 polymorphism is associated with rheumatoid arthritis in Mexican population. Clin. Chim. Acta 2010, 411, 725–728. [Google Scholar] [CrossRef] [PubMed]
- Slavik, J.M.; Hutchcroft, J.E.; Bierer, B.E. CD80 and CD86 are not equivalent in their ability to induce the tyrosine phosphorylation of CD28. J. Biol. Chem. 1999, 274, 3116–3124. [Google Scholar] [CrossRef] [PubMed]

| Variable | Baseline | ||
|---|---|---|---|
| N | % | Mean ± Standard Deviation | |
| Sex | |||
| Female | 78 | 71.56 | - |
| Male | 31 | 28.44 | - |
| Smoking | |||
| Smokers | 15 | 13.76 | - |
| Former-smokers | 11 | 10.09 | - |
| Non smokers | 83 | 76.15 | - |
| Age at RA diagnosis | 109 | - | 44.94 ± 14.46 |
| Disease duration (years) | 109 | - | 16 (8–22) |
| Age at ABA start | 109 | - | 56.37 ± 13.05 |
| Duration of ABA (months) | 109 | - | 28 (14–46) |
| ABA administration | |||
| Subcutaneous | 60 | 55.05 | - |
| Intravenous | 49 | 44.95 | - |
| Concomitant csDMARDs | |||
| Methotrexate | 38 | 34.86 | - |
| Leflunomide | 14 | 12.84 | - |
| Others | 2 | 1.83 | - |
| None | 55 | 50.45 | |
| Concomitant glucocorticoids | |||
| No | 18 | 16.51 | - |
| Yes | 91 | 83.49 | - |
| Monotherapy | |||
| No | 103 | 94.50 | - |
| Yes | 6 | 5.50 | - |
| Number of previous BTs | 109 | - | 2 (1–3) |
| Duration of previous BTs (months) | 109 | - | 36 (12–60) |
| Previous BTs | |||
| Bionaive | 16 | 14.68 | - |
| 1 TNFi | 29 | 26.61 | - |
| 2 TNFis | 31 | 28.44 | - |
| 3 or more TNFis | 33 | 30.28 | - |
| Suspension reason of ABA | |||
| Primary failure | 25 | 22.94 | - |
| Secondary failure | 9 | 8.26 | - |
| Adverse reaction | 6 | 5.50 | - |
| No suspension | 69 | 63.30 | - |
| Rheumatoid factor | |||
| Negative | 22 | 20.18 | - |
| Positive | 87 | 79.82 | - |
| ACPAs | |||
| Negative | 30 | 27.52 | - |
| Positive | 79 | 72.48 | - |
| DAS28 | 109 | - | 4.77 ± 1.43 |
| NPJ | 109 | - | 7 (3–10) |
| NIJ | 109 | - | 3 (1–6) |
| PVAS | 109 | - | 70 (50–80) |
| CRP | 109 | - | 2.42 (1.40–5.00) |
| ESR | 109 | - | 22 (10–38) |
| HAQ | 109 | - | 1.75 (1.25–2.00) |
| Non-Bionaive Patients | ||||
| Response variable | 6 months | 12 months | ||
| N | % | N | % | |
| EULAR response | ||||
| Satisfactory | 36 | 34.29 | 43 | 46.74 |
| Unsatisfactory | 69 | 65.71 | 49 | 53.26 |
| Remission (DAS28 < 2.6) | 18 | 17.14 | 28 | 30.43 |
| LDA (2.6 ≥ DAS28 ≥ 3.2) | 22 | 20.95 | 20 | 21.74 |
| ABA-Bionaive Patients | ||||
| Response variable | 6 months | 12 months | ||
| N | % | N | % | |
| EULAR response | ||||
| Satisfactory | 8 | 53.33 | 11 | 78.57 |
| Unsatisfactory | 7 | 46.67 | 3 | 21.43 |
| Remission (DAS28 < 2.6) | 3 | 20 | 9 | 64.29 |
| LDA (2.6 ≥ DAS28 ≥ 3.2) | 5 | 33.33 | 3 | 21.43 |
| Response Variable | Independent Variable | B | Odds Ratio | p-Value (Variable) | 95% Confidence INTERVAL | R2 | Goodness of Fit |
|---|---|---|---|---|---|---|---|
| 6 months | |||||||
| EULAR response | |||||||
| Duration of previous BTs (months) | −0.026 | 0.97 | 0.004 | 0.95–0.99 | R2 Cox Snell = 0.382 R2 Nagelkerke = 0.528 | X2 = 10.396 p = 0.238 | |
| PVAS | −0.052 | 0.95 | 0.003 | 0.91–0.98 | |||
| DAS28 | −0.651 | 0.52 | 0.015 | 0.30–0.87 | |||
| LDA | |||||||
| DAS28 | −0.367 | 0.69 | 0.032 | 0.49–0.96 | R2 Cox Snell = 0.045 R2 Nagelkerke = 0.070 | X2 = 7.062 p = 0.529 | |
| Remission | |||||||
| Duration of ABA (months) | 0.047 | 1.05 | 0.002 | 1.01–1.08 | R2 Cox Snell = 0.349 R2 Nagelkerke = 0.581 | X2 = 62.774 p < 0.001 | |
| Concomitant glucocorticoids | 1.993 | 7.34 | 0.031 | 1.33–54.79 | |||
| NPJ | −0.436 | 0.65 | 0.001 | 0.48–0.81 | |||
| ESR | −0.109 | 0.89 | 0.004 | 0.82–0.95 | |||
| 12 months | |||||||
| EULAR response | |||||||
| PVAS | −0.069 | 0.93 | <0.001 | 0.90–0.96 | R2 Cox Snell = 0.335 R2 Nagelkerke = 0.447 | X2 = 2.509 p = 0.961 | |
| CTLA-4 rs5742909 (T vs. CC) | 1.772 | 5.88 | 0.012 | 1.48–23.29 | |||
| CTLA-4 rs231775 (G vs. AA) | 1.247 | 3.48 | 0.022 | 1.20–10.09 | |||
| LDA | |||||||
| CTLA-4 rs5742909 (T vs. CC) | 1.556 | 4.75 | 0.016 | 1.35–17.94 | R2 Cox Snell = 0.117 R2 Nagelkerke = 0.180 | X2 = 0.156 p = 1 | |
| CTLA-4 rs231775 (G vs. AA) | 1.540 | 4.67 | 0.013 | 1.49–17.94 | |||
| Remission | |||||||
| Age at ABA start | −0.044 | 0.96 | 0.027 | 0.92–0.99 | R2 Cox Snell = 0.239 R2 Nagelkerke = 0.339 | X2 = 6.561 p = 0.585 | |
| Number of previous BTs | −0.574 | 0.56 | 0.023 | 0.34–0.92 | |||
| PVAS | −0.051 | 0.95 | <0.001 | 0.93–0.98 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marquez Pete, N.; Maldonado Montoro, M.d.M.; Pérez Ramírez, C.; Sánchez Martín, A.; Martínez de la Plata, J.E.; Martínez Martínez, F.; Caliz Caliz, R.; Daddaoua, A.; Ramírez Tortosa, M.d.C.; Jiménez Morales, A. Impact of Single-Nucleotide Polymorphisms of CTLA-4, CD80 and CD86 on the Effectiveness of Abatacept in Patients with Rheumatoid Arthritis. J. Pers. Med. 2020, 10, 220. https://doi.org/10.3390/jpm10040220
Marquez Pete N, Maldonado Montoro MdM, Pérez Ramírez C, Sánchez Martín A, Martínez de la Plata JE, Martínez Martínez F, Caliz Caliz R, Daddaoua A, Ramírez Tortosa MdC, Jiménez Morales A. Impact of Single-Nucleotide Polymorphisms of CTLA-4, CD80 and CD86 on the Effectiveness of Abatacept in Patients with Rheumatoid Arthritis. Journal of Personalized Medicine. 2020; 10(4):220. https://doi.org/10.3390/jpm10040220
Chicago/Turabian StyleMarquez Pete, Noelia, María del Mar Maldonado Montoro, Cristina Pérez Ramírez, Almudena Sánchez Martín, Juan Enrique Martínez de la Plata, Fernando Martínez Martínez, Rafael Caliz Caliz, Abdelali Daddaoua, María del Carmen Ramírez Tortosa, and Alberto Jiménez Morales. 2020. "Impact of Single-Nucleotide Polymorphisms of CTLA-4, CD80 and CD86 on the Effectiveness of Abatacept in Patients with Rheumatoid Arthritis" Journal of Personalized Medicine 10, no. 4: 220. https://doi.org/10.3390/jpm10040220
APA StyleMarquez Pete, N., Maldonado Montoro, M. d. M., Pérez Ramírez, C., Sánchez Martín, A., Martínez de la Plata, J. E., Martínez Martínez, F., Caliz Caliz, R., Daddaoua, A., Ramírez Tortosa, M. d. C., & Jiménez Morales, A. (2020). Impact of Single-Nucleotide Polymorphisms of CTLA-4, CD80 and CD86 on the Effectiveness of Abatacept in Patients with Rheumatoid Arthritis. Journal of Personalized Medicine, 10(4), 220. https://doi.org/10.3390/jpm10040220