Establishment and Characterization of NCC-DDLPS4-C1: A Novel Patient-Derived Cell Line of Dedifferentiated Liposarcoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient History
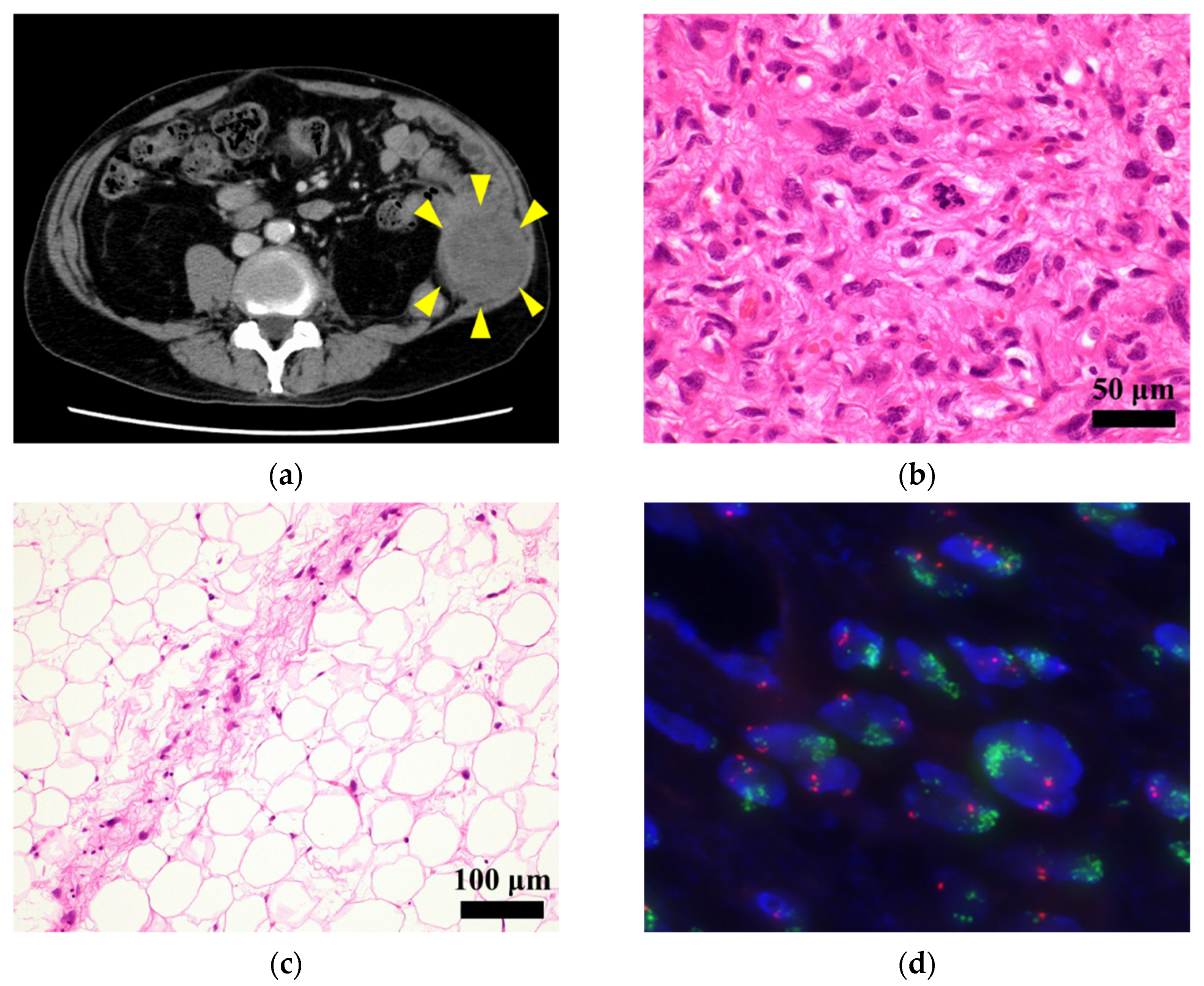
2.2. Pathological Examination
2.3. FISH Analysis
2.4. Primary Cell Culture Procedure
2.5. Authentication and Quality Control
2.6. Single-Nucleotide Polymorphism Array Analysis
2.7. Western Blotting
2.8. Cell Proliferation Assay
2.9. Spheroid Formation Assay
2.10. Invasion Assay by Real-Time Cell Analyzer
2.11. Tumorigenesis Assessment in Nude Mice
2.12. Drug Screening Test
3. Results
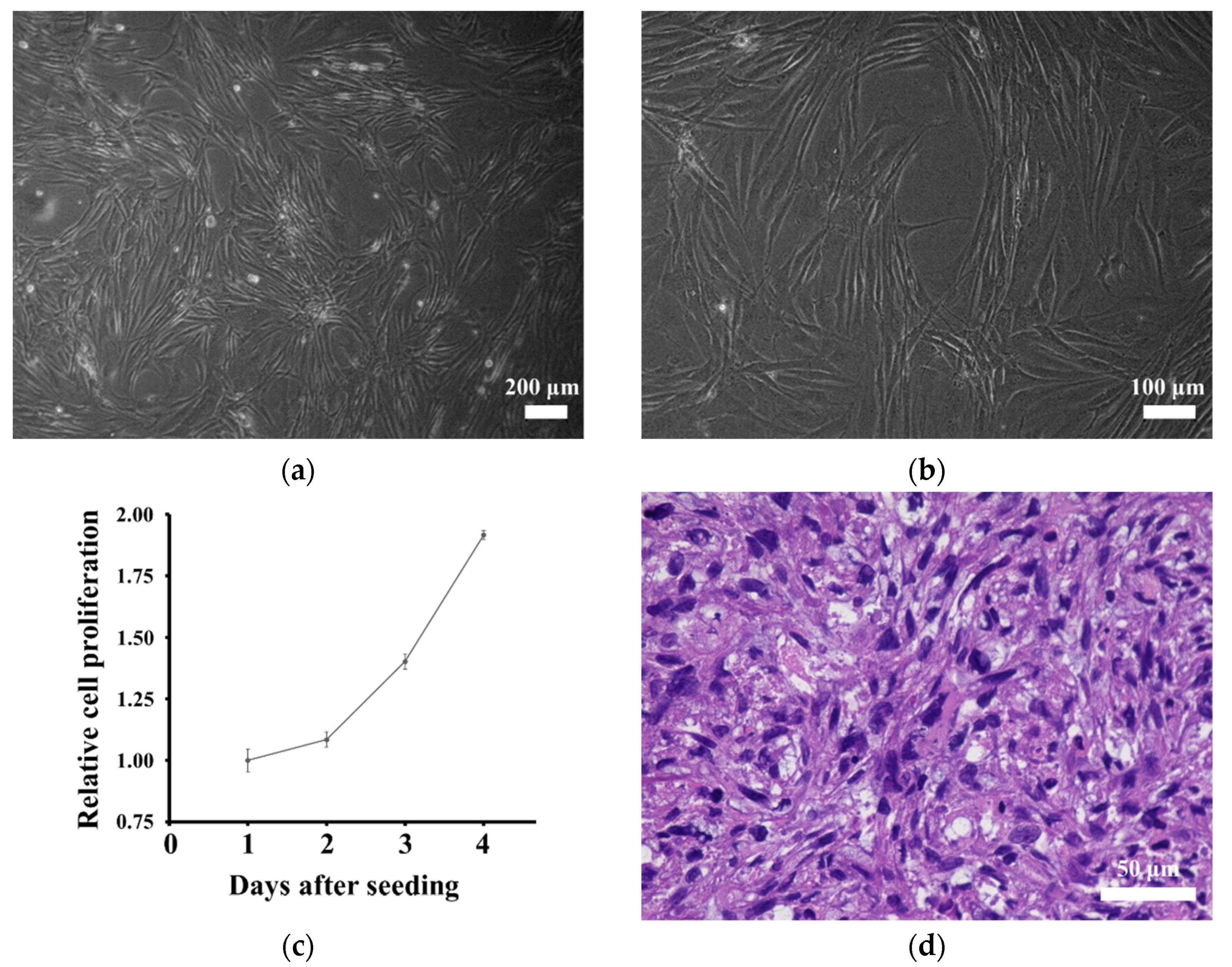
3.1. Authentication of the Established Cell Line
3.2. Genomic Landscape of the Cell Line
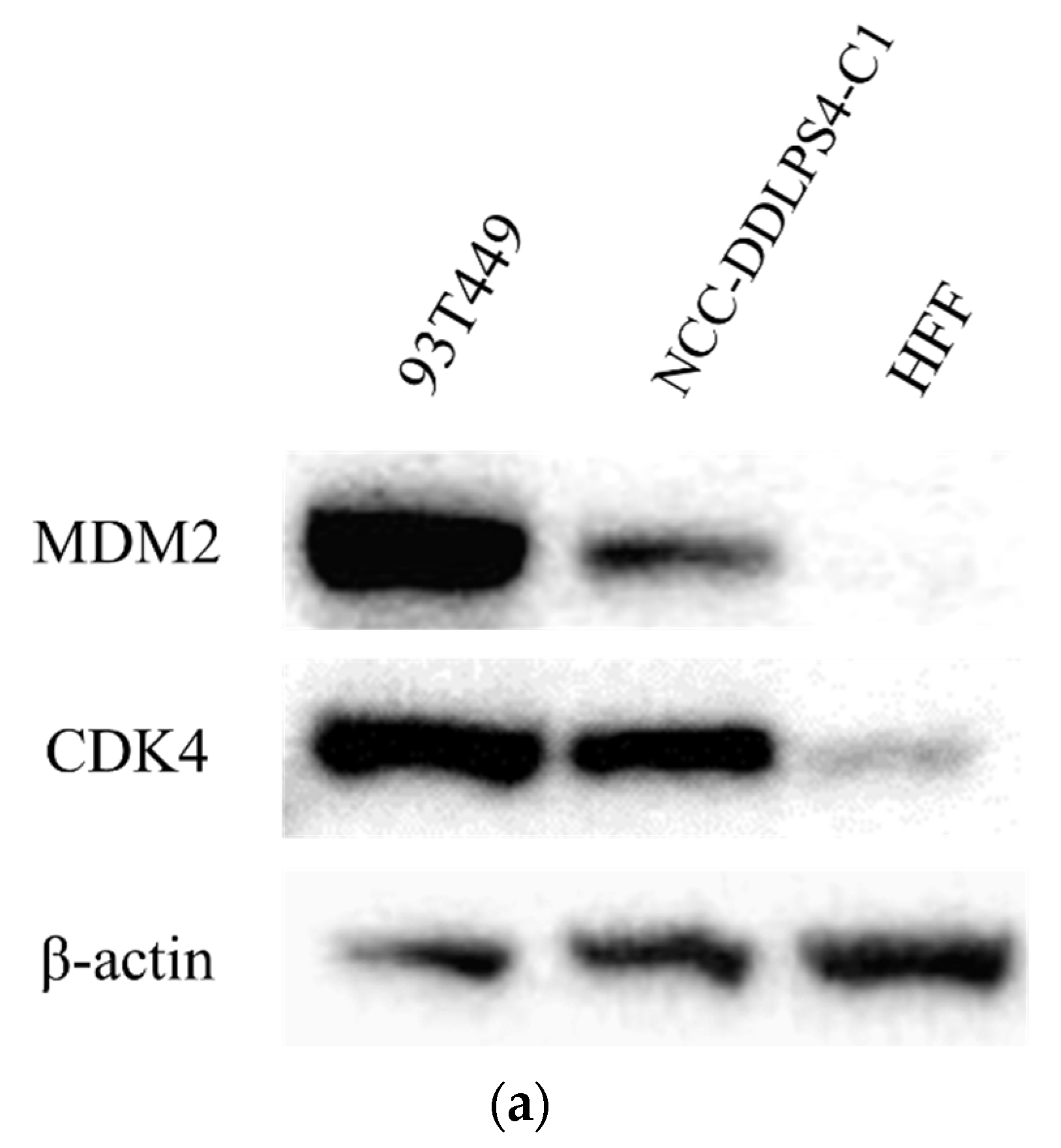
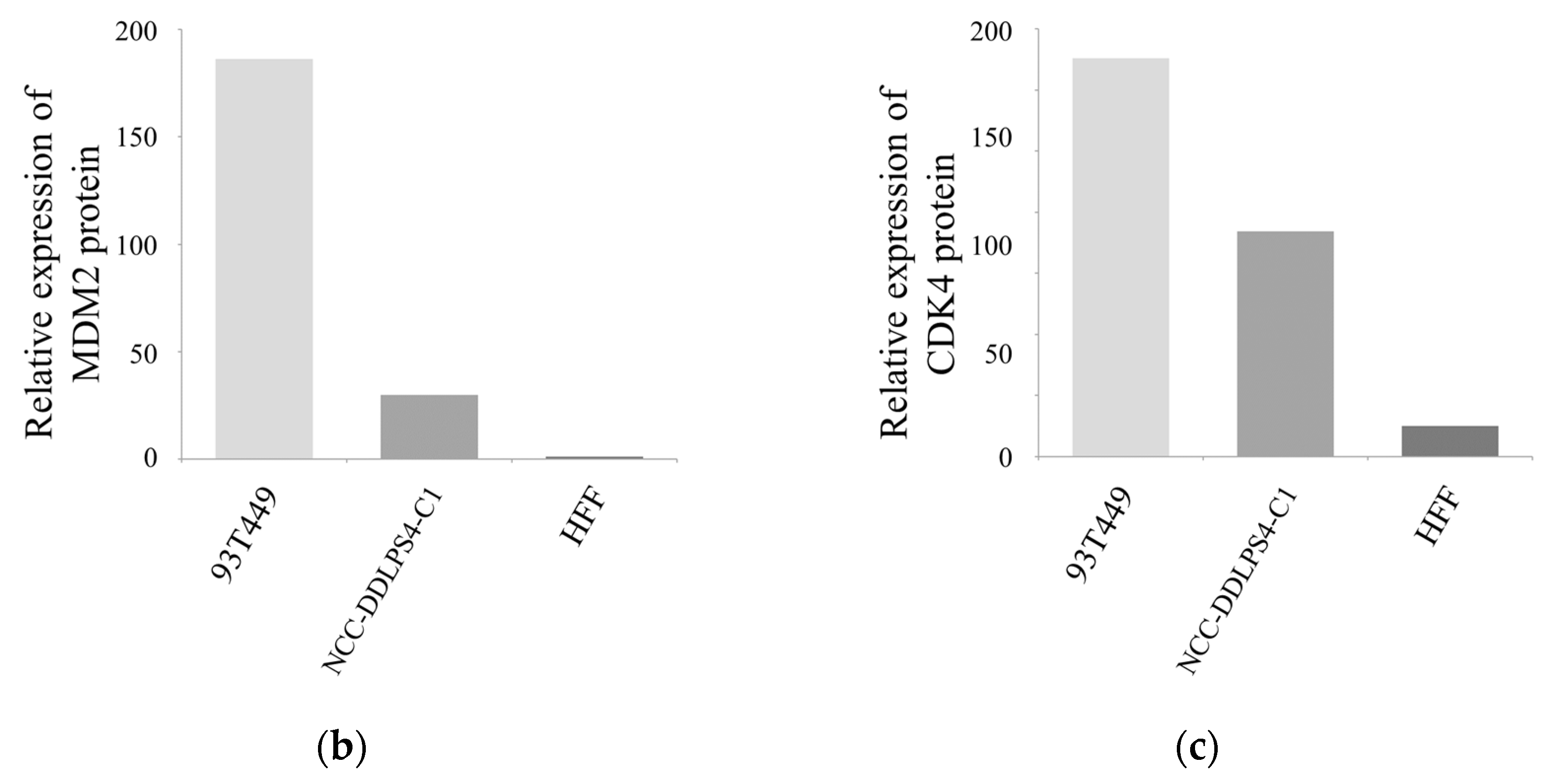
3.3. MDM2 and CDK4 Expressions of the Cell Line
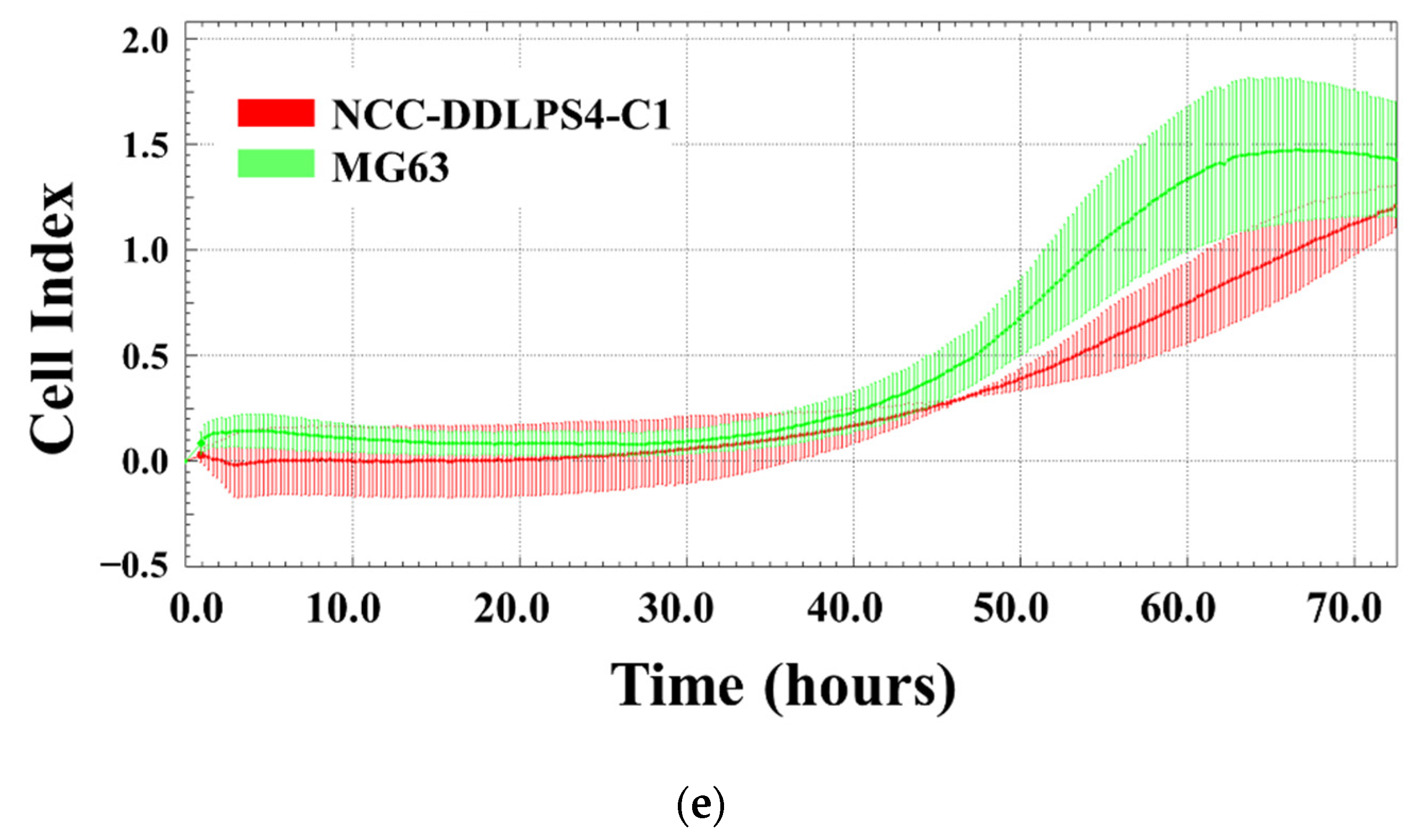
3.4. In Vitro Characteristics of the Cell Line
3.5. Tumorigenesis in Nude Mice
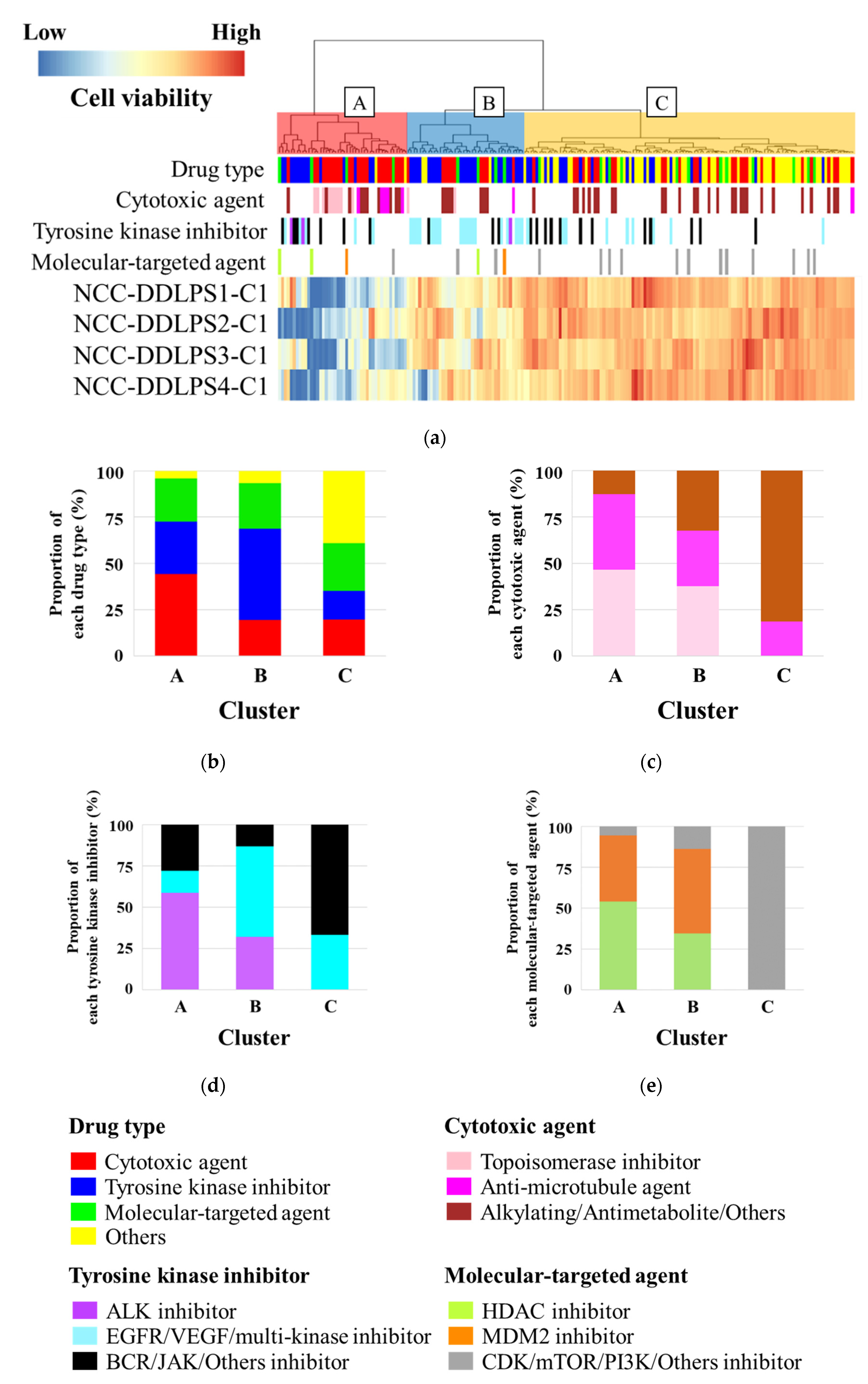
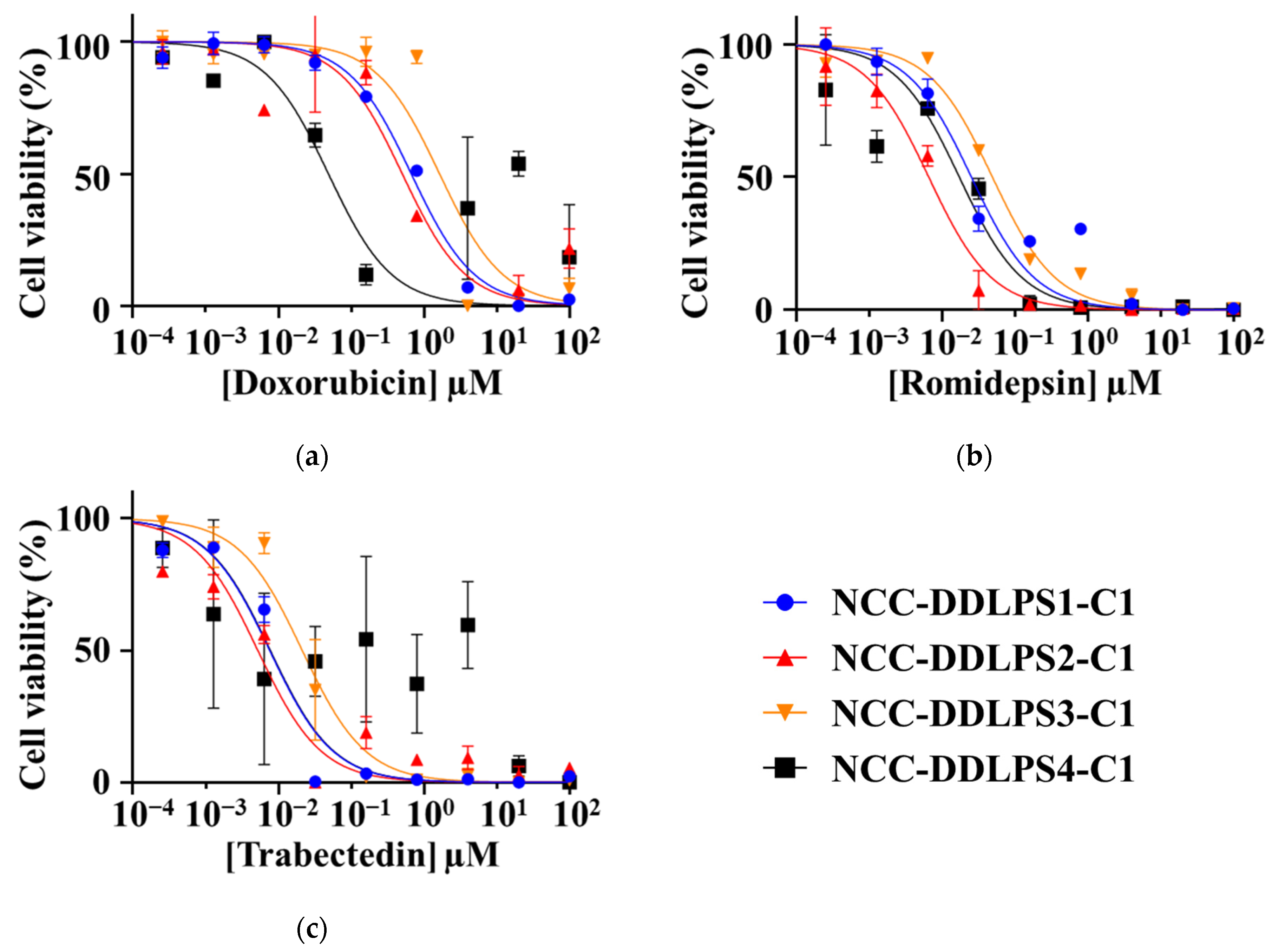
3.6. Sensitivity to Anticancer Drugs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Classification of Tumours of Soft Tissue and Bone, 5th ed.; IARC: Lyon, France, 2020. [Google Scholar]
- Nishio, J.; Nakayama, S.; Nabeshima, K.; Yamamoto, T. Biology and Management of Dedifferentiated Liposarcoma: State of the Art and Perspectives. J. Clin. Med. 2021, 10, 3230. [Google Scholar] [CrossRef]
- Gootee, J.; Aurit, S.; Curtin, C.; Silberstein, P. Primary anatomical site, adjuvant therapy, and other prognostic variables for dedifferentiated liposarcoma. J. Cancer Res. Clin. Oncol. 2019, 145, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Tyler, R.; Wanigasooriya, K.; Taniere, P.; Almond, M.; Ford, S.; Desai, A.; Beggs, A. A review of retroperitoneal liposarcoma genomics. Cancer Treat Rev. 2020, 86, 102013. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Wood, D.; Ingley, E.; Koks, S.; Wong, D. Update on genomic and molecular landscapes of well-differentiated liposarcoma and dedifferentiated liposarcoma. Mol. Biol. Rep. 2021, 48, 3637–3647. [Google Scholar] [CrossRef] [PubMed]
- Sciot, R. MDM2 Amplified Sarcomas: A Literature Review. Diagnostics 2021, 11, 496. [Google Scholar] [CrossRef] [PubMed]
- Italiano, A.; Bianchini, L.; Keslair, F.; Bonnafous, S.; Cardot-Leccia, N.; Coindre, J.-M.; Dumollard, J.-M.; Hofman, P.; Leroux, A.; Mainguené, C.; et al. HMGA2 is the partner of MDM2 in well-differentiated and dedifferentiated liposarcomas whereas CDK4 belongs to a distinct inconsistent amplicon. Int. J. Cancer 2008, 122, 2233–2241. [Google Scholar] [CrossRef] [PubMed]
- Snyder, E.L.; Sandstrom, D.J.; Law, K.; Fiore, C.; Sicinska, E.; Brito, J.; Bailey, D.; A Fletcher, J.; Loda, M.; Rodig, S.J.; et al. c-Jun amplification and overexpression are oncogenic in liposarcoma but not always sufficient to inhibit the adipocytic differentiation programme. J. Pathol. 2009, 218, 292–300. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive and Integrated Genomic Characterization of Adult Soft Tissue Sarcomas. Cell 2017, 171, 950–965.e28. [Google Scholar] [CrossRef] [Green Version]
- Crago, A.; Socci, N.D.; Decarolis, P.; O’Connor, R.; Taylor, B.S.; Qin, L.-X.; Antonescu, C.R.; Singer, S. Copy number losses define subgroups of dedifferentiated liposarcoma with poor prognosis and genomic instability. Clin. Cancer Res. 2012, 18, 1334–1340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirata, M.; Asano, N.; Katayama, K.; Yoshida, A.; Tsuda, Y.; Sekimizu, M.; Mitani, S.; Kobayashi, E.; Komiyama, M.; Fujimoto, H.; et al. Integrated exome and RNA sequencing of dedifferentiated liposarcoma. Nat. Commun. 2019, 10, 5683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amer, K.M.; Congiusta, D.V.; Thomson, J.E.; Elsamna, S.; Chaudhry, I.; Bozzo, A.; Amer, R.; Siracuse, B.; Ghert, M.; Beebe, K.S.; et al. Epidemiology and survival of liposarcoma and its subtypes: A dual database analysis. J. Clin. Orthop. Trauma 2020, 11, S479–S484. [Google Scholar] [CrossRef]
- Haddox, C.L.; Riedel, R.F. Recent advances in the understanding and management of liposarcoma. Fac. Rev. 2021, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Gahvari, Z.; Parkes, A. Dedifferentiated Liposarcoma: Systemic Therapy Options. Curr. Treat Options Oncol. 2020, 21, 15. [Google Scholar] [CrossRef] [PubMed]
- Livingston, J.A.; Bugano, D.; Barbo, A.; Lin, H.; Madewell, J.E.; Wang, W.L.; Lazar, A.J.; Tseng, W.W.; Roland, C.L.; Feig, B.W.; et al. Role of chemotherapy in dedifferentiated liposarcoma of the retroperitoneum: Defining the benefit and challenges of the standard. Sci. Rep. 2017, 7, 11836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ray-Coquard, I.; Blay, J.-Y.; Italiano, A.; Le Cesne, A.; Penel, N.; Zhi, J.; Heil, F.; Rueger, R.; Graves, B.; Ding, M.; et al. Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients with MDM2-amplified, well-differentiated or dedifferentiated liposarcoma: An exploratory proof-of-mechanism study. Lancet Oncol. 2012, 13, 1133–1140. [Google Scholar] [CrossRef]
- Wagner, A.J.; Banerji, U.; Mahipal, A.; Somaiah, N.; Hirsch, H.; Fancourt, C.; Johnson-Levonas, A.O.; Lam, R.; Meister, A.K.; Russo, G.; et al. Phase I Trial of the Human Double Minute 2 Inhibitor MK-8242 in Patients with Advanced Solid Tumors. J. Clin. Oncol. 2017, 35, 1304–1311. [Google Scholar] [CrossRef] [PubMed]
- Dickson, M.A.; Koff, A.; D’Angelo, S.P.; Gounder, M.M.; Keohan, M.L.; Kelly, C.M.; Chi, P.; Antonescu, C.R.; Landa, J.; Qin, L.-X.; et al. Phase 2 study of the CDK4 inhibitor abemaciclib in dedifferentiated liposarcoma. J. Clin. Oncol. 2019, 37, 11004. [Google Scholar] [CrossRef]
- Dickson, M.A.; Schwartz, G.K.; Keohan, M.L.; D’Angelo, S.P.; Gounder, M.M.; Chi, P.; Antonescu, C.R.; Landa, J.; Qin, L.-X.; Aimee, M.C.; et al. Progression-Free Survival Among Patients with Well-Differentiated or Dedifferentiated Liposarcoma Treated with CDK4 Inhibitor Palbociclib: A Phase 2 Clinical Trial. JAMA Oncol. 2016, 2, 937–940. [Google Scholar] [CrossRef] [Green Version]
- Saerens, M.; Brusselaers, N.; Rottey, S.; Decruyenaere, A.; Creytens, D.; Lapeire, L. Immune checkpoint inhibitors in treatment of soft-tissue sarcoma: A systematic review and meta-analysis. Eur. J Cancer. 2021, 152, 165–182. [Google Scholar] [CrossRef] [PubMed]
- Saito, S.; Morita, K.; Kohara, A.; Masui, T.; Sasao, M.; Ohgushi, H.; Hirano, T. Use of BAC array CGH for evaluation of chromosomal stability of clinically used human mesenchymal stem cells and of cancer cell lines. Hum. Cell 2011, 24, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Ben-David, U.; Beroukhim, R.; Golub, T.R. Genomic evolution of cancer models: Perils and opportunities. Nat. Rev. Cancer 2019, 19, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Tseng, Y.Y.; Boehm, J.S. From cell lines to living biosensors: New opportunities to prioritize cancer dependencies using ex vivo tumor cultures. Curr. Opin. Genet. Dev. 2019, 54, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, R.H. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer 2006, 6, 813–823. [Google Scholar] [CrossRef]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.; Bodycombe, N.E.; Cheah, J.H.; Price, E.V.; Liu, K.; Schaefer, G.I.; Ebright, R.Y.; Stewart, M.L.; Ito, D.; Wang, S.; et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell 2013, 154, 1151–1161. [Google Scholar] [PubMed] [Green Version]
- Yang, W.; Soares, J.; Greninger, P.; Edelman, E.; Lightfoot, H.; Forbes, S.; Sridhar, R.; Futreal, P.A.; Haber, D.; Stratton, M.; et al. Genomics of Drug Sensitivity in Cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013, 41, D955–D961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hattori, E.; Oyama, R.; Kondo, T. Systematic Review of the Current Status of Human Sarcoma Cell Lines. Cells 2019, 8, 157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodspeed, A.; Heiser, L.M.; Gray, J.W.; Costello, J.C. Tumor-Derived Cell Lines as Molecular Models of Cancer Pharmacogenomics. Mol. Cancer Res. 2016, 14, 3–13. [Google Scholar] [CrossRef] [Green Version]
- Tsuchiya, R.; Yoshimatsu, Y.; Noguchi, R.; Ono, T.; Sei, A.; Takeshita, F.; Sugaya, J.; Fukushima, S.; Yoshida, A.; Ohtori, S.; et al. Establishment and characterization of NCC-DDLPS1-C1: A novel patient-derived cell line of dedifferentiated liposarcoma. Hum. Cell 2021, 34, 260–270. [Google Scholar]
- Noguchi, R.; Yoshimatsu, Y.; Ono, T.; Sei, A.; Hirabayashi, K.; Ozawa, I.; Kikuta, K.; Kondo, T. Establishment and characterization of a novel cell line, NCC-DDLPS2-C1, derived from a patient with dedifferentiated liposarcoma. Hum. Cell 2021, 34, 990–997. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, R.; Yoshimatsu, Y.; Noguchi, R.; Sin, Y.; Ono, T.; Sei, A.; Takeshita, F.; Sugaya, J.; Iwata, S.; Yoshida, A.; et al. Establishment and characterization of NCC-DDLPS3-C1: A novel patient-derived cell line of dedifferentiated liposarcoma. Hum. Cell 2021, 34, 1008–1018. [Google Scholar] [CrossRef] [PubMed]
- Bairoch, A. The Cellosaurus, a Cell-Line Knowledge Resource. J. Biomol. Tech. 2018, 29, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Billiau, A.; Edy, V.G.; Heremans, H.; Van Damme, J.; Desmyter, J.; Georgiades, J.A.; De Somer, P. Human interferon: Mass production in a newly established cell line, MG-63. Antimicrob. Agents Chemother. 1977, 12, 11–15. [Google Scholar] [CrossRef] [Green Version]
- Tanabe, H.; Takada, Y.; Minegishi, D.; Kurematsu, M.; Masui, T.; Mizusawa, H. Cell line individualization by STR multiplex system in the cell bank found cross-contamination between ECV304 and EJ-1/T24. Tiss. Cult. Res. Commun. 1999, 18, 329–338. [Google Scholar]
- Capes-Davis, A.; Reid, Y.A.; Kline, M.C.; Storts, D.R.; Strauss, E.; Dirks, W.G.; Drexler, H.G.; MacLeod, R.A.F.; Sykes, G.; Kohara, A.; et al. Match criteria for human cell line authentication: Where do we draw the line? Int. J. Cancer 2013, 132, 2510–2519. [Google Scholar] [CrossRef]
- Creytens, D.; Van Gorp, J.; Speel, E.J.; Ferdinande, L. Characterization of the 12q amplicons in lipomatous soft tissue tumors by multiplex ligation-dependent probe amplification-based copy number analysis. Anticancer Res. 2015, 35, 1835–1842. [Google Scholar] [PubMed]
- Kanojia, D.; Nagata, Y.; Garg, M.; Lee, D.H.; Sato, A.; Yoshida, K.; Sato, Y.; Sanada, M.; Mayakonda, A.; Bartenhagen, C.; et al. Genomic landscape of liposarcoma. Oncotarget 2015, 6, 42429–42444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dadone-Montaudié, B.; Laroche-Clary, A.; Mongis, A.; Chamorey, E.; Di Mauro, I.; Chaire, V.; Finetti, P.; Schiappa, R.; Le Loarer, F.; Birtwisle-Peyrottes, I.; et al. Novel Therapeutic Insights in Dedifferentiated Liposarcoma: A Role for FGFR and MDM2 Dual Targeting. Cancers 2020, 12, 3058. [Google Scholar] [CrossRef]
- Hanes, R.; Munthe, E.; Grad, I.; Han, J.; Karlsen, I.; McCormack, E.; Meza-Zepeda, L.A.; Stratford, E.W.; Myklebost, O. Preclinical Evaluation of the Pan-FGFR Inhibitor LY2874455 in FRS2-Amplified Liposarcoma. Cells 2019, 8, 189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grad, I.; Hanes, R.; Ayuda-Durán, P.; Kuijjer, M.L.; Enserink, J.M.; Meza-Zepeda, L.A.; Myklebost, O. Discovery of novel candidates for anti-liposarcoma therapies by medium-scale high-throughput drug screening. PLoS ONE 2021, 16, e0248140. [Google Scholar] [CrossRef] [PubMed]
- Que, Y.; Zhang, X.-L.; Liu, Z.-X.; Zhao, J.-J.; Pan, Q.-Z.; Wen, X.-Z.; Xiao, W.; Xu, B.-S.; Hong, D.-C.; Guo, T.-H.; et al. Frequent amplification of HDAC genes and efficacy of HDAC inhibitor chidamide and PD-1 blockade combination in soft tissue sarcoma. J. Immunother. Cancer 2021, 9, e001696. [Google Scholar] [CrossRef] [PubMed]
- Seligson, N.D.; Stets, C.W.; Demoret, B.; Awasthi, A.; Grosenbacher, N.; Shakya, R.; Hays, J.L.; Chen, J.L.; Stenzinger, A.; Stephan-Falkenau, S.; et al. Inhibition of histone deacetylase 2 reduces MDM2 expression and reduces tumor growth in dedifferentiated liposarcoma. Oncotarget 2019, 10, 5671–5679. [Google Scholar] [CrossRef] [PubMed]
- Wilding, J.L.; Bodmer, W.F. Cancer cell lines for drug discovery and development. Cancer Res. 2014, 74, 2377–2384. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Microsatellite | NCC-DDLPS4-C1 | Original Tumor Tissue |
|---|---|---|
| (Chromosome) | ||
| Amelogenin (X Y) | X, Y | X, Y |
| TH01 (3) | 7 | 7, 9 |
| D21S11 (21) | 29, 30 | 29, 30 |
| D5S818 (5) | 11, 12 | 11, 12 |
| D13S317 (13) | 12 | 12 |
| D7S820 (7) | 8, 11 | 8, 11 |
| D16S539 (16) | 9 | 9 |
| CSF1PO (5) | 10, 12 | 10, 12 |
| vWA (12) | 17 | 14, 17 |
| TPOX (2) | 8, 11 | 8, 11 |
| Gene Symbol | Chromosome Region | Copy Number | Type |
|---|---|---|---|
| ATF4P4 | 11q23.2 | 0.1 | Loss |
| LRRC37A13P | 11q23.2 | 0.1 | Loss |
| USP28 | 11q23.2 | 0.1 | Loss |
| TSPAN31 | 12q14.1 | 3.0 | Amp |
| CDK4 | 12q14.1 | 3.0 | Amp |
| HMGA2 | 12q14.3 | 3.4 | Amp |
| SLC35E3 | 12q15 | 3.5 | Amp |
| MDM2 | 12q15 | 3.5 | Amp |
| CPM | 12q15 | 3.5 | Amp |
| YEATS4 | 12q15 | 3.4 | Amp |
| FRS2 | 12q15 | 3.4 | Amp |
| CAS# | Drug | NCC-DDLPS1-C1 | NCC-DDLPS2-C1 | NCC-DDLPS3-C1 | NCC-DDLPS4-C1 |
|---|---|---|---|---|---|
| 25316-40-9 | Doxorubicin | 0.6706 | 0.53 | 1.642 | 0.04878 |
| 128517-07-7 | Romidepsin | 0.02538 | 0.0068 | 0.0493 | 0.01734 |
| 114899-77-3 | Trabectedin | 0.007564 | 0.0049 | 0.0212 | 0.007721 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsuchiya, R.; Yoshimatsu, Y.; Noguchi, R.; Sin, Y.; Ono, T.; Akiyama, T.; Hirose, T.; Iwata, S.; Yoshida, A.; Ohtori, S.; et al. Establishment and Characterization of NCC-DDLPS4-C1: A Novel Patient-Derived Cell Line of Dedifferentiated Liposarcoma. J. Pers. Med. 2021, 11, 1075. https://doi.org/10.3390/jpm11111075
Tsuchiya R, Yoshimatsu Y, Noguchi R, Sin Y, Ono T, Akiyama T, Hirose T, Iwata S, Yoshida A, Ohtori S, et al. Establishment and Characterization of NCC-DDLPS4-C1: A Novel Patient-Derived Cell Line of Dedifferentiated Liposarcoma. Journal of Personalized Medicine. 2021; 11(11):1075. https://doi.org/10.3390/jpm11111075
Chicago/Turabian StyleTsuchiya, Ryuto, Yuki Yoshimatsu, Rei Noguchi, Yooksil Sin, Takuya Ono, Taro Akiyama, Takeshi Hirose, Shintaro Iwata, Akihiko Yoshida, Seiji Ohtori, and et al. 2021. "Establishment and Characterization of NCC-DDLPS4-C1: A Novel Patient-Derived Cell Line of Dedifferentiated Liposarcoma" Journal of Personalized Medicine 11, no. 11: 1075. https://doi.org/10.3390/jpm11111075
APA StyleTsuchiya, R., Yoshimatsu, Y., Noguchi, R., Sin, Y., Ono, T., Akiyama, T., Hirose, T., Iwata, S., Yoshida, A., Ohtori, S., Kawai, A., & Kondo, T. (2021). Establishment and Characterization of NCC-DDLPS4-C1: A Novel Patient-Derived Cell Line of Dedifferentiated Liposarcoma. Journal of Personalized Medicine, 11(11), 1075. https://doi.org/10.3390/jpm11111075








