Microbiome Research and Multi-Omics Integration for Personalized Medicine in Asthma
Abstract
:1. Introduction
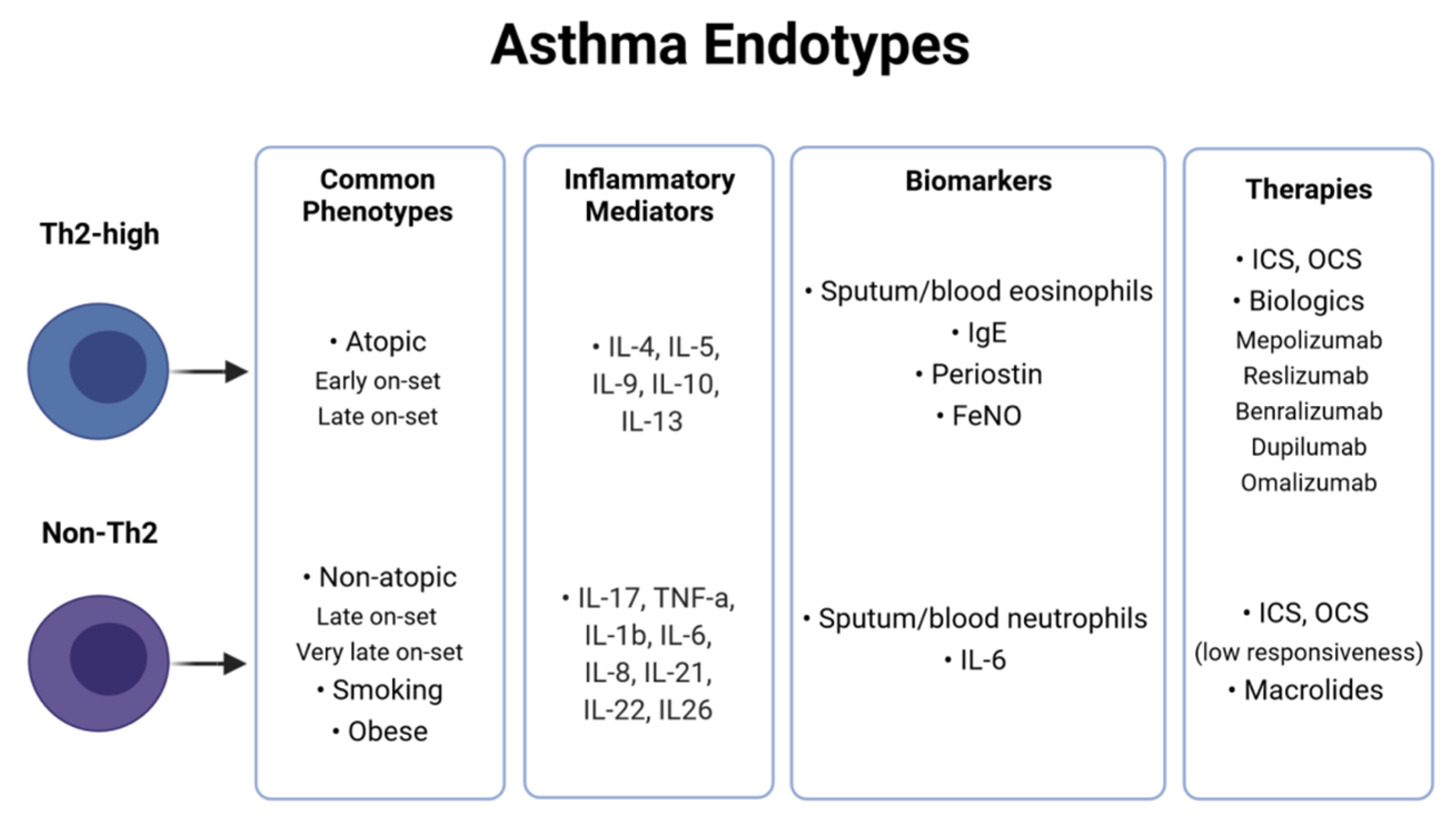
2. Asthma Heterogeneity and Classification
3. Insights from Host-Omics Research
3.1. Genomics-Epigenomics
3.2. Transcriptomics–Proteomics
3.3. Metabolomics
4. The Human Microbiome in Asthma Research
4.1. The Importance of the Initial Host Microbial Colonization
4.2. The Key Role of the Gut and Airway Microbiota Cross-Talk in Asthma
4.3. Microbiome and Crucial Asthma Risk Factors
4.3.1. Tobacco Smoking
4.3.2. Diet and Obesity
5. Airway Microbiome Correlations with Asthma Subtypes
6. Relationships between Asthmatics Airway Microbiome and Treatment
7. Integration of Microbiome and Multi-Omics Data from Asthma Patients
7.1. Microbiome and Host Gene Expression Data Integration
7.2. Microbiome and Host Metabolome/Proteome Data Integration
7.3. Large Consortia Integrative Multi-Omics Studies
8. Challenges
9. Future Directions
10. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Asthma. Available online: www.who.int/news-room/fact-sheets/detail/asthma (accessed on 20 May 2021).
- Wenzel, S.E. Asthma Phenotypes: The Evolution from Clinical to Molecular Approaches. Nat. Med. 2012, 18, 716–725. [Google Scholar] [CrossRef] [PubMed]
- Loza, M.J.; Djukanovic, R.; Chung, K.F.; Horowitz, D.; Ma, K.; Branigan, P.; Barnathan, E.S.; Susulic, V.S.; Silkoff, P.E.; Sterk, P.J.; et al. Validated and Longitudinally Stable Asthma Phenotypes Based on Cluster Analysis of the ADEPT Study. Respir. Res. 2016, 17, 165. [Google Scholar] [CrossRef] [PubMed]
- Kuruvilla, M.E.; Lee, F.E.-H.; Lee, G.B. Understanding Asthma Phenotypes, Endotypes, and Mechanisms of Disease. Clin. Rev. Allergy Immunol. 2019, 56, 219–233. [Google Scholar] [CrossRef]
- Papi, A.; Brightling, C.; Pedersen, S.E.; Reddel, H.K. Asthma. Lancet 2018, 391, 783–800. [Google Scholar] [CrossRef]
- Global Initiative for Asthma. GINA 2021 Asthma Management and Prevention; Global Initiative for Asthma: Fontana, WI, USA, 2021. [Google Scholar]
- Muraro, A.; Lemanske, R.F.; Hellings, P.W.; Akdis, C.A.; Bieber, T.; Casale, T.B.; Jutel, M.; Ong, P.Y.; Poulsen, L.K.; Schmid-Grendelmeier, P.; et al. Precision Medicine in Patients with Allergic Diseases: Airway Diseases and Atopic Dermatitis—PRACTALL Document of the European Academy of Allergy and Clinical Immunology and the American Academy of Allergy, Asthma & Immunology. J. Allergy Clin. Immunol. 2016, 137, 1347–1358. [Google Scholar]
- Chung, K.F. Precision Medicine in Asthma. Curr. Opin. Pulm. Med. 2018, 24, 4–10. [Google Scholar] [CrossRef]
- Cazzola, M.; Ora, J.; Cavalli, F.; Rogliani, P.; Matera, M.G. Treatable Mechanisms in Asthma. Mol. Diagn. Ther. 2021, 25, 111–121. [Google Scholar] [CrossRef]
- Chiu, C.-J.; Huang, M.-T. Asthma in the Precision Medicine Era: Biologics and Probiotics. Int. J. Mol. Sci. 2021, 22, 4528. [Google Scholar] [CrossRef] [PubMed]
- Silkoff, P.E.; Moore, W.C.; Sterk, P.J. Three Major Efforts to Phenotype Asthma: Severe Asthma Research Program, Asthma Disease Endotyping for Personalized Therapeutics, and Unbiased Biomarkers for the Prediction of Respiratory Disease Outcome. Clin. Chest Med. 2019, 40, 13–28. [Google Scholar] [CrossRef] [PubMed]
- Colas, L.; Hassoun, D.; Magnan, A. Needs for Systems Approaches to Better Treat Individuals With Severe Asthma: Predicting Phenotypes and Responses to Treatments. Front. Med. 2020, 7, 98. [Google Scholar] [CrossRef] [PubMed]
- Hufnagl, K.; Pali-Schöll, I.; Roth-Walter, F.; Jensen-Jarolim, E. Dysbiosis of the Gut and Lung Microbiome Has a Role in Asthma. Semin. Immunopathol. 2020, 42, 75–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.-C.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome Definition Re-Visited: Old Concepts and New Challenges. Microbiome 2020, 8, 103. [Google Scholar] [CrossRef]
- Escobar-Zepeda, A.; Vera-Ponce de León, A.; Sanchez-Flores, A. The Road to Metagenomics: From Microbiology to DNA Sequencing Technologies and Bioinformatics. Front. Genet. 2015, 6, 348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davidson, R.M.; Epperson, L.E. Microbiome Sequencing Methods for Studying Human Diseases. Methods Mol. Biol. 2018, 1706, 77–90. [Google Scholar]
- Breitwieser, F.P.; Lu, J.; Salzberg, S.L. A Review of Methods and Databases for Metagenomic Classification and Assembly. Brief. Bioinform. 2019, 20, 1125–1136. [Google Scholar] [CrossRef]
- Holgate, S.T.; Wenzel, S.; Postma, D.S.; Weiss, S.T.; Renz, H.; Sly, P.D. Asthma. Nat. Rev. Dis. Prim. 2015, 1, 15025. [Google Scholar] [CrossRef]
- Barnes, P.J. Corticosteroid Resistance in Patients with Asthma and Chronic Obstructive Pulmonary Disease. J. Allergy Clin. Immunol. 2013, 131, 636–645. [Google Scholar] [CrossRef] [PubMed]
- Kiboneka, A.; Kibuule, D. The immunology of asthma and allergic rhinitis. In Rhinosinusitis; Balwant, S.G., Turkalj, M., Eds.; IntechOpen: London, UK, 2019. [Google Scholar]
- Raphael, I.; Nalawade, S.; Eagar, T.N.; Forsthuber, T.G. T Cell Subsets and Their Signature Cytokines in Autoimmune and Inflammatory Diseases. Cytokine 2015, 74, 5–17. [Google Scholar] [CrossRef] [Green Version]
- Fahy, J.V. Type 2 Inflammation in Asthma—Present in Most, Absent in Many. Nat. Rev. Immunol. 2015, 15, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; Chupp, G. Phenotypes and Endotypes of Adult Asthma: Moving toward Precision Medicine. J. Allergy Clin. Immunol. 2019, 144, 1–12. [Google Scholar] [CrossRef]
- Akar-Ghibril, N.; Casale, T.; Custovic, A.; Phipatanakul, W. Allergic Endotypes and Phenotypes of Asthma. J. Allergy Clin. Immunol. Pract. 2020, 8, 429–440. [Google Scholar] [CrossRef]
- Katsaounou, P.; Buhl, R.; Brusselle, G.; Pfister, P.; Martínez, R.; Wahn, U.; Bousquet, J. Omalizumab as Alternative to Chronic Use of Oral Corticosteroids in Severe Asthma. Respir. Med. 2019, 150, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Martinez, G.J.; Nurieva, R.I.; Yang, X.O.; Dong, C. Regulation and Function of Proinflammatory TH17 Cells. Ann. N. Y. Acad. Sci. 2008, 1143, 188–211. [Google Scholar] [CrossRef] [Green Version]
- Newcomb, D.C.; Peebles, R.S. Th17-Mediated Inflammation in Asthma. Curr. Opin. Immunol. 2013, 25, 755–760. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Cui, J.; Yi, L.; Qin, J.; Tulake, W.; Teng, F.; Tang, W.; Wei, Y.; Dong, J. The Role of T Cells and Macrophages in Asthma Pathogenesis: A New Perspective on Mutual Crosstalk. Mediat. Inflamm. 2020, 2020, 1–14. [Google Scholar] [CrossRef]
- Sze, E.; Bhalla, A.; Nair, P. Mechanisms and Therapeutic Strategies for Non-T2 Asthma. Allergy 2020, 75, 311–325. [Google Scholar] [CrossRef] [Green Version]
- Nair, P.; Surette, M.G.; Virchow, J.C. Neutrophilic Asthma: Misconception or Misnomer? Lancet Respir. Med. 2021, 9, 441–443. [Google Scholar] [CrossRef]
- Israel, E.; Reddel, H.K. Severe and Difficult-to-Treat Asthma in Adults. N. Engl. J. Med. 2017, 377, 965–976. [Google Scholar] [CrossRef]
- Chau-Etchepare, F.; Hoerger, J.L.; Kuhn, B.T.; Zeki, A.A.; Haczku, A.; Louie, S.; Kenyon, N.J.; Davis, C.E.; Schivo, M. Viruses and Non-Allergen Environmental Triggers in Asthma. J. Investig. Med. 2019, 67, 1029–1041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bardin, P.G. Escalating Inhaled Glucocorticoids to Prevent Asthma Exacerbations. N. Engl. J. Med. 2018, 378, 950–952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tyler, S.R.; Bunyavanich, S. Leveraging -Omics for Asthma Endotyping. J. Allergy Clin. Immunol. 2019, 144, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, O.; Richards, L.B.; Vijverberg, S.J.; Neerincx, A.H.; Sinha, A.; Sterk, P.J.; Maitland-van der Zee, A.H. What Did We Learn from Multiple Omics Studies in Asthma? Allergy 2019, 74, 2129–2145. [Google Scholar] [CrossRef] [Green Version]
- Abdel-Aziz, M.I.; Neerincx, A.H.; Vijverberg, S.J.; Kraneveld, A.D.; Maitland-van der Zee, A.H. Omics for the Future in Asthma. Semin. Immunopathol. 2020, 42, 111–126. [Google Scholar] [CrossRef]
- Tang, H.H.F.; Sly, P.D.; Holt, P.G.; Holt, K.E.; Inouye, M. Systems Biology and Big Data in Asthma and Allergy: Recent Discoveries and Emerging Challenges. Eur. Respir. J. 2020, 55, 1900844. [Google Scholar] [CrossRef]
- Shaw, D.E.; Sousa, A.R.; Fowler, S.J.; Fleming, L.J.; Roberts, G.; Corfield, J.; Pandis, I.; Bansal, A.T.; Bel, E.H.; Auffray, C.; et al. Clinical and Inflammatory Characteristics of the European U-BIOPRED Adult Severe Asthma Cohort. Eur. Respir. J. 2015, 46, 1308–1321. [Google Scholar] [CrossRef] [Green Version]
- Fleming, L.; Murray, C.; Bansal, A.T.; Hashimoto, S.; Bisgaard, H.; Bush, A.; Frey, U.; Hedlin, G.; Singer, F.; van Aalderen, W.M.; et al. The Burden of Severe Asthma in Childhood and Adolescence: Results from the Paediatric U-BIOPRED Cohorts. Eur. Respir. J. 2015, 46, 1322–1333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teague, W.G.; Phillips, B.R.; Fahy, J.V.; Wenzel, S.E.; Fitzpatrick, A.M.; Moore, W.C.; Hastie, A.T.; Bleecker, E.R.; Meyers, D.A.; Peters, S.P.; et al. Baseline Features of the Severe Asthma Research Program (SARP III) Cohort: Differences with Age. J. Allergy Clin. Immunol. Pract. 2018, 6, 545–554.e4. [Google Scholar] [CrossRef] [PubMed]
- Moore, W.C.; Bleecker, E.R.; Curran-Everett, D.; Erzurum, S.C.; Ameredes, B.T.; Bacharier, L.; Calhoun, W.J.; Castro, M.; Chung, K.F.; Clark, M.P.; et al. Characterization of the Severe Asthma Phenotype by the National Heart, Lung, and Blood Institute’s Severe Asthma Research Program. J. Allergy Clin. Immunol. 2007, 119, 405–413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moffatt, M.F.; Gut, I.G.; Demenais, F.; Strachan, D.P.; Bouzigon, E.; Heath, S.; von Mutius, E.; Farrall, M.; Lathrop, M.; Cookson, W.O.C.M. A Large-Scale, Consortium-Based Genomewide Association Study of Asthma. N. Engl. J. Med. 2010, 363, 1211–1221. [Google Scholar] [CrossRef] [Green Version]
- Shrine, N.; Portelli, M.A.; John, C.; Soler Artigas, M.; Bennett, N.; Hall, R.; Lewis, J.; Henry, A.P.; Billington, C.K.; Ahmad, A.; et al. Moderate-to-Severe Asthma in Individuals of European Ancestry: A Genome-Wide Association Study. Lancet Respir. Med. 2019, 7, 20–34. [Google Scholar] [CrossRef] [Green Version]
- Ober, C.; Nicolae, D.L. Meta-Analysis of Genome-Wide Association Studies of Asthma in Ethnically Diverse North American Populations. Nat. Genet. 2011, 43, 887–892. [Google Scholar] [CrossRef] [PubMed]
- Farzan, N.; Vijverberg, S.J.; Hernandez-Pacheco, N.; Bel, E.H.D.; Berce, V.; Bønnelykke, K.; Bisgaard, H.; Burchard, E.G.; Canino, G.; Celedón, J.C.; et al. 17q21 Variant Increases the Risk of Exacerbations in Asthmatic Children despite Inhaled Corticosteroids Use. Allergy 2018, 73, 2083–2088. [Google Scholar] [CrossRef] [Green Version]
- Turner, S.; Francis, B.; Vijverberg, S.; Pino-Yanes, M.; Maitland-van der Zee, A.H.; Basu, K.; Bignell, L.; Mukhopadhyay, S.; Tavendale, R.; Palmer, C.; et al. Childhood Asthma Exacerbations and the Arg16 Β2-Receptor Polymorphism: A Meta-Analysis Stratified by Treatment. J. Allergy Clin. Immunol. 2016, 138, 107–113.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slob, E.M.A.; Vijverberg, S.J.H.; Palmer, C.N.A.; Zazuli, Z.; Farzan, N.; Oliveri, N.M.B.; Pijnenburg, M.W.; Koppelman, G.H.; Maitland-van der Zee, A.H. Pharmacogenetics of Inhaled Long-Acting Beta2-Agonists in Asthma: A Systematic Review. Pediatr. Allergy Immunol. 2018, 29, 705–714. [Google Scholar] [CrossRef]
- Vijverberg, S.J.H.; Farzan, N.; Slob, E.M.A.; Neerincx, A.H.; Maitland-van der Zee, A.H. Treatment Response Heterogeneity in Asthma: The Role of Genetic Variation. Expert Rev. Respir. Med. 2018, 12, 55–65. [Google Scholar] [CrossRef] [Green Version]
- Joubert, B.R.; Felix, J.F.; Yousefi, P.; Bakulski, K.M.; Just, A.C.; Breton, C.; Reese, S.E.; Markunas, C.A.; Richmond, R.C.; Xu, C.-J.; et al. DNA Methylation in Newborns and Maternal Smoking in Pregnancy: Genome-Wide Consortium Meta-Analysis. Am. J. Hum. Genet. 2016, 98, 680–696. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.J.; Söderhäll, C.; Bustamante, M.; Baïz, N.; Gruzieva, O.; Gehring, U.; Mason, D.; Chatzi, L.; Basterrechea, M.; Llop, S.; et al. DNA Methylation in Childhood Asthma: An Epigenome-Wide Meta-Analysis. Lancet Respir. Med. 2018, 6, 379–388. [Google Scholar] [CrossRef] [Green Version]
- Reese, S.E.; Xu, C.-J.; den Dekker, H.T.; Lee, M.K.; Sikdar, S.; Ruiz-Arenas, C.; Merid, S.K.; Rezwan, F.I.; Page, C.M.; Ullemar, V.; et al. Epigenome-Wide Meta-Analysis of DNA Methylation and Childhood Asthma. J. Allergy Clin. Immunol. 2019, 143, 2062–2074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicodemus-Johnson, J.; Myers, R.A.; Sakabe, N.J.; Sobreira, D.R.; Hogarth, D.K.; Naureckas, E.T.; Sperling, A.I.; Solway, J.; White, S.R.; Nobrega, M.A.; et al. DNA Methylation in Lung Cells Is Associated with Asthma Endotypes and Genetic Risk. JCI Insight 2016, 1, 1–15. [Google Scholar] [CrossRef]
- Woodruff, P.G.; Modrek, B.; Choy, D.F.; Jia, G.; Abbas, A.R.; Ellwanger, A.; Arron, J.R.; Koth, L.L.; Fahy, J.V. T-Helper Type 2-Driven Inflammation Defines Major Subphenotypes of Asthma. Am. J. Respir. Crit. Care Med. 2009, 180, 388–395. [Google Scholar] [CrossRef]
- Howrylak, J.A.; Moll, M.; Weiss, S.T.; Raby, B.A.; Wu, W.; Xing, E.P. Gene Expression Profiling of Asthma Phenotypes Demonstrates Molecular Signatures of Atopy and Asthma Control. J. Allergy Clin. Immunol. 2016, 137, 1390–1397.e6. [Google Scholar] [CrossRef] [Green Version]
- Lefaudeux, D.; De Meulder, B.; Loza, M.J.; Peffer, N.; Rowe, A.; Baribaud, F.; Bansal, A.T.; Lutter, R.; Sousa, A.R.; Corfield, J.; et al. U-BIOPRED Clinical Adult Asthma Clusters Linked to a Subset of Sputum Omics. J. Allergy Clin. Immunol. 2017, 139, 1797–1807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuo, C.-H.S.; Pavlidis, S.; Loza, M.; Baribaud, F.; Rowe, A.; Pandis, I.; Hoda, U.; Rossios, C.; Sousa, A.; Wilson, S.J.; et al. A Transcriptome-Driven Analysis of Epithelial Brushings and Bronchial Biopsies to Define Asthma Phenotypes in U-BIOPRED. Am. J. Respir. Crit. Care Med. 2017, 195, 443–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuo, C.-H.S.; Pavlidis, S.; Loza, M.; Baribaud, F.; Rowe, A.; Pandis, I.; Sousa, A.; Corfield, J.; Djukanovic, R.; Lutter, R.; et al. T-Helper Cell Type 2 (Th2) and Non-Th2 Molecular Phenotypes of Asthma Using Sputum Transcriptomics in U-BIOPRED. Eur. Respir. J. 2017, 49, 1602135. [Google Scholar] [CrossRef]
- Bigler, J.; Boedigheimer, M.; Schofield, J.P.R.; Skipp, P.J.; Corfield, J.; Rowe, A.; Sousa, A.R.; Timour, M.; Twehues, L.; Hu, X.; et al. A Severe Asthma Disease Signature from Gene Expression Profiling of Peripheral Blood from U-BIOPRED Cohorts. Am. J. Respir. Crit. Care Med. 2017, 195, 1311–1320. [Google Scholar] [CrossRef] [PubMed]
- Hekking, P.-P.; Loza, M.J.; Pavlidis, S.; de Meulder, B.; Lefaudeux, D.; Baribaud, F.; Auffray, C.; Wagener, A.H.; Brinkman, P.; Lutter, R.; et al. Pathway Discovery Using Transcriptomic Profiles in Adult-Onset Severe Asthma. J. Allergy Clin. Immunol. 2018, 141, 1280–1290. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, K.; Pavlidis, S.; Ng Kee Kwong, F.; Hoda, U.; Rossios, C.; Sun, K.; Loza, M.; Baribaud, F.; Chanez, P.; Fowler, S.J.; et al. Sputum Proteomics and Airway Cell Transcripts of Current and Ex-Smokers with Severe Asthma in U-BIOPRED: An Exploratory Analysis. Eur. Respir. J. 2018, 51, 1702173. [Google Scholar] [CrossRef]
- Schofield, J.P.R.; Burg, D.; Nicholas, B.; Strazzeri, F.; Brandsma, J.; Staykova, D.; Folisi, C.; Bansal, A.T.; Xian, Y.; Guo, Y.; et al. Stratification of Asthma Phenotypes by Airway Proteomic Signatures. J. Allergy Clin. Immunol. 2019, 144, 70–82. [Google Scholar] [CrossRef] [Green Version]
- Modena, B.D.; Bleecker, E.R.; Busse, W.W.; Erzurum, S.C.; Gaston, B.M.; Jarjour, N.N.; Meyers, D.A.; Milosevic, J.; Tedrow, J.R.; Wu, W.; et al. Gene Expression Correlated with Severe Asthma Characteristics Reveals Heterogeneous Mechanisms of Severe Disease. Am. J. Respir. Crit. Care Med. 2017, 195, 1449–1463. [Google Scholar] [CrossRef]
- Brasier, A.R.; Victor, S.; Ju, H.; Busse, W.W.; Curran-Everett, D.; Bleecker, E.; Castro, M.; Chung, K.F.; Gaston, B.; Israel, E.; et al. Predicting Intermediate Phenotypes in Asthma Using Bronchoalveolar Lavage-Derived Cytokines. Clin. Transl. Sci. 2010, 3, 147–157. [Google Scholar] [CrossRef]
- Hastie, A.T.; Moore, W.C.; Meyers, D.A.; Vestal, P.L.; Li, H.; Peters, S.P.; Bleecker, E.R.; National Heart, Lung, and Blood Institute Severe Asthma Research Program. Analyses of Asthma Severity Phenotypes and Inflammatory Proteins in Subjects Stratified by Sputum Granulocytes. J. Allergy Clin. Immunol. 2010, 125, 1028–1036.e13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hastie, A.T.; Steele, C.; Dunaway, C.W.; Moore, W.C.; Rector, B.M.; Ampleford, E.; Li, H.; Denlinger, L.C.; Jarjour, N.; Meyers, D.A.; et al. Complex Association Patterns for Inflammatory Mediators in Induced Sputum from Subjects with Asthma. Clin. Exp. Allergy 2018, 48, 787–797. [Google Scholar] [CrossRef]
- Kelly, R.S.; Dahlin, A.; McGeachie, M.J.; Qiu, W.; Sordillo, J.; Wan, E.S.; Wu, A.C.; Lasky-Su, J. Asthma Metabolomics and the Potential for Integrative Omics in Research and the Clinic. Chest 2017, 151, 262–277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montuschi, P.; Mores, N.; Trové, A.; Mondino, C.; Barnes, P.J. The Electronic Nose in Respiratory Medicine. Respiration 2013, 85, 72–84. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, B.; Basanta, M.; Cadden, P.; Singh, D.; Douce, D.; Woodcock, A.; Fowler, S.J. Non-Invasive Phenotyping Using Exhaled Volatile Organic Compounds in Asthma. Thorax 2011, 66, 804–809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinha, A.; Desiraju, K.; Aggarwal, K.; Kutum, R.; Roy, S.; Lodha, R.; Kabra, S.K.; Ghosh, B.; Sethi, T.; Agrawal, A. Exhaled Breath Condensate Metabolome Clusters for Endotype Discovery in Asthma. J. Transl. Med. 2017, 15, 262. [Google Scholar] [CrossRef] [Green Version]
- Van der Schee, M.P.; Palmay, R.; Cowan, J.O.; Taylor, D.R. Predicting Steroid Responsiveness in Patients with Asthma Using Exhaled Breath Profiling. Clin. Exp. Allergy 2013, 43, 1217–1225. [Google Scholar] [CrossRef] [PubMed]
- Brinkman, P.; Wagener, A.H.; Hekking, P.-P.; Bansal, A.T.; Maitland-van der Zee, A.-H.; Wang, Y.; Weda, H.; Knobel, H.H.; Vink, T.J.; Rattray, N.J.; et al. Identification and Prospective Stability of Electronic Nose (ENose)–Derived Inflammatory Phenotypes in Patients with Severe Asthma. J. Allergy Clin. Immunol. 2019, 143, 1811–1820.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, J.; Kim, S.H.; Lee, H.S.; Choi, G.S.; Jung, Y.S.; Ryu, D.H.; Park, H.S.; Hwang, G.S. Serum Metabolomics Reveals Pathways and Biomarkers Associated with Asthma Pathogenesis. Clin. Exp. Allergy 2013, 43, 425–433. [Google Scholar] [CrossRef]
- Reinke, S.N.; Gallart-Ayala, H.; Gómez, C.; Checa, A.; Fauland, A.; Naz, S.; Kamleh, M.A.; Djukanović, R.; Hinks, T.S.C.; Wheelock, C.E. Metabolomics Analysis Identifies Different Metabotypes of Asthma Severity. Eur. Respir. J. 2017, 49, 1601740. [Google Scholar] [CrossRef] [Green Version]
- Kelly, R.S.; Sordillo, J.E.; Lasky-Su, J.; Dahlin, A.; Perng, W.; Rifas-Shiman, S.L.; Weiss, S.T.; Gold, D.R.; Litonjua, A.A.; Hivert, M.-F.; et al. Plasma Metabolite Profiles in Children with Current Asthma. Clin. Exp. Allergy 2018, 48, 1297–1304. [Google Scholar] [CrossRef] [PubMed]
- Durack, J.; Boushey, H.A.; Huang, Y.J. Incorporating the Airway Microbiome into Asthma Phenotyping: Moving toward Personalized Medicine for Noneosinophilic Asthma. J. Allergy Clin. Immunol. 2018, 141, 82–83. [Google Scholar] [CrossRef]
- Wang, G.; McDonald, V.M. Contemporary Concise Review 2020: Asthma. Respirology 2021, 26, 804–811. [Google Scholar] [CrossRef]
- Tang, H.H.F.; Teo, S.M.; Sly, P.D.; Holt, P.G.; Inouye, M. The Intersect of Genetics, Environment, and Microbiota in Asthma—Perspectives and Challenges. J. Allergy Clin. Immunol. 2021, 147, 781–793. [Google Scholar] [CrossRef] [PubMed]
- Sokolowska, M.; Frei, R.; Lunjani, N.; Akdis, C.A.; O’Mahony, L. Microbiome and Asthma. Asthma Res. Pract. 2018, 4, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.J.; Marsland, B.J.; Bunyavanich, S.; O’mahony, L.; Leung, D.Y.M.; Muraro, A.; Fleisher, T.A. The Microbiome in Allergic Disease: Current Understanding and Future Opportunities. J. Allergy Clin. Immunol. 2018, 139, 1099–1110. [Google Scholar] [CrossRef] [Green Version]
- Stiemsma, L.T.; Turvey, S.E. Asthma and the Microbiome: Defining the Critical Window in Early Life. Allergy Asthma Clin. Immunol. 2017, 13, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chotirmall, S.H.; Gellatly, S.L.; Budden, K.F.; Mac Aogáin, M.; Shukla, S.D.; Wood, D.L.A.; Hugenholtz, P.; Pethe, K.; Hansbro, P.M. Microbiomes in Respiratory Health and Disease: An Asia-Pacific Perspective. Respirology 2017, 22, 240–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ege, M.J. The Hygiene Hypothesis in the Age of the Microbiome. Ann. Am. Thorac. Soc. 2017, 14, S348–S353. [Google Scholar] [CrossRef] [PubMed]
- Garn, H.; Potaczek, D.P.; Pfefferle, P.I. The Hygiene Hypothesis and New Perspectives—Current Challenges Meeting an Old Postulate. Front. Immunol. 2021, 12, 637087. [Google Scholar] [CrossRef]
- Sbihi, H.; Boutin, R.C.T.; Cutler, C.; Suen, M.; Finlay, B.B.; Turvey, S.E. Thinking Bigger: How Early-Life Environmental Exposures Shape the Gut Microbiome and Influence the Development of Asthma and Allergic Disease. Allergy 2019, 74, 2103–2115. [Google Scholar] [CrossRef] [Green Version]
- Ege, M.J.; Mayer, M.; Normand, A.-C.; Genuneit, J.; Cookson, W.O.C.M.; Braun-Fahrländer, C.; Heederik, D.; Piarroux, R.; von Mutius, E. Exposure to Environmental Microorganisms and Childhood Asthma. N. Engl. J. Med. 2011, 364, 701–709. [Google Scholar] [CrossRef]
- Hanski, I.; von Hertzen, L.; Fyhrquist, N.; Koskinen, K.; Torppa, K.; Laatikainen, T.; Karisola, P.; Auvinen, P.; Paulin, L.; Makela, M.J.; et al. Environmental Biodiversity, Human Microbiota, and Allergy Are Interrelated. Proc. Natl. Acad. Sci. USA 2012, 109, 8334–8339. [Google Scholar] [CrossRef] [Green Version]
- Jartti, T.; Bønnelykke, K.; Elenius, V.; Feleszko, W. Role of Viruses in Asthma. Semin. Immunopathol. 2020, 42, 61–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandenborght, L.-E.; Enaud, R.; Urien, C.; Coron, N.; Girodet, P.-O.; Ferreira, S.; Berger, P.; Delhaes, L. Type 2–High Asthma Is Associated with a Specific Indoor Mycobiome and Microbiome. J. Allergy Clin. Immunol. 2021, 147, 1296–1305.e6. [Google Scholar] [CrossRef] [PubMed]
- Du Toit, A. The Gut Microbiome and Mental Health. Nat. Rev. Microbiol. 2019, 17, 196. [Google Scholar] [CrossRef] [PubMed]
- Frati, F.; Salvatori, C.; Incorvaia, C.; Bellucci, A.; Di Cara, G.; Marcucci, F.; Esposito, S. The Role of the Microbiome in Asthma: The Gut–Lung Axis. Int. J. Mol. Sci. 2018, 20, 123. [Google Scholar] [CrossRef] [Green Version]
- Durack, J.; Lynch, S.V. The Gut Microbiome: Relationships with Disease and Opportunities for Therapy. J. Exp. Med. 2019, 216, 20–40. [Google Scholar] [CrossRef] [Green Version]
- Gu, S.; Chen, Y.; Wu, Z.; Chen, Y.; Gao, H.; Lv, L.; Guo, F.; Zhang, X.; Luo, R.; Huang, C.; et al. Alterations of the Gut Microbiota in Patients With Coronavirus Disease 2019 or H1N1 Influenza. Clin. Infect. Dis. 2020, 71, 2669–2678. [Google Scholar] [CrossRef]
- Chattopadhyay, I.; Dhar, R.; Pethusamy, K.; Seethy, A.; Srivastava, T.; Sah, R.; Sharma, J.; Karmakar, S. Exploring the Role of Gut Microbiome in Colon Cancer. Appl. Biochem. Biotechnol. 2021, 193, 1780–1799. [Google Scholar] [CrossRef] [PubMed]
- Fujimura, K.E.; Lynch, S.V. Microbiota in Allergy and Asthma and the Emerging Relationship with the Gut Microbiome. Cell Host Microbe 2015, 17, 592–602. [Google Scholar] [CrossRef] [Green Version]
- Schirmer, M.; Smeekens, S.P.; Vlamakis, H.; Jaeger, M.; Oosting, M.; Franzosa, E.A.; ter Horst, R.; Jansen, T.; Jacobs, L.; Bonder, M.J.; et al. Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell 2016, 167, 1125–1136.e8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geva-Zatorsky, N.; Sefik, E.; Kua, L.; Pasman, L.; Tan, T.G.; Ortiz-Lopez, A.; Yanortsang, T.B.; Yang, L.; Jupp, R.; Mathis, D.; et al. Mining the Human Gut Microbiota for Immunomodulatory Organisms. Cell 2017, 168, 928–943.e11. [Google Scholar] [CrossRef] [Green Version]
- Zimmermann, M.; Zimmermann-Kogadeeva, M.; Wegmann, R.; Goodman, A.L. Mapping Human Microbiome Drug Metabolism by Gut Bacteria and Their Genes. Nature 2019, 570, 462–467. [Google Scholar] [CrossRef] [PubMed]
- Klünemann, M.; Andrejev, S.; Blasche, S.; Mateus, A.; Phapale, P.; Devendran, S.; Vappiani, J.; Simon, B.; Scott, T.A.; Kafkia, E.; et al. Bioaccumulation of Therapeutic Drugs by Human Gut Bacteria. Nature 2021, 597, 533–538. [Google Scholar] [CrossRef]
- Arrieta, M.-C.; Stiemsma, L.T.; Dimitriu, P.A.; Thorson, L.; Russell, S.; Yurist-Doutsch, S.; Kuzeljevic, B.; Gold, M.J.; Britton, H.M.; Lefebvre, D.L.; et al. Early Infancy Microbial and Metabolic Alterations Affect Risk of Childhood Asthma. Sci. Transl. Med. 2015, 7, 307ra152. [Google Scholar] [CrossRef] [PubMed]
- Fujimura, K.E.; Sitarik, A.R.; Havstad, S.; Lin, D.L.; Levan, S.; Fadrosh, D.; Panzer, A.R.; LaMere, B.; Rackaityte, E.; Lukacs, N.W.; et al. Neonatal Gut Microbiota Associates with Childhood Multisensitized Atopy and T Cell Differentiation. Nat. Med. 2016, 22, 1187–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stokholm, J.; Blaser, M.J.; Thorsen, J.; Rasmussen, M.A.; Waage, J.; Vinding, R.K.; Schoos, A.-M.M.; Kunøe, A.; Fink, N.R.; Chawes, B.L.; et al. Maturation of the Gut Microbiome and Risk of Asthma in Childhood. Nat. Commun. 2018, 9, 141. [Google Scholar] [CrossRef] [PubMed]
- Martín-Orozco, E.; Norte-Muñoz, M.; Martínez-García, J. Regulatory T Cells in Allergy and Asthma. Front. Pediatr. 2017, 5, 117. [Google Scholar] [CrossRef]
- Pandiyan, P.; Bhaskaran, N.; Zou, M.; Schneider, E.; Jayaraman, S.; Huehn, J. Microbiome Dependent Regulation of Tregs and Th17 Cells in Mucosa. Front. Immunol. 2019, 10, 426. [Google Scholar] [CrossRef] [Green Version]
- McLoughlin, R.M.; Mills, K.H.G. Influence of Gastrointestinal Commensal Bacteria on the Immune Responses That Mediate Allergy and Asthma. J. Allergy Clin. Immunol. 2011, 127, 1097–1107. [Google Scholar] [CrossRef]
- Lee-Sarwar, K.A.; Lasky-Su, J.; Kelly, R.S.; Litonjua, A.A.; Weiss, S.T. Gut Microbial-Derived Metabolomics of Asthma. Metabolites 2020, 10, 97. [Google Scholar] [CrossRef] [Green Version]
- MacFabe, D.F. Short-Chain Fatty Acid Fermentation Products of the Gut Microbiome: Implications in Autism Spectrum Disorders. Microb. Ecol. Health Dis. 2012, 23, 19260. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Oshima, K.; Suda, W.; Nagano, Y.; Nishikawa, H.; Fukuda, S.; Saito, T.; Narushima, S.; Hase, K.; et al. Treg Induction by a Rationally Selected Mixture of Clostridia Strains from the Human Microbiota. Nature 2013, 500, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Sefik, E.; Geva-Zatorsky, N.; Oh, S.; Konnikova, L.; Zemmour, D.; McGuire, A.M.; Burzyn, D.; Ortiz-Lopez, A.; Lobera, M.; Yang, J.; et al. MUCOSAL IMMUNOLOGY. Individual Intestinal Symbionts Induce a Distinct Population of RORγ+ Regulatory T Cells. Science 2015, 349, 993–997. [Google Scholar] [CrossRef] [Green Version]
- Roduit, C.; Frei, R.; Ferstl, R.; Loeliger, S.; Westermann, P.; Rhyner, C.; Schiavi, E.; Barcik, W.; Rodriguez-Perez, N.; Wawrzyniak, M.; et al. High Levels of Butyrate and Propionate in Early Life Are Associated with Protection against Atopy. Allergy 2019, 74, 799–809. [Google Scholar] [CrossRef]
- Cui, L.; Morris, A.; Huang, L.; Beck, J.M.; Twigg, H.L.; von Mutius, E.; Ghedin, E. The Microbiome and the Lung. Ann. Am. Thorac. Soc. 2014, 11, S227–S232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lynch, S.V. The Lung Microbiome and Airway Disease. Ann. Am. Thorac. Soc. 2016, 13, S462–S465. [Google Scholar] [CrossRef] [PubMed]
- Dima, E.; Kyriakoudi, A.; Kaponi, M.; Vasileiadis, I.; Stamou, P.; Koutsoukou, A.; Koulouris, N.G.; Rovina, N. The Lung Microbiome Dynamics between Stability and Exacerbation in Chronic Obstructive Pulmonary Disease (COPD): Current Perspectives. Respir. Med. 2019, 157, 1–6. [Google Scholar] [CrossRef]
- Hakansson, A.P.; Orihuela, C.J.; Bogaert, D. Bacterial-Host Interactions: Physiology and Pathophysiology of Respiratory Infection. Physiol. Rev. 2018, 98, 781–811. [Google Scholar] [CrossRef]
- Loverdos, K.; Bellos, G.; Kokolatou, L.; Vasileiadis, I.; Giamarellos, E.; Pecchiari, M.; Koulouris, N.; Koutsoukou, A.; Rovina, N. Lung Microbiome in Asthma: Current Perspectives. J. Clin. Med. 2019, 8, 1967. [Google Scholar] [CrossRef] [Green Version]
- Barcik, W.; Boutin, R.C.T.; Sokolowska, M.; Finlay, B.B. The Role of Lung and Gut Microbiota in the Pathology of Asthma. Immunity 2020, 52, 241–255. [Google Scholar] [CrossRef] [Green Version]
- Hilty, M.; Burke, C.; Pedro, H.; Cardenas, P.; Bush, A.; Bossley, C.; Davies, J.; Ervine, A.; Poulter, L.; Pachter, L.; et al. Disordered Microbial Communities in Asthmatic Airways. PLoS ONE 2010, 5, e8578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Green, B.J.; Wiriyachaiporn, S.; Grainge, C.; Rogers, G.B.; Kehagia, V.; Lau, L.; Carroll, M.P.; Bruce, K.D.; Howarth, P.H. Potentially Pathogenic Airway Bacteria and Neutrophilic Inflammation in Treatment Resistant Severe Asthma. PLoS ONE 2014, 9, e100645. [Google Scholar] [CrossRef] [PubMed]
- Durack, J.; Lynch, S.V.; Nariya, S.; Bhakta, N.R.; Beigelman, A.; Castro, M.; Dyer, A.-M.; Israel, E.; Kraft, M.; Martin, R.J.; et al. Features of the Bronchial Bacterial Microbiome Associated with Atopy, Asthma, and Responsiveness to Inhaled Corticosteroid Treatment. J. Allergy Clin. Immunol. 2017, 140, 63–75. [Google Scholar] [CrossRef] [Green Version]
- Budden, K.F.; Gellatly, S.L.; Wood, D.L.A.; Cooper, M.A.; Morrison, M.; Hugenholtz, P.; Hansbro, P.M. Emerging Pathogenic Links between Microbiota and the Gut-Lung Axis. Nat. Rev. Microbiol. 2017, 15, 55–63. [Google Scholar] [CrossRef]
- Zhang, D.; Li, S.; Wang, N.; Tan, H.-Y.; Zhang, Z.; Feng, Y. The Cross-Talk Between Gut Microbiota and Lungs in Common Lung Diseases. Front. Microbiol. 2020, 11, 301. [Google Scholar] [CrossRef] [PubMed]
- Laforest-Lapointe, I.; Arrieta, M.-C. Patterns of Early-Life Gut Microbial Colonization during Human Immune Development: An Ecological Perspective. Front. Immunol. 2017, 8, 788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Tilburg Bernardes, E.; Gutierrez, M.W.; Arrieta, M.-C. The Fungal Microbiome and Asthma. Front. Cell. Infect. Microbiol. 2020, 10, 583418. [Google Scholar] [CrossRef]
- Guillien, A.; Cadiou, S.; Slama, R.; Siroux, V. The Exposome Approach to Decipher the Role of Multiple Environmental and Lifestyle Determinants in Asthma. Int. J. Environ. Res. Public Health 2021, 18, 1138. [Google Scholar] [CrossRef]
- Shimoda, T.; Obase, Y.; Kishikawa, R.; Iwanaga, T. Influence of Cigarette Smoking on Airway Inflammation and Inhaled Corticosteroid Treatment in Patients with Asthma. Allergy Asthma Proc. 2016, 37, 50–58. [Google Scholar] [CrossRef]
- Katsaounou, P.; Ioannou, M.; Hyland, M.E.; Odemyr, M.; Spranger, O.; Lindberg, A.; Gasser, M.; Conde, L.G.; Jaumont, X.; Kasujee, I. Smoking Asthmatics, a Neglected Large Phenotype of Asthmatic Patients. Open J. Asthma 2019, 3, 1–8. [Google Scholar]
- Chatkin, J.M.; Dullius, C.R. The Management of Asthmatic Smokers. Asthma Res. Pract. 2016, 2, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Morris, A.; Beck, J.M.; Schloss, P.D.; Campbell, T.B.; Crothers, K.; Curtis, J.L.; Flores, S.C.; Fontenot, A.P.; Ghedin, E.; Huang, L.; et al. Comparison of the Respiratory Microbiome in Healthy Nonsmokers and Smokers. Am. J. Respir. Crit. Care Med. 2013, 187, 1067–1075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Peters, B.A.; Dominianni, C.; Zhang, Y.; Pei, Z.; Yang, L.; Ma, Y.; Purdue, M.P.; Jacobs, E.J.; Gapstur, S.M.; et al. Cigarette Smoking and the Oral Microbiome in a Large Study of American Adults. ISME J. 2016, 10, 2435–2446. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Rabassa, M.; López, P.; Rodríguez-Santiago, R.E.; Cases, A.; Felici, M.; Sánchez, R.; Yamamura, Y.; Rivera-Amill, V. Cigarette Smoking Modulation of Saliva Microbial Composition and Cytokine Levels. Int. J. Environ. Res. Public Health 2018, 15, 2479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savin, Z.; Kivity, S.; Yonath, H.; Yehuda, S. Smoking and the Intestinal Microbiome. Arch. Microbiol. 2018, 200, 677–684. [Google Scholar] [CrossRef]
- Huang, C.; Shi, G. Smoking and Microbiome in Oral, Airway, Gut and Some Systemic Diseases. J. Transl. Med. 2019, 17, 225. [Google Scholar] [CrossRef] [Green Version]
- Nolan-Kenney, R.; Wu, F.; Hu, J.; Yang, L.; Kelly, D.; Li, H.; Jasmine, F.; Kibriya, M.G.; Parvez, F.; Shaheen, I.; et al. The Association Between Smoking and Gut Microbiome in Bangladesh. Nicotine Tob. Res. 2020, 22, 1339–1346. [Google Scholar] [CrossRef]
- Biedermann, L.; Zeitz, J.; Mwinyi, J.; Sutter-Minder, E.; Rehman, A.; Ott, S.J.; Steurer-Stey, C.; Frei, A.; Frei, P.; Scharl, M.; et al. Smoking Cessation Induces Profound Changes in the Composition of the Intestinal Microbiota in Humans. PLoS ONE 2013, 8, e59260. [Google Scholar]
- Çolak, Y.; Afzal, S.; Nordestgaard, B.G.; Lange, P. Characteristics and Prognosis of Never-Smokers and Smokers with Asthma in the Copenhagen General Population Study. A Prospective Cohort Study. Am. J. Respir. Crit. Care Med. 2015, 192, 172–181. [Google Scholar] [CrossRef] [PubMed]
- Munck, C.; Helby, J.; Westergaard, C.G.; Porsbjerg, C.; Backer, V.; Hansen, L.H. Smoking Cessation and the Microbiome in Induced Sputum Samples from Cigarette Smoking Asthma Patients. PLoS ONE 2016, 11, e0158622. [Google Scholar] [CrossRef] [Green Version]
- Wood, L.G. Diet, Obesity, and Asthma. Ann. Am. Thorac. Soc. 2017, 14, S332–S338. [Google Scholar] [CrossRef]
- Tashiro, H.; Shore, S.A. Obesity and Severe Asthma. Allergol. Int. 2019, 68, 135–142. [Google Scholar] [CrossRef]
- Mukadam, S.; Zacharias, J.; Henao, M.P.; Kraschnewski, J.; Ishmael, F. Differential Effects of Obesity on Eosinophilic vs. Non-Eosinophilic Asthma Subtypes. J. Asthma 2018, 55, 720–725. [Google Scholar] [CrossRef] [PubMed]
- Miethe, S.; Karsonova, A.; Karaulov, A.; Renz, H. Obesity and Asthma. J. Allergy Clin. Immunol. 2020, 146, 685–693. [Google Scholar] [CrossRef]
- Andrianasolo, R.M.; Kesse-Guyot, E.; Adjibade, M.; Hercberg, S.; Galan, P.; Varraso, R. Associations between Dietary Scores with Asthma Symptoms and Asthma Control in Adults. Eur. Respir. J. 2018, 52, 1702572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; He, X.; Huang, J. Diet Effects in Gut Microbiome and Obesity. J. Food Sci. 2014, 79, R442–R451. [Google Scholar] [CrossRef] [Green Version]
- Hullar, M.A.J.; Fu, B.C. Diet, the Gut Microbiome, and Epigenetics. Cancer J. 2014, 20, 170–175. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.J.; Gerasimidis, K.; Edwards, C.A.; Shaikh, M.G. Role of Gut Microbiota in the Aetiology of Obesity: Proposed Mechanisms and Review of the Literature. J. Obes. 2016, 2016, 7353642. [Google Scholar] [CrossRef] [Green Version]
- Nagpal, R.; Kumar, M.; Yadav, A.K.; Hemalatha, R.; Yadav, H.; Marotta, F.; Yamashiro, Y. Gut Microbiota in Health and Disease: An Overview Focused on Metabolic Inflammation. Benef. Microbes 2016, 7, 181–194. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.; Shore, S.A. Obesity, Asthma, and the Microbiome. Physiology 2016, 31, 108–116. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.J.; Nariya, S.; Harris, J.M.; Lynch, S.V.; Choy, D.F.; Arron, J.R.; Boushey, H. The Airway Microbiome in Patients with Severe Asthma: Associations with Disease Features and Severity. J. Allergy Clin. Immunol. 2015, 136, 874–884. [Google Scholar] [CrossRef] [Green Version]
- Michalovich, D.; Rodriguez-Perez, N.; Smolinska, S.; Pirozynski, M.; Mayhew, D.; Uddin, S.; Van Horn, S.; Sokolowska, M.; Altunbulakli, C.; Eljaszewicz, A.; et al. Obesity and Disease Severity Magnify Disturbed Microbiome-Immune Interactions in Asthma Patients. Nat. Commun. 2019, 10, 5711. [Google Scholar] [CrossRef] [Green Version]
- Simpson, J.L.; Daly, J.; Baines, K.J.; Yang, I.A.; Upham, J.W.; Reynolds, P.N.; Hodge, S.; James, A.L.; Hugenholtz, P.; Willner, D.; et al. Airway Dysbiosis: Haemophilus Influenzae and Tropheryma in Poorly Controlled Asthma. Eur. Respir. J. 2016, 47, 792–800. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Cox, M.; Liang, Z.; Brinkmann, F.; Cardenas, P.A.; Duff, R.; Bhavsar, P.; Cookson, W.; Moffatt, M.; Chung, K.F. Airway Microbiota in Severe Asthma and Relationship to Asthma Severity and Phenotypes. PLoS ONE 2016, 11, e0152724. [Google Scholar] [CrossRef] [Green Version]
- Sverrild, A.; Kiilerich, P.; Brejnrod, A.; Pedersen, R.; Porsbjerg, C.; Bergqvist, A.; Erjefält, J.S.; Kristiansen, K.; Backer, V. Eosinophilic Airway Inflammation in Asthmatic Patients Is Associated with an Altered Airway Microbiome. J. Allergy Clin. Immunol. 2017, 140, 407–417.e11. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Qiu, R.; Yang, Z.; Li, J.; Chung, K.F.; Zhong, N.; Zhang, Q. Sputum Microbiota in Severe Asthma Patients: Relationship to Eosinophilic Inflammation. Respir. Med. 2017, 131, 192–198. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Li, H.; Ma, Q.; Zhang, Q.; Wang, C. Neutrophilic Asthma Is Associated with Increased Airway Bacterial Burden and Disordered Community Composition. Biomed. Res. Int. 2018, 2018, 9230234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, S.L.; Leong, L.E.X.; Choo, J.M.; Wesselingh, S.; Yang, I.A.; Upham, J.W.; Reynolds, P.N.; Hodge, S.; James, A.L.; Jenkins, C.; et al. Inflammatory Phenotypes in Patients with Severe Asthma Are Associated with Distinct Airway Microbiology. J. Allergy Clin. Immunol. 2018, 141, 94–103.e15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghebre, M.A.; Pang, P.H.; Diver, S.; Desai, D.; Bafadhel, M.; Haldar, K.; Kebadze, T.; Cohen, S.; Newbold, P.; Rapley, L.; et al. Biological Exacerbation Clusters Demonstrate Asthma and Chronic Obstructive Pulmonary Disease Overlap with Distinct Mediator and Microbiome Profiles. J. Allergy Clin. Immunol. 2018, 141, 2027–2036.e12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, Z.; Wang, G.; Gibson, P.; Guan, X.; Zhang, W.; Zheng, R.; Chen, F.; Wang, Z.; Wang, F. Airway Microbiome in Different Inflammatory Phenotypes of Asthma: A Cross-Sectional Study in Northeast China. Int. J. Med. Sci. 2019, 16, 477–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durack, J.; Christian, L.S.; Nariya, S.; Gonzalez, J.; Bhakta, N.R.; Ansel, K.M.; Beigelman, A.; Castro, M.; Dyer, A.-M.; Israel, E.; et al. Distinct Associations of Sputum and Oral Microbiota with Atopic, Immunologic, and Clinical Features in Mild Asthma. J. Allergy Clin. Immunol. 2020, 146, 1016–1026. [Google Scholar] [CrossRef]
- Abdel-Aziz, M.I.; Brinkman, P.; Vijverberg, S.J.H.; Neerincx, A.H.; Riley, J.H.; Bates, S.; Hashimoto, S.; Kermani, N.Z.; Chung, K.F.; Djukanovic, R.; et al. Sputum Microbiome Profiles Identify Severe Asthma Phenotypes of Relative Stability at 12 to 18 Months. J. Allergy Clin. Immunol. 2021, 147, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Frenette, P.S. Cross Talk between Neutrophils and the Microbiota. Blood 2019, 133, 2168–2177. [Google Scholar] [CrossRef]
- Chen, J.; Zheng, P.; Liu, Y.; Zhong, X.; Wang, H.; Guo, Y.; Xie, P. Sex Differences in Gut Microbiota in Patients with Major Depressive Disorder. Neuropsychiatr. Dis. Treat. 2018, 14, 647–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simpson, J.L.; Powell, H.; Boyle, M.J.; Scott, R.J.; Gibson, P.G. Clarithromycin Targets Neutrophilic Airway Inflammation in Refractory Asthma. Am. J. Respir. Crit. Care Med. 2008, 177, 148–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, A.; Laxman, B.; Naureckas, E.T.; Hogarth, D.K.; Sperling, A.I.; Solway, J.; Ober, C.; Gilbert, J.A.; White, S.R. Associations between Fungal and Bacterial Microbiota of Airways and Asthma Endotypes. J. Allergy Clin. Immunol. 2019, 144, 1214–1227.e7. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Yu, Y.; Du, W.; Liu, Y.; Dai, R.; Tang, W.; Wang, P.; Zhang, C.; Shi, G. Fungal and Bacterial Microbiome Dysbiosis and Imbalance of Trans-Kingdom Network in Asthma. Clin. Transl. Allergy 2020, 10, 42. [Google Scholar] [CrossRef] [PubMed]
- Denner, D.R.; Sangwan, N.; Becker, J.B.; Hogarth, D.K.; Oldham, J.; Castillo, J.; Sperling, A.I.; Solway, J.; Naureckas, E.T.; Gilbert, J.A.; et al. Corticosteroid Therapy and Airflow Obstruction Influence the Bronchial Microbiome, Which Is Distinct from That of Bronchoalveolar Lavage in Asthmatic Airways. J. Allergy Clin. Immunol. 2016, 137, 1398–1405.e3. [Google Scholar] [CrossRef] [Green Version]
- Jung, J.-W.; Choi, J.-C.; Shin, J.-W.; Kim, J.-Y.; Park, I.-W.; Choi, B.W.; Park, H.-W.; Cho, S.-H.; Kim, K.; Kang, H.-R. Lung Microbiome Analysis in Steroid-Naïve Asthma Patients by Using Whole Sputum. Tuberc. Respir. Dis. 2016, 79, 165–178. [Google Scholar] [CrossRef] [Green Version]
- McCauley, K.; Durack, J.; Valladares, R.; Fadrosh, D.W.; Lin, D.L.; Calatroni, A.; LeBeau, P.K.; Tran, H.T.; Fujimura, K.E.; LaMere, B.; et al. Distinct Nasal Airway Bacterial Microbiotas Differentially Relate to Exacerbation in Pediatric Patients with Asthma. J. Allergy Clin. Immunol. 2019, 144, 1187–1197. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.J.; Zain, N.M.M.; Hearson, G.; Rivett, D.W.; Koller, G.; Wooldridge, D.J.; Rose, G.; Gharbia, S.E.; Forbes, B.; Bruce, K.D.; et al. The Airways Microbiome of Individuals with Asthma Treated with High and Low Doses of Inhaled Corticosteroids. PLoS ONE 2020, 15, e0244681. [Google Scholar] [CrossRef]
- Goleva, E.; Jackson, L.P.; Harris, J.K.; Robertson, C.E.; Sutherland, E.R.; Hall, C.F.; Good, J.T.; Gelfand, E.W.; Martin, R.J.; Leung, D.Y.M. The Effects of Airway Microbiome on Corticosteroid Responsiveness in Asthma. Am. J. Respir. Crit. Care Med. 2013, 188, 1193–1201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thorsen, J.; Stokholm, J.; Rasmussen, M.A.; Mortensen, M.S.; Brejnrod, A.D.; Hjelmsø, M.; Shah, S.; Chawes, B.; Bønnelykke, K.; Sørensen, S.J.; et al. The Airway Microbiota Modulates Effect of Azithromycin Treatment for Episodes of Recurrent Asthma-like Symptoms in Preschool Children: A Randomized Clinical Trial. Am. J. Respir. Crit. Care Med. 2021, 204, 149–158. [Google Scholar] [CrossRef]
- Benton, A.S.; Wang, Z.; Lerner, J.; Foerster, M.; Teach, S.J.; Freishtat, R.J. Overcoming Heterogeneity in Pediatric Asthma: Tobacco Smoke and Asthma Characteristics within Phenotypic Clusters in an African American Cohort. J. Asthma 2010, 47, 728–734. [Google Scholar] [CrossRef]
- Castro-Nallar, E.; Shen, Y.; Freishtat, R.J.; Pérez-Losada, M.; Manimaran, S.; Liu, G.; Johnson, W.E.; Crandall, K.A. Integrating Microbial and Host Transcriptomics to Characterize Asthma-Associated Microbial Communities. BMC Med. Genom. 2015, 8, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Losada, M.; Castro-Nallar, E.; Bendall, M.L.; Freishtat, R.J.; Crandall, K.A. Dual Transcriptomic Profiling of Host and Microbiota during Health and Disease in Pediatric Asthma. PLoS ONE 2015, 10, e0131819. [Google Scholar]
- Chun, Y.; Do, A.; Grishina, G.; Grishin, A.; Fang, G.; Rose, S.; Spencer, C.; Vicencio, A.; Schadt, E.; Bunyavanich, S. Integrative Study of the Upper and Lower Airway Microbiome and Transcriptome in Asthma. JCI Insight 2020, 5, 1–16. [Google Scholar] [CrossRef]
- Chiu, C.-Y.; Chou, H.-C.; Chang, L.-C.; Fan, W.-L.; Dinh, M.C.V.; Kuo, Y.-L.; Chung, W.-H.; Lai, H.-C.; Hsieh, W.-P.; Su, S.-C. Integration of Metagenomics-Metabolomics Reveals Specific Signatures and Functions of Airway Microbiota in Mite-Sensitized Childhood Asthma. Allergy 2020, 75, 2846–2857. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Llorente, M.A.; Martínez-Cañavate, A.; Chueca, N.; Rico, M. de la C.; Romero, R.; Anguita-Ruiz, A.; Aguilera, C.M.; Gil-Campos, M.; Mesa, M.D.; Khakimov, B.; et al. A Multi-Omics Approach Reveals New Signatures in Obese Allergic Asthmatic Children. Biomedicines 2020, 8, 359. [Google Scholar]
- Perez-Garcia, J.; Hernández-Pérez, J.M.; González-Pérez, R.; Sardón, O.; Martin-Gonzalez, E.; Espuela-Ortiz, A.; Mederos-Luis, E.; Callero, A.; Herrera-Luis, E.; Corcuera, P.; et al. The Genomics and Metagenomics of Asthma Severity (GEMAS) Study: Rationale and Design. J. Pers. Med. 2020, 10, 123. [Google Scholar] [CrossRef] [PubMed]
- Igartua, C.; Davenport, E.R.; Gilad, Y.; Nicolae, D.L.; Pinto, J.; Ober, C. Host Genetic Variation in Mucosal Immunity Pathways Influences the Upper Airway Microbiome. Microbiome 2017, 5, 16. [Google Scholar] [CrossRef] [Green Version]
- Abdel-Aziz, M.; Neerincx, A.; Vijverberg, S.; Hashimoto, S.; Brinkman, P.; Gorenjak, M.; Toncheva, A.; Harner, S.; Brandstetter, S.; Wolff, C.; et al. A System Pharmacology Multi-Omics Approach toward Uncontrolled Pediatric Asthma. J. Pers. Med. 2021, 11, 484. [Google Scholar] [CrossRef]
- Abdel-Aziz, M.I.; Kermani, N.Z.; Neerincx, A.H.; Vijverberg, S.J.H.; Guo, Y.; Howarth, P.; Dahlen, S.; Djukanovic, R.; Sterk, P.J.; Kraneveld, A.D.; et al. Association of Endopeptidases, Involved in SARS-CoV-2 Infection, with Microbial Aggravation in Sputum of Severe Asthma. Allergy 2021, 76, 1917–1921. [Google Scholar] [CrossRef]
- Tyler, S.R.; Chun, Y.; Ribeiro, V.M.; Grishina, G.; Grishin, A.; Hoffman, G.E.; Do, A.N.; Bunyavanich, S. Merged Affinity Network Association Clustering: Joint Multi-Omic/Clinical Clustering to Identify Disease Endotypes. Cell Rep. 2021, 35, 108975. [Google Scholar]
- Do, A.N.; Chun, Y.; Grishina, G.; Grishin, A.; Rogers, A.J.; Raby, B.A.; Weiss, S.T.; Vicencio, A.; Schadt, E.E.; Bunyavanich, S. Network Study of Nasal Transcriptome Profiles Reveals Master Regulator Genes of Asthma. J. Allergy Clin. Immunol. 2021, 147, 879–893. [Google Scholar] [CrossRef] [PubMed]
- Carney, S.M.; Clemente, J.C.; Cox, M.J.; Dickson, R.P.; Huang, Y.J.; Kitsios, G.D.; Kloepfer, K.M.; Leung, J.M.; Levan, T.D.; Molyneaux, P.L.; et al. Methods in Lung Microbiome Research. Am. J. Respir. Cell Mol. Biol. 2020, 62, 283–299. [Google Scholar] [CrossRef]
- Bharti, R.; Grimm, D.G. Current Challenges and Best-Practice Protocols for Microbiome Analysis. Brief. Bioinform. 2021, 22, 178–193. [Google Scholar] [PubMed] [Green Version]
- Prodan, A.; Tremaroli, V.; Brolin, H.; Zwinderman, A.H.; Nieuwdorp, M.; Levin, E. Comparing Bioinformatic Pipelines for Microbial 16S RRNA Amplicon Sequencing. PLoS ONE 2020, 15, e0227434. [Google Scholar]
- Ye, S.H.; Siddle, K.J.; Park, D.J.; Sabeti, P.C. Benchmarking Metagenomics Tools for Taxonomic Classification. Cell 2019, 178, 779–794. [Google Scholar] [CrossRef] [PubMed]
- Bersanelli, M.; Mosca, E.; Remondini, D.; Giampieri, E.; Sala, C.; Castellani, G.; Milanesi, L. Methods for the Integration of Multi-Omics Data: Mathematical Aspects. BMC Bioinform. 2016, 17, 167–177. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Chaudhary, K.; Garmire, L.X. More Is Better: Recent Progress in Multi-Omics Data Integration Methods. Front. Genet. 2017, 8, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramanian, I.; Verma, S.; Kumar, S.; Jere, A.; Anamika, K. Multi-Omics Data Integration, Interpretation, and Its Application. Bioinform. Biol. Insights 2020, 14, 117793221989905. [Google Scholar] [CrossRef] [Green Version]
- Graw, S.; Chappell, K.; Washam, C.L.; Gies, A.; Bird, J.; Robeson, M.S.; Byrum, S.D. Multi-Omics Data Integration Considerations and Study Design for Biological Systems and Disease. Mol. Omics 2021, 17, 170–185. [Google Scholar] [CrossRef]
- Narayana, J.K.; Mac Aogáin, M.; Ali, N.A. tika. B.M.; Tsaneva-Atanasova, K.; Chotirmall, S.H. Similarity Network Fusion for the Integration of Multi-Omics and Microbiomes in Respiratory Disease. Eur. Respir. J. 2021, 58, 2101016. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Perez, D.; Lugo-Martinez, J.; Bourguignon, N.; Mathee, K.; Lerner, B.; Bar-Joseph, Z.; Narasimhan, G. Dynamic Bayesian Networks for Integrating Multi-Omics Time Series Microbiome Data. mSystems 2021, 6, e01105-20. [Google Scholar] [CrossRef]
- Jiang, D.; Armour, C.R.; Hu, C.; Mei, M.; Tian, C.; Sharpton, T.J.; Jiang, Y. Microbiome Multi-Omics Network Analysis: Statistical Considerations, Limitations, and Opportunities. Front. Genet. 2019, 10, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Wang, K.; Wu, W.; Giannoulatou, E.; Ho, J.W.K.; Li, L. Host and Microbiome Multi-Omics Integration: Applications and Methodologies. Biophys. Rev. 2019, 11, 55–65. [Google Scholar] [CrossRef]
- Pedersen, H.K.; Forslund, S.K.; Gudmundsdottir, V.; Petersen, A.Ø.; Hildebrand, F.; Hyötyläinen, T.; Nielsen, T.; Hansen, T.; Bork, P.; Ehrlich, S.D.; et al. A Computational Framework to Integrate High-Throughput “-Omics” Datasets for the Identification of Potential Mechanistic Links. Nat. Protoc. 2018, 13, 2781–2800. [Google Scholar] [CrossRef]
- Zhang, X.; Li, L.; Butcher, J.; Stintzi, A.; Figeys, D. Advancing Functional and Translational Microbiome Research Using Meta-Omics Approaches. Microbiome 2019, 7, 154. [Google Scholar] [CrossRef]
- Daliri, E.B.-M.; Ofosu, F.K.; Chelliah, R.; Lee, B.H.; Oh, D.-H. Challenges and Perspective in Integrated Multi-Omics in Gut Microbiota Studies. Biomolecules 2021, 11, 300. [Google Scholar] [CrossRef]
- Liu, Z.; Ma, A.; Mathé, E.; Merling, M.; Ma, Q.; Liu, B. Network Analyses in Microbiome Based on High-Throughput Multi-Omics Data. Brief. Bioinform. 2021, 22, 1639–1655. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Hyeon, D.Y.; Hwang, D. Single-Cell Multiomics: Technologies and Data Analysis Methods. Exp. Mol. Med. 2020, 52, 1428–1442. [Google Scholar] [CrossRef]
- Nyholm, L.; Koziol, A.; Marcos, S.; Botnen, A.B.; Aizpurua, O.; Gopalakrishnan, S.; Limborg, M.T.; Gilbert, M.T.P.; Alberdi, A. Holo-Omics: Integrated Host-Microbiota Multi-Omics for Basic and Applied Biological Research. iScience 2020, 23, 101414. [Google Scholar] [CrossRef] [PubMed]
- Leitao Filho, F.S.; Alotaibi, N.M.; Ngan, D.; Tam, S.; Yang, J.; Hollander, Z.; Chen, V.; FitzGerald, J.M.; Nislow, C.; Leung, J.M.; et al. Sputum Microbiome Is Associated with 1-Year Mortality after Chronic Obstructive Pulmonary Disease Hospitalizations. Am. J. Respir. Crit. Care Med. 2019, 199, 1205–1213. [Google Scholar] [CrossRef]
- Lynch, J.P.; Sikder, M.A.A.; Curren, B.F.; Werder, R.B.; Simpson, J.; Cuív, P.Ó.; Dennis, P.G.; Everard, M.L.; Phipps, S. The Influence of the Microbiome on Early-Life Severe Viral Lower Respiratory Infections and Asthma-Food for Thought? Front. Immunol. 2017, 8, 156. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Year | Participants | Sample Type | Key Findings | Reference |
|---|---|---|---|---|
| 2014 | -28 severe asthmatics | Sputum | -Neutrophilic asthmatics: ↑ abundance of pathogenic bacterial species (Haemophilus sp., Streptococcus sp., Moraxella catarrhalis) | [117] |
| 2015 | -40 severe asthmatics | Bronchial (Brushings) | -Eosinophils: negative correlation with relative abundance of Proteobacteria (Moraxellaceae, Helicobacteraceae families), positive correlation with Actinobacteria (Streptomyces and Propionicimonas species) | [146] |
| 2016 | -30 asthmatics | Sputum | -Neutrophilic vs. non-neutrophilic asthmatics: ↓ evenness and richness of bacterial species, ↑ Proteobacteria (Haemophilus influenzae) ↓ Actinobacteria, Firmicutes -Eosinophilic asthmatics: ↑ abundance of Actinobacteria (Tropheryma whipplei) | [148] |
| 2016 | -26 severe asthmatics -18 non-severe asthmatics -12 healthy controls | Sputum | -Eosinophils: ↑ Firmicutes (Streptococcus sp.) | [149] |
| 2017 | -23 steroid-free asthmatics -10 healthy controls | BAL 1 | -Eosinophilic asthmatics vs. healthy controls: ↑ Neisseria, Bacteroides and Rothia ↓ Sphingomonas, Halomonas, Aeribacillus -Neutrophilic asthmatics vs. healthy controls: differences in Flavobacterium, Phenylobacterium, Brevundimonas, Bradyrhizobium, Sediminibacterium, Gemella | [150] |
| 2017 | -25 severe asthmatics -24 non-severe asthmatics -15 healthy controls | Sputum | -Eosinophilic vs. non-eosinophilic asthmatics: ↑ Actinomycetaceae, Enterobacteriaceae family members | [151] |
| 2017 | -42 atopic asthmatics -21 atopic non-asthmatics -21 non-atopic healthy controls | Bronchial (Brushings) Oral wash | -T2-high vs. non-Th2: ↓ bronchial bacterial burden | [118] |
| 2018 | -20 neutrophilic asthmatics -34 non-neutrophilic asthmatics | Sputum | -Neutrophilic vs. non-neutrophilic asthmatics: ↑ total bacterial burden, ↓ Firmicutes, Actinobacteria, Saccharibacteria, ↑ Bacteroidetes phyla (Porphyromonas spp., Capnocytophaga spp.), Proteobacteria (Haemophilus spp., Moraxella spp.) | [152] |
| 2018 | -84 eosinophilic asthmatics -14 neutrophilic asthmatics -60 paucigranulocytic asthmatics -9 mixed neutrophilic and eosinophilic asthmatics | Sputum | -Neutrophilic asthmatics vs. all other endotypes: ↓ diversity, richness and evenness, ↑ high relative abundance in pathogenic taxa (Haemophilus and Moraxella), ↓ Streptococcus, Gemella and Porphyromonas -Eosinophilic vs other endotypes: ↓ Haemophilus, Gemella, Rothia and Porphyromonas | [153] |
| 2018 | -32 asthmatics -73 COPD 2 patients | Sputum | -Neutrophilic asthmatics: ↑ Proteobacteria phyla -Eosinophilic asthmatics: ↑ Bacteroidetes | [154] |
| 2019 | -10 eosinophilic asthmatics -14 non-eosinophilic asthmatics -12 healthy controls | Sputum | -Eosinophilic vs. non-eosinophilic asthmatics: ↑ richness, evenness and diversity, ↑ Glaciecola, Helicobacter ↓ Scardovia, Bifidobacterium, Desulfobulbus, Deinococcus | [155] |
| 2020 | -32 atopic asthmatics -18 atopic non-asthmatics -16 non-atopic healthy controls | Sputum BAL Oral wash | -T2-high vs. non-Th2: ↓ Sputum bacterial burden | [156] |
| 2021 | -100 severe asthmatics | Sputum | -High neutrophilic vs. low neutrophilic asthmatics: ↓ richness and diversity, ↑ increased relative abundance of pathogenic species (Haemophilus influenzae, Moraxella catarrhalis, Streptococcus pseudopneumoniae) ↓ Veillonella, Prevotella and Neisseria | [157] |
| Year | Asthma Type | Sample Types | Multi-Omics Data | Reference |
|---|---|---|---|---|
| 2015 | Pediatric vs. Healthy | Nasal (Brushings) | Metatrascriptome Transcriptome (RNA sequencing, in silico separation) | [170] |
| 2015 | Pediatric vs. Healthy | Nasal (Brushings) | Metatrascriptome Transcriptome (RNA sequencing, in silico separation) | [171] |
| 2015 | Severe vs. Healthy and Mild-to-Moderate | Bronchial (Brushings) | Metagenome (16S rRNA gene array) Transcriptome (Gene expression microarray) | [146] |
| 2020 | Severe Persistent Childhood vs. Healthy | Nasal (Brushings-swabs) Bronchial (Brushings-BAL 1) | Metagenome (16S rRNA gene sequencing) Transcriptome (RNA sequencing) | [172] |
| 2020 | Mite-sensitized Childhood vs. Healthy | Airway (Throat swabs) Blood (Serum) | Metagenome (Shotgun metagenome sequencing) Metabolome (NMR) | [173] |
| 2020 | Allergic Childhood Obese vs. Non-obese and Persistent vs. Non-persistent | Fecal (Stool) Blood (Plasma) | Metagenome (16S rRNA gene sequencing) Metabolome (NMR and GC-MS) Proteome (Luminex and ELISA) | [174] |
| 2021 | Severe vs. Healthy and Mild-to-Moderate | Sputum | Metagenome (16S rRNA gene sequencing) Transcriptome (Gene expression microarray) Proteome (SomaScan assay proteomics) | [178] |
| 2021 | Severe vs. Healthy and Mild-to-Moderate | Nasal (Brushings) | Metagenome (16S rRNA gene sequencing) Transcriptome (RNA sequencing) Methylome (Infinium MethylationEPIC array) | [179] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Logotheti, M.; Agioutantis, P.; Katsaounou, P.; Loutrari, H. Microbiome Research and Multi-Omics Integration for Personalized Medicine in Asthma. J. Pers. Med. 2021, 11, 1299. https://doi.org/10.3390/jpm11121299
Logotheti M, Agioutantis P, Katsaounou P, Loutrari H. Microbiome Research and Multi-Omics Integration for Personalized Medicine in Asthma. Journal of Personalized Medicine. 2021; 11(12):1299. https://doi.org/10.3390/jpm11121299
Chicago/Turabian StyleLogotheti, Marianthi, Panagiotis Agioutantis, Paraskevi Katsaounou, and Heleni Loutrari. 2021. "Microbiome Research and Multi-Omics Integration for Personalized Medicine in Asthma" Journal of Personalized Medicine 11, no. 12: 1299. https://doi.org/10.3390/jpm11121299
APA StyleLogotheti, M., Agioutantis, P., Katsaounou, P., & Loutrari, H. (2021). Microbiome Research and Multi-Omics Integration for Personalized Medicine in Asthma. Journal of Personalized Medicine, 11(12), 1299. https://doi.org/10.3390/jpm11121299







