Defects in MMR Genes as a Seminal Example of Personalized Medicine: From Diagnosis to Therapy
Abstract
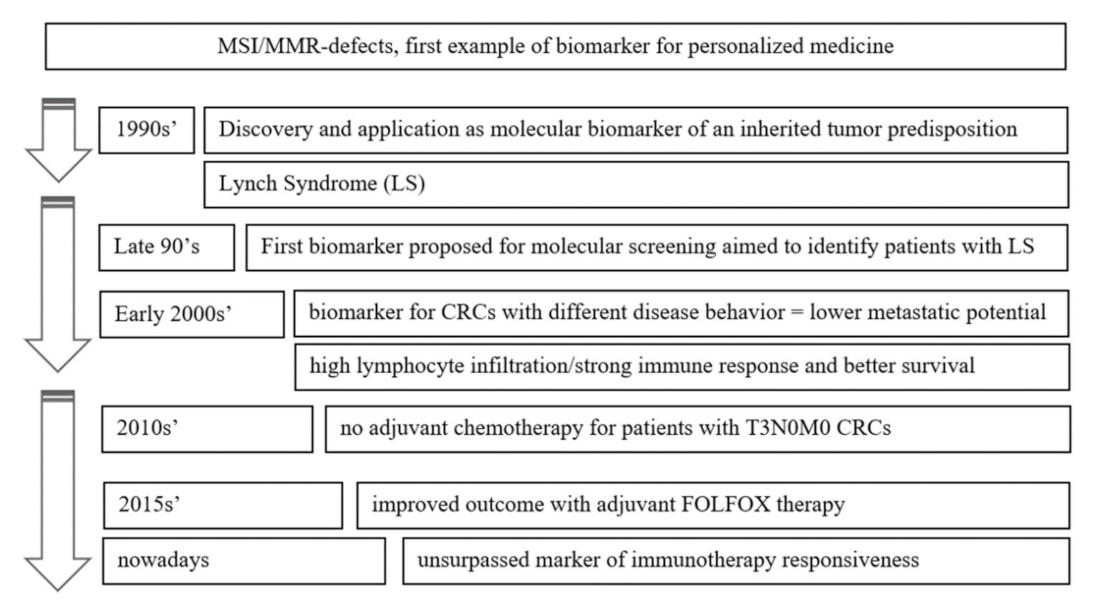
:1. Introduction
2. Discovery of MSI, Its Relevance in Lynch Syndrome and Understanding the Different Molecular Pathogenesis of CRC
2.1. Parallel Discovery
2.2. Unraveling the Pool of Genes Involved in DNA MMR and Deranged in Lynch Syndrome
2.3. Sporadic MSI Cancers and Hypermethylation
2.4. Lynch Syndrome versus Lynch-Like Syndrome
3. Prognostic Value of MSI in CRC
3.1. Lower Metastatic Potential and Better Survival of MSI CRC
3.2. Adaptive Immune Response and the Relevance of Immune Parameters
4. Predictive Value of MSI
4.1. Implication for the Adjuvant Treatment: Stage 2 vs. Stage 3
4.2. Removing the Breaks from the Immune Response: Immunotherapy
4.3. Silencing Map Kinases in Sporadic MSI
5. Discussion and Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
| CRC | colorectal carcinoma |
| MMR | mismatch repair |
| MSI | microsatellite instability |
| LS | Lynch syndrome |
| TILs | tumor-infiltrating lymphocytes |
References
- Biller, L.H.; Schrag, D. Diagnosis and Treatment of Metastatic Colorectal Cancer: A Review. JAMA 2021, 325, 669–685. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Goding Sauer, A.; Fedewa, S.A.; Butterly, L.F.; Anderson, J.C.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 145–164. [Google Scholar] [CrossRef] [Green Version]
- National Cancer Institute Surveillance, Epidemiology, and End Results Program. Cancer Stat Facts: Colorectal Cancer. Available online: https://seer.cancer.gov/statfacts/html/colorect.html (accessed on 9 September 2021).
- Boland, C.R.; Goel, A. Microsatellite instability in colorectal cancer. Gastroenterology 2010, 138, 2073–2087. [Google Scholar] [CrossRef]
- Ward, R.; Meagher, A.; Tomlinson, I.; O’Connor, T.; Norrie, M.; Wu, R.; Hawkins, N. Microsatellite instability and the clinico-pathological features of sporadic colorectal cancer. Gut 2001, 48, 821–882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taieb, J.; Shi, Q.; Pederson, L.; Alberts, S.; Wolmark, N.; Van Cutsem, E.; de Gramont, A.; Kerr, R.; Grothey, A.; Lonardi, S.; et al. Prognosis of microsatellite instability and/or mismatch repair deficiency stage III colon cancer patients after disease recurrence following adjuvant treatment: Results of an ACCENT pooled analysis of seven studies. Ann. Oncol. 2019, 30, 1466–1471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, Z.; Sinicrope, F.A. Prognostic and Predictive Values of Mismatch Repair Deficiency in Non-Metastatic Colorectal Cancer. Cancers 2021, 13, 300. [Google Scholar] [CrossRef]
- Ganesh, K.; Stadler, Z.K.; Cercek, A.; Mendelsohn, R.B.; Shia, J.; Segal, N.H.; Diaz, L.A., Jr. Immunotherapy in colorectal cancer: Rationale, challenges and potential. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 361–375. [Google Scholar] [CrossRef] [PubMed]
- Messersmith, W.A. NCCN Guidelines Updates: Management of Metastatic Colorectal Cancer. J. Natl. Compr. Canc. Net. 2019, 17, 599–601. [Google Scholar]
- Peinado, M.A.; Malkhosyan, S.; Velazquez, A.; Perucho, M. Isolation and characterization of allelic losses and gains in colorectal tumors by arbitrarily primed polymerase chain reaction. Proc. Natl. Acad. Sci. USA 1992, 89, 10065–10069. [Google Scholar] [CrossRef] [Green Version]
- Ionov, Y.; Peinado, M.A.; Malkhosyan, S.; Shibata, D.; Perucho, M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature 1993, 363, 558–561. [Google Scholar] [CrossRef]
- Thibodeau, S.N.; Bren, G.; Schaid, D. Microsatellite Instability in Cancer of the Proximal Colon. Science 1993, 260, 816–819. [Google Scholar] [CrossRef] [PubMed]
- Aaltonen, L.A.; Peltomäki, P.; Leach, F.S.; Sistonen, P.; Pylkkänen, L.; Mecklin, J.-P.; Järvinen, H.; Powell, S.M.; Jen, J.; Hamilton, S.R.; et al. Clues to the Pathogenesis of Familial Colorectal Cancer. Science 1993, 260, 812–816. [Google Scholar] [CrossRef] [PubMed]
- Peltomäki, P.; Aaltonen, L.A.; Sistonen, P.; Pylkkänen, L.; Mecklin, J.-P.; Järvinen, H.; Green, J.S.; Jass, J.R.; Weber, J.L.; Leach, F.S.; et al. Genetic Mapping of a Locus Predisposing to Human Colorectal Cancer. Science 1993, 260, 810–812. [Google Scholar] [CrossRef]
- Maddox, J. Competition and the death of science. Nature 1993, 363, 667. [Google Scholar] [CrossRef]
- Fishel, R.; Lescoe, M.K.; Rao, M.R.; Copeland, N.G.; Jenkins, N.A.; Garber, J.; Kane, M.; Kolodner, R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell 1993, 75, 1027–1038. [Google Scholar] [CrossRef]
- Leach, F.S.; Nicolaides, N.C.; Papadopoulos, N.; Liu, B.; Jen, J.; Parsons, R.; Peltomäki, P.; Sistonen, P.; Aaltonen, L.; Nyström-Lahti, M.; et al. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell 1993, 75, 1215–1225. [Google Scholar] [CrossRef]
- Kinzler, K.W.; Vogelstein, B. Lessons from Hereditary Colorectal Cancer. Cell 1996, 87, 159–170. [Google Scholar] [CrossRef] [Green Version]
- Malkhosyan, S.; Rampino, N.; Yamamoto, H.; Perucho, M. Frameshift mutator mutations. Nature 1996, 382, 499–500. [Google Scholar] [CrossRef] [PubMed]
- Perucho, M. Microsatellite instability: The mutator that mutates the other mutator. Nat. Med. 1996, 2, 630–631. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Jen, J.; Vogelstein, B.; Hamilton, S.R. Clinical and pathological characteristics of sporadic colorectal carcinomas with DNA replication errors in microsatellite sequences. Am. J. Pathol. 1994, 145, 148–156. [Google Scholar] [PubMed]
- Guidoboni, M.; Gafà, R.; Viel, A.; Doglioni, C.; Russo, A.; Santini, A.; Del Tin, L.; Macrì, E.; Lanza, G.; Boiocchi, M.; et al. Microsatellite instability and high content of activated cytotoxic lymphocytes identify colon cancer patients with a favorable prognosis. Am. J. Pathol. 2001, 159, 297–304. [Google Scholar] [CrossRef] [Green Version]
- Aaltonen, L.A.; Salovaara, R.; Kristo, P.; Canzian, F.; Hemminki, A.; Peltomäki, P.; Chadwick, R.B.; Kääriäinen, H.; Eskelinen, M.; Järvinen, H.; et al. Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. N. Engl. J. Med. 1998, 338, 1481–1487. [Google Scholar] [CrossRef] [PubMed]
- De’ Angelis, G.L.; Bottarelli, L.; Azzoni, C.; De’Angelis, N.; Leandro, G.; Di Mario, F.; Gaiani, F.; Negri, F. Microsatellite instability in colorectal cancer. Acta Biomed. 2018, 89, 97–101. [Google Scholar]
- Peltomäki, P. Update on Lynch syndrome genomics. Fam. Cancer 2016, 15, 385–393. [Google Scholar] [CrossRef] [Green Version]
- Tutlewska, K.; Lubinski, J.; Kurzawski, G. Germline deletions in the EPCAM gene as a cause of Lynch syndrome—Literature review. Hered. Cancer Clin. Pr. 2013, 11, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiricny, J. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 2006, 7, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Boland, C.R.; Thibodeau, S.N.; Hamilton, S.R.; Sidransky, D.; Eshleman, J.R.; Burt, R.W.; Meltzer, S.J.; Rodriguez-Bigas, M.A.; Fodde, R.; Ranzani, G.N.; et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: Development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998, 58, 5248–5257. [Google Scholar] [PubMed]
- Umar, A.; Boland, C.R.; Terdiman, J.P.; Syngal, S.; Chapelle, A.D.L.; Rüschoff, J.; Srivastava, S. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J. Natl. Cancer Inst. 2004, 96, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Vasen, H.F.; Mecklin, J.P.; Khan, P.M.; Lynch, H.T. The International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (ICG-HNPCC). Dis. Colon Rectum 1991, 34, 424–425. [Google Scholar] [CrossRef]
- Vasen, H.F.; Watson, P.; Mecklin, J.P.; Lynch, H.T. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology 1999, 116, 1453–1456. [Google Scholar] [CrossRef]
- Jass, J.R. HNPCC and sporadic MSI-H colorectal cancer: A review of the morphological similarities and differences. Fam Cancer 2004, 3, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Laiho, P.; Launonen, V.; Lahermo, P.; Esteller, M.; Guo, M.; Herman, J.G.; Mecklin, J.P.; Järvinen, H.; Sistonen, P.; Kim, K.M.; et al. Low-level microsatellite instability in most colorectal carcinomas. Cancer Res. 2002, 62, 1166–1170. [Google Scholar]
- Tomlinson, I.; Halford, S.; Aaltonen, L.; Hawkins, N.; Ward, R. Does MSI-low exist? J. Pathol. 2002, 197, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Hampel, H.; Frankel, W.L.; Martin, E.; Arnold, M.; Khanduja, K.; Kuebler, P.; Nakagawa, H.; Sotamaa, K.; Prior, T.W.; Westman, J.; et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer). N. Engl. J. Med. 2005, 352, 1851–1860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laghi, L.; Negri, F.; Gaiani, F.; Cavalleri, T.; Grizzi, F.; De’ Angelis, G.L.; Malesci, A. Prognostic and Predictive Cross-Roads of Microsatellite Instability and Immune Response to Colon Cancer. Int. J. Mol. Sci. 2020, 21, 9680. [Google Scholar] [CrossRef] [PubMed]
- Domingo, E.; Laiho, P.; Ollikainen, M.; Pinto, M.; Wang, L.; French, A.J.; Westra, J.; Frebourg, T.; Espín, E.; Armengol, M.; et al. BRAF screening as a low-cost effective strategy for simplifying HNPCC genetic testing. J. Med. Genet. 2004, 41, 664–668. [Google Scholar] [CrossRef] [PubMed]
- Kane, M.F.; Loda, M.; Gaida, G.M.; Lipman, J.; Mishra, R.; Goldman, H.; Jessup, J.M.; Kolodner, R. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997, 57, 808–811. [Google Scholar] [PubMed]
- Cunningham, J.M.; Christensen, E.R.; Tester, D.J.; Kim, C.Y.; Roche, P.C.; Burgart, L.J.; Thibodeau, S.N. Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer Res. 1998, 58, 3455–3460. [Google Scholar]
- Herman, J.G.; Umar, A.; Polyak, K.; Graff, J.R.; Ahuja, N.; Issa, J.P.; Markowitz, S.; Willson, J.K.; Hamilton, S.R.; Kinzler, K.W.; et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc. Natl. Acad. Sci. USA 1998, 95, 6870–6875. [Google Scholar] [CrossRef] [Green Version]
- Gallois, C.; Laurent-Puig, P.; Taieb, J. Methylator phenotype in colorectal cancer: A prognostic factor or not? Crit. Rev. Oncol. Hematol. 2016, 99, 74–80. [Google Scholar] [CrossRef]
- Toyota, M.; Ahuja, N.; Ohe-Toyota, M.; Herman, J.G.; Baylin, S.B.; Issa, J.-P. CpG island methylator phenotype in colorectal cancer. Proc. Natl. Acad. Sci. USA 1999, 96, 8681–8686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogino, S.; Kawasaki, T.; Kirkner, G.J.; Kraft, P.; Loda, M.; Fuchs, C.S. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J. Mol. Diagn. 2007, 9, 305–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samowitz, W.S.; Albertsen, H.; Herrick, J.; Levin, T.R.; Sweeney, C.; Murtaugh, M.A.; Wolff, R.K.; Slattery, M.L. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology 2005, 129, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Gaiani, F.; Marchesi, F.; Negri, F.; Greco, L.; Malesci, A.; de’Angelis, G.L.; Laghi, L. Heterogeneity of Colorectal Cancer Progression: Molecular Gas and Brakes. Int. J. Mol. Sci. 2021, 22, 5246. [Google Scholar] [CrossRef] [PubMed]
- Kambara, T.; Simms, L.A.; Whitehall, V.L.; Spring, K.J.; Wynter, C.V.; Walsh, M.D.; Barker, M.A.; Arnold, S.; McGivern, A.; Matsubara, N.; et al. BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut 2004, 53, 1137–1144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nosho, K.; Irahara, N.; Shima, K.; Kure, S.; Kirkner, G.J.; Schernhammer, E.S.; Hazra, A.; Hunter, D.J.; Quackenbush, J.; Spiegelman, D.; et al. Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample. PLoS ONE 2008, 3, e3698. [Google Scholar] [CrossRef] [PubMed]
- Ref Merlo, A.; Herman, J.G.; Mao, L.; Lee, D.J.; Gabrielson, E.; Burger, P.C.; Baylin, S.B.; Sidransky, D. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat. Med. 1995, 1, 686–692. [Google Scholar] [CrossRef] [PubMed]
- Warthin, A.S. Heredity with reference to carcinoma as shown by the study of the cases examined in the Pathological Laboratory of the University of Michigan, 1895−1912. Arch. Int. Med. 1913, 12, 546–555. [Google Scholar] [CrossRef] [Green Version]
- Boland, C.R.; Lynch, H.T. The history of Lynch syndrome. Fam. Cancer 2013, 12, 145–157. [Google Scholar] [CrossRef] [Green Version]
- Lynch, H.T.; Kimberling, W.; Albano, W.A.; Lynch, J.F.; Biscone, K.; Schuelke, G.S.; Sandberg, A.A.; Lipkin, M.; Deschner, E.E.; Mikol, Y.B.; et al. Hereditary nonpolyposis colorectal cancer (Lynch syndromes I and II). I. Clinical description of resource. Cancer 1985, 56, 934–938. [Google Scholar] [CrossRef]
- Carethers, J.M.; Stoffel, E.M. Lynch syndrome and Lynch syndrome mimics: The growing complex landscape of hereditary colon cancer. World J. Gastroenterol. 2015, 21, 9253–9261. [Google Scholar] [CrossRef]
- Pico, M.D.; Castillejo, A.; Murcia, O.; Giner-Calabuig, M.; Alustiza, M.; Sanchez, A.; Moreira, L.; Pellise, M.; Castells, A.; Carrillo-Palau, M.; et al. Clinical and Pathological Characterization of Lynch-Like Syndrome. Clin. Gastroenterol. Hepatol. 2020, 18, 368–374. [Google Scholar] [CrossRef]
- Rodriguez-Soler, M.; Perez-Carbonell, L.; Guarinos, C.; Zapater, P.; Castillejo, A.; Barbera, V.M.; Juarez, M.; Bessa, X.; Xicola, R.M.; Clofent, J.; et al. Risk of cancer in cases of suspected lynch syndrome without germline mutation. Gastroenterology 2013, 144, 926.e921–932.e921, quiz e913–e924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mensenkamp, A.R.; Vogelaar, I.P.; van Zelst-Stams, W.A.; Goossens, M.; Ouchene, H.; Hendriks-Cornelissen, S.J.; Kwint, M.P.; Hoogerbrugge, N.; Nagtegaal, I.D.; Ligtenberg, M.J. Somatic mutations in MLH1 and MSH2 are a frequent cause of mismatch-repair deficiency in Lynch syndrome-like tumors. Gastroenterology 2014, 146, 643.e648–646.e648. [Google Scholar] [CrossRef] [PubMed]
- Sankila, R.; Aaltonen, L.A.; Järvinen, H.J.; Mecklin, J.P. Better survival rates in patients with MLH1-associated hereditary colorectal cancer. Gastroenterology 1996, 110, 682–687. [Google Scholar] [CrossRef]
- Gryfe, R.; Gallinger, S. Microsatellite instability, mismatch repair deficiency, and colorectal cancer. Surgery 2001, 130, 17–20. [Google Scholar] [CrossRef] [PubMed]
- Dienstmann, R.; Mason, M.J.; Sinicrope, F.A.; Phipps, A.I.; Tejpar, S.; Nesbakken, A.; Danielsen, S.A.; Sveen, A.; Buchanan, D.D.; Clendenning, M.; et al. Prediction of overall survival in stage II and III colon cancer beyond TNM system: A retrospective, pooled biomarker study. Ann. Oncol. 2017, 28, 1023–1031. [Google Scholar] [CrossRef]
- Sargent, D.J.; Shi, Q.; Yothers, G.; Tejpar, S.; Bertagnolli, M.M.; Thibodeau, S.N.; Andre, T.; Labianca, R.; Gallinger, S.; Hamilton, S.R.; et al. Prognostic impact of deficient mismatch repair (dMMR) in 7,803 stage II/III colon cancer (CC) patients (pts): A pooled individual pt data analysis of 17 adjuvant trials in the ACCENT database. J. Clin. Oncol. 2014, 32, 3507. [Google Scholar] [CrossRef]
- Klingbiel, D.; Saridaki, Z.; Roth, A.D.; Bosman, F.T.; Delorenzi, M.; Tejpar, S. Prognosis of stage II and III colon cancer treated with adjuvant 5-fluorouracil or FOLFIRI in relation to microsatellite status: Results of the PETACC−3 trial. Ann. Oncol. 2015, 26, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Li, F.; Zhou, X.; Ma, Y.; Fu, W. Is microsatellite instability-high really a favorable prognostic factor for advanced colorectal cancer? A meta-analysis. World J. Surg. Oncol. 2019, 17, 169. [Google Scholar] [CrossRef] [Green Version]
- Buckowitz, A.; Knaebel, H.P.; Benner, A.; Blaker, H.; Gebert, J.; Kienle, P.; von Doeberitz, K.M.; Kloor, M. Microsatellite instability in colorectal cancer is associated with local lymphocyte infiltration and low frequency of distant metastases. Br. J. Cancer 2005, 92, 1746–1753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venderbosch, S.; Nagtegaal, I.D.; Maughan, T.S.; Smith, C.G.; Cheadle, J.P.; Fisher, D.; Kaplan, R.; Quirke, P.; Seymour, M.T.; Richman, S.D.; et al. Mismatch repair status and BRAF mutation status in metastatic colorectal cancer patients: A pooled analysis of the CAIRO, CAIRO2, COIN, and FOCUS studies. Clin. Cancer Res. 2014, 20, 5322–5330. [Google Scholar] [CrossRef] [Green Version]
- Laghi, L.; Bianchi, P.; Miranda, E.; Balladore, E.; Pacetti, V.; Grizzi, F.; Allavena, P.; Torri, V.; Repici, A.; Santoro, A.; et al. CD3+ cells at the invasive margin of deeply invading (pT3-T4) colorectal cancer and risk of post-surgical metastasis: A longitudinal study. Lancet Oncol. 2009, 10, 877–884. [Google Scholar] [CrossRef]
- Pagès, F.; Kirilovsky, A.; Mlecnik, B.; Asslaber, M.; Tosolini, M.; Bindea, G.; Galon, J. In situ cytotoxic and memory T cells predict outcome in patients with early stage colorectal cancer. J. Clin. Oncol. 2009, 27, 5944–5951. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.; Zhang, J.; Luo, P. Crosstalk Between the MSI Status and Tumor Microenvironment in Colorectal Cancer. Front Immunol. 2020, 11, 2039. [Google Scholar] [CrossRef]
- Galon, J.; Mlecnik, B.; Bindea, G.; Angell, H.K.; Berger, A.; Lagorce, C.; Pagès, F. Towards the introduction of the ‘Immunoscore’ in the classification of malignant tumours. J. Pathol. 2014, 232, 199–209. [Google Scholar] [CrossRef] [Green Version]
- Wirta, E.V.; Seppälä, T.; Friman, M.; Väyrynen, J.; Ahtiainen, M.; Kautiainen, H.; Kuopio, T.; Kellokumpu, I.; Mecklin, J.P.; Böhm, J. Immunoscore in mismatch repair-proficient and -deficient colon cancer. J. Pathol. Clin. Res. 2017, 3, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Noepel-Duennebacke, S.; Juette, H.; Schulmann, K.; Graeven, U.; Porschen, R.; Stoehlmacher, J.; Hegewisch-Becker, S.; Raulf, A.; Arnold, D.; Reinacher-Schick, A.; et al. Microsatellite instability (MSI-H) is associated with a high immunoscore but not with PD-L1 expression or increased survival in patients (pts.) with metastatic colorectal cancer (mCRC) treated with oxaliplatin (ox) and fluoropyrimidine (FP) with and without bevacizumab (bev): A pooled analysis of the AIO KRK 0207 and RO91 trials. J. Cancer Res. Clin. Oncol. 2021, 147, 3063–3072. [Google Scholar]
- Ribic, C.M.; Sargent, D.J.; Moore, M.J.; Thibodeau, S.N.; French, A.J.; Goldberg, R.M.; Gallinger, S. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N. Engl. J. Med. 2003, 349, 247–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sargent, D.J.; Marsoni, S.; Monges, G.; Thibodeau, S.N.; Labianca, R.; Hamilton, S.R.; French, A.J.; Kabat, B.; Foster, N.R.; Torri, V.; et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J. Clin. Oncol. 2010, 28, 3219–3226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinicrope, F.A.; Foster, N.R.; Thibodeau, S.N.; Marsoni, S.; Monges, G.; Labianca, R.; Kim, G.P.; Yothers, G.; Allegra, C.; Moore, M.J.; et al. DNA mismatch repair status and colon cancer recurrence and survival in clinical trials of 5-fluorouracil-based adjuvant therapy. J. Natl. Cancer Inst. 2011, 103, 863–875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hutchins, G.; Southward, K.; Handley, K.; Magill, L.; Beaumont, C.; Stahlschmidt, J.; Richman, S.; Chambers, P.; Seymour, M.; Kerr, D.; et al. Value of mismatch repair, KRAS, and BRAF mutations in predicting recurrence and benefits from chemotherapy in colorectal cancer. J. Clin. Oncol. 2011, 29, 1261–1270. [Google Scholar] [CrossRef]
- Petrelli, F.; Ghidini, M.; Cabiddu, M.; Pezzica, E.; Corti, D.; Turati, L.; Costanzo, A.; Varricchio, A.; Ghidini, A.; Barni, S.; et al. Microsatellite Instability and Survival in Stage II Colorectal Cancer: A Systematic Review and Meta-analysis. Anticancer Res. 2019, 39, 6431–6441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fomiti, A.; Rulli, E.; Pilozzi, E.; Gerardi, C.; Roberto, M.; Legramandi, L.; Falcone, R.; Pacchetti, I.; Marchetti, P.; Floriani, I. Exploring the prognostic role of microsatellite instability in patients with stage II colorectal cancer: A systematic review and meta-analysis. Clin. Colorectal Cancer 2017, 16, e55–e59. [Google Scholar]
- Laghi, L.; Malesci, A. Microsatellite instability and therapeutic consequences in colorectal cancer. Dig. Dis. 2012, 30, 304–309. [Google Scholar] [CrossRef] [PubMed]
- Koopman, M.; Kortman, G.A.; Mekenkamp, L.; Ligtenberg, M.J.; Hoogerbrugge, N.; Antonini, N.F.; Punt, C.J.; van Krieken, J.H. Deficient mismatch repair system in patients with sporadic advanced colorectal cancer. Br. J. Cancer 2009, 100, 266–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinicrope, F.A.; Mahoney, M.R.; Smyrk, T.C.; Thibodeau, S.N.; Warren, R.S.; Bertagnolli, M.M.; Nelson, G.D.; Goldberg, R.M.; Sargent, D.J.; Alberts, S.R. Prognostic impact of deficient DNA mismatch repair in patients with stage III colon cancer from a randomized trial of FOLFOX-based adjuvant chemotherapy. J. Clin. Oncol. 2013, 31, 3664–3672. [Google Scholar] [CrossRef] [PubMed]
- Emile, J.F.; Julié, C.; Le Malicot, K.; Lepage, C.; Tabernero, J.; Mini, E.; Budnik, T.M. Prospective validation of a lymphocyte infiltration prognostic test in stage III colon cancer patients treated with adjuvant FOLFOX. Eur. J. Cancer 2017, 82, 16–24. [Google Scholar] [CrossRef]
- Zaanan, A.; Shi, Q.; Taieb, J.; Alberts, S.R.; Meyers, J.P.; Smyrk, T.C.; Julie, C.; Zawadi, A.; Tabernero, J.; Mini, E.; et al. Role of Deficient DNA Mismatch Repair Status in Patients with Stage III Colon Cancer Treated with FOLFOX Adjuvant Chemotherapy: A Pooled Analysis From 2 Randomized Clinical Trials. JAMA Oncol. 2018, 4, 379–383. [Google Scholar] [CrossRef] [PubMed]
- Kucherlapati, M.H.; Lee, K.; Nguyen, A.A.; Clark, A.B.; Hou, H., Jr.; Rosulek, A.; Li, H.; Yang, K.; Fan, K.; Lipkin, M.; et al. An Msh2 conditional knockout mouse for studying intestinal cancer and testing anticancer agents. Gastroenterology 2010, 138, 993.e1–1002.e1. [Google Scholar] [CrossRef] [Green Version]
- Gavin, P.G.; Colangelo, L.H.; Fumagalli, D.; Tanaka, N.; Remillard, M.Y.; Yothers, G.; Kim, C.; Taniyama, Y.; Kim, S.I.; Choi, H.J.; et al. Mutation profiling and microsatellite instability in stage II and III colon cancer: An assessment of their prognostic and oxaliplatin predictive value. Clin. Cancer Res. 2012, 18, 6531–6541. [Google Scholar] [CrossRef] [Green Version]
- Taieb, J.; Le Malicot, K.; Shi, Q.; Penault-Llorca, F.; Bouché, O.; Tabernero, J.; Mini, E.; Goldberg, R.M.; Folprecht, G.; Luc Van Laethem, J.; et al. Prognostic Value of BRAF and KRAS Mutations in MSI and MSS Stage III Colon Cancer. J. Natl. Cancer Inst. 2016, 109, 272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rizvi, N.A.; Hellmann, M.D.; Snyder, A.; Kvistborg, P.; Makarov, V.; Havel, J.J.; Lee, W.; Yuan, J.; Wong, P.; Ho, T.S.; et al. Cancer immunology. Mutational landscape determines sensitivity to PD−1 blockade in non-small cell lung cancer. Science 2015, 348, 124–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giannakis, M.; Mu, X.J.; Shukla, S.A.; Qian, Z.R.; Cohen, O.; Nishihara, R.; Garraway, L.A. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep. 2016, 15, 857–865. [Google Scholar] [CrossRef] [Green Version]
- Le, D.T.; Durham, J.N.; Smith, K.N.; Wang, H.; Bartlett, B.R.; Aulakh, L.K.; Diaz, L.A. Mismatch repair deficiency predicts response of solid tumors to PD−1 blockade. Science 2017, 357, 409–413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roudko, V.; Bozkus, C.C.; Orfanelli, T.; McClain, C.B.; Carr, C.; O’Donnell, T.; Chakraborty, L.; Samstein, R.; Huang, K.L.; Blank, S.V.; et al. Shared Immunogenic Poly-Epitope Frameshift Mutations in Microsatellite Unstable Tumors. Cell 2020, 183, 1634–1649.e17. [Google Scholar] [CrossRef] [PubMed]
- Llosa, N.J.; Cruise, M.; Tam, A.; Wicks, E.C.; Hechenbleikner, E.M.; Taube, J.M.; Blosser, R.L.; Fan, H.; Wang, H.; Luber, B.S.; et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015, 5, 43–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Diaz, L.A., Jr. PD−1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Overman, M.J.; Lonardi, S.; Wong, K.Y.M.; Lenz, H.J.; Gelsomino, F.; Aglietta, M.; André, T. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J. Clin. Oncol. 2018, 36, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Ursem, C.; Atreya, C.E.; Van Loon, K. Emerging treatment options for BRAF-mutant colorectal cancer. Gastrointest. Cancer 2018, 8, 13–23. [Google Scholar] [CrossRef] [Green Version]
- The Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tran, B.; Kopetz, S.; Tie, J.; Gibbs, P.; Jiang, Z.Q.; Lieu, C.H.; Desai, J. Impact of BRAF mutation and microsatellite instability on the pattern of metastatic spread and prognosis in metastatic colorectal cancer. Cancer 2011, 117, 4623–4632. [Google Scholar] [CrossRef] [Green Version]
- Hyman, D.M.; Puzanov, I.; Subbiah, V.; Faris, J.E.; Chau, I.; Blay, J.Y.; Baselga, J. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. N. Engl. J. Med. 2015, 373, 726–736. [Google Scholar] [CrossRef]
- Corcoran, R.B.; Atreya, C.E.; Falchook, G.S.; Kwak, E.L.; Ryan, D.P.; Bendell, J.C.; Hamid, O.; Messersmith, W.A.; Daud, A.; Kurzrock, R.; et al. Combined BRAF and MEK Inhibition with Dabrafenib and Trametinib in BRAF V600-Mutant Colorectal Cancer. J. Clin. Oncol. 2015, 33, 4023–4031. [Google Scholar] [CrossRef] [Green Version]
- Corcoran, R.B.; André, T.; Atreya, C.E.; Schellens, J.H.M.; Yoshino, T.; Bendell, J.C.; Hollebecque, A.; McRee, A.J.; Siena, S.; Middleton, G.; et al. Combined BRAF, EGFR, and MEK Inhibition in Patients with BRAFV600E-Mutant Colorectal Cancer. Cancer Discov. 2018, 8, 428–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tahara, T.; Yamamoto, E.; Suzuki, H.; Maruyama, R.; Chung, W.; Garriga, J.; Issa, J.P.J. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 2014, 74, 1311–1318. [Google Scholar] [CrossRef] [Green Version]
- Mima, K.; Cao, Y.; Chan, A.T.; Qian, Z.R.; Nowak, J.A.; Masugi, Y.; Ogino, S. Fusobacterium nucleatum in colorectal carcinoma tissue according to tumor location. Gut 2016, 65, 1973–1980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamada, T.; Zhang, X.; Mima, K.; Bullman, S.; Sukawa, Y.; Nowak, J.A.; Kosumi, K.; Masugi, Y.; Twombly, T.S.; Cao, Y.; et al. Fusobacterium nucleatum in Colorectal Cancer Relates to Immune Response Differentially by Tumor Microsatellite Instability Status. Cancer Immunol. Res. 2018, 6, 1327–1336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Compton, C.C.; Sullivan, D.C.; Byrd, D.R.; Greene, F.L.; Gershenwald, J.E.; Hess, K.R.; Gaspar, L.E.; Washington, M.K.; Schilsky, R.L.; Edge, S. AJCC Cancer Staging Manual, 8th ed.; Amin, M.B., Edge, S.B., Greene, F.L., Byrd, D.R., Brookland, R.K., Washington, M.K., Gershenwald, J.E., Compton, C.C., Hess, K.R., Sullivan, D.C., Jessup, J.M., Brierley, J.D., Gaspar, L.E., Schilsky, R.L., Balch, C.M., Winchester, D.P., Asare, E.A., Madera, M., Gress, D.M., Meyer, L.R., Eds.; Springer International Publishing: New York, NY, USA, 2017. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dal Buono, A.; Gaiani, F.; Poliani, L.; Correale, C.; Laghi, L. Defects in MMR Genes as a Seminal Example of Personalized Medicine: From Diagnosis to Therapy. J. Pers. Med. 2021, 11, 1333. https://doi.org/10.3390/jpm11121333
Dal Buono A, Gaiani F, Poliani L, Correale C, Laghi L. Defects in MMR Genes as a Seminal Example of Personalized Medicine: From Diagnosis to Therapy. Journal of Personalized Medicine. 2021; 11(12):1333. https://doi.org/10.3390/jpm11121333
Chicago/Turabian StyleDal Buono, Arianna, Federica Gaiani, Laura Poliani, Carmen Correale, and Luigi Laghi. 2021. "Defects in MMR Genes as a Seminal Example of Personalized Medicine: From Diagnosis to Therapy" Journal of Personalized Medicine 11, no. 12: 1333. https://doi.org/10.3390/jpm11121333
APA StyleDal Buono, A., Gaiani, F., Poliani, L., Correale, C., & Laghi, L. (2021). Defects in MMR Genes as a Seminal Example of Personalized Medicine: From Diagnosis to Therapy. Journal of Personalized Medicine, 11(12), 1333. https://doi.org/10.3390/jpm11121333





