Towards Multimodal Machine Learning Prediction of Individual Cognitive Evolution in Multiple Sclerosis
Abstract
:1. Introduction
2. An Introduction to Machine Learning
- Classical approach. The first approach is to analyze the brain MR images, yielding a set of features that describe the image such as volumetric quantifications of brain structures. This allows for the use of more classical supervised learning algorithms such as linear/logistic regression, support vector machines (SVM) and random forests (RF). Table 1 summarizes some frequently used supervised learning algorithms;
- Deep learning. The second option is to use the raw brain MR images as input and use a technique called deep learning, which recently gained popularity as a subtype of machine learning. The major difference compared to classical machine learning is that it mitigates the necessity to manually transform raw data in a meaningful feature representation, the so-called “feature engineering” step, relying on human domain-specific knowledge [14]. Deep learning will automatically create meaningful representations from raw data, thus achieving representation learning [14]. This will typically yield “latent features”, which are hard to interpret by humans, but are deemed by the machine to be relevant. The advantage of deep learning lies in the more complex relationships that can be learned, while a major drawback is the need for large datasets, time and computational power.
3. Caveats for Machine Learning and Potential Solutions
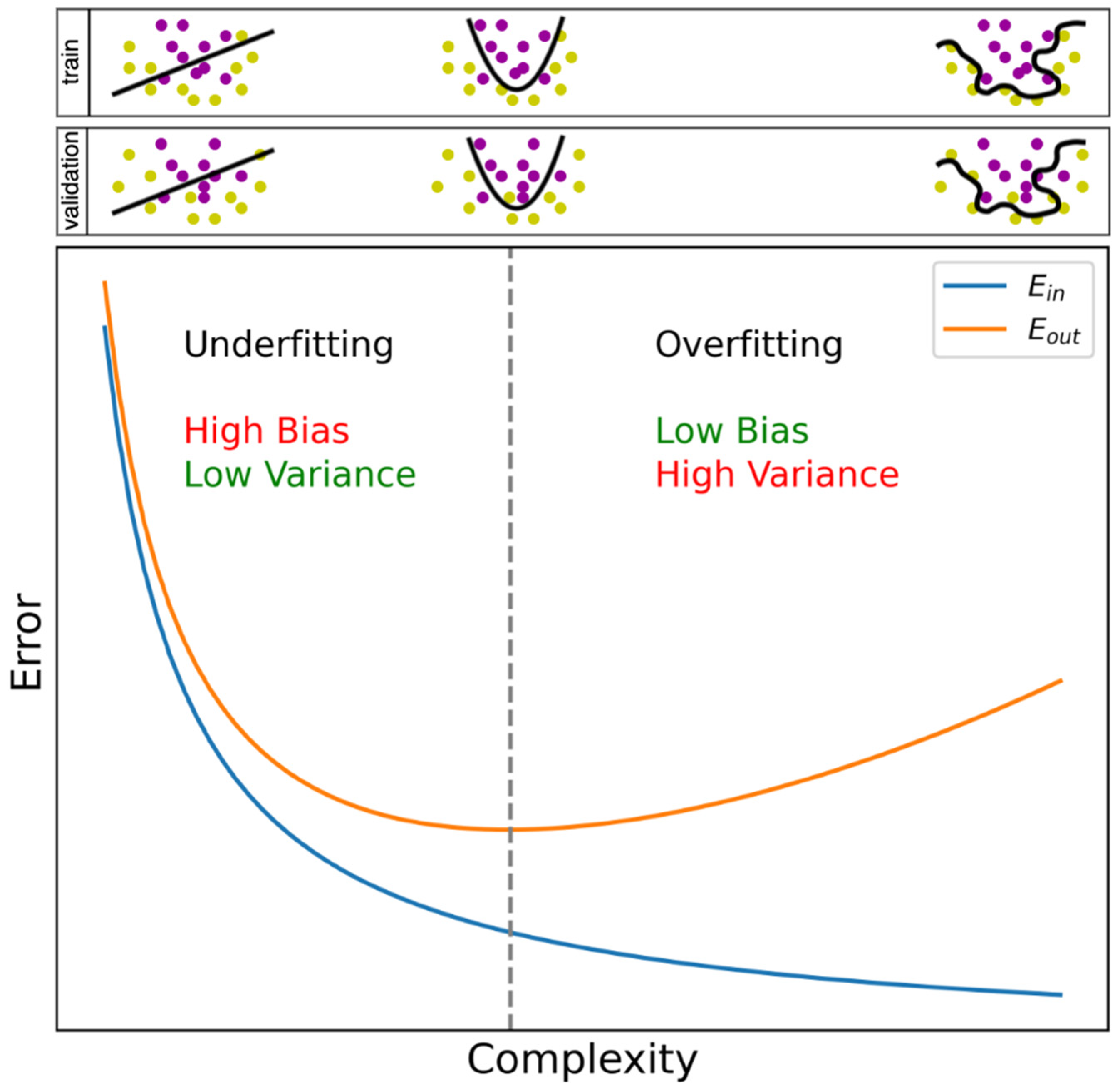
3.1. General Pitfalls in Machine Learning
- Addressing the observations. Upscaling the number of observations is one way of tackling overfitting, but researchers often possess a database with a fixed number of observations. Nonetheless, several techniques exist to increase the number of observations based on those already present, e.g., using data augmentation. Although numerous variants exist, an easy-to-grasp data augmentation method is the insertion of random noise into an observation [18], and can be interpreted as a similar, yet different subject record. A generative adversarial network (GAN) [19] serves the same purpose, which we will explain by means of a metaphor. Imagine a game-like situation in which a radiologist has to find out whether an image is a true MR image of the brain or was produced by a computer, i.e., a “villain”, trying to fool the radiologist. Initially, the radiologist will easily identify which images were produced by the villain, since it had no clue how to generate a representative image. However, since the villain receives feedback on its effort, it will gradually start to understand how to create an image that will give the radiologist a hard time in telling whether it is a true image or a fake one. The radiologist on the other hand is forced to keep on improving classification skills, since it gradually becomes harder to distinguish them, in turn stimulating the villain to propose better images. Hence, the radiologist and the villain will infinitely stimulate each other to perform better. Ultimately, MR images are produced by the villain that could in fact have been the true ones, and which can subsequently be used to expand a dataset. Similar to deep learning, a GAN needs, besides time and computational power, large amounts and diversity of data to create qualitative new observations;
- Addressing the features. The second option to restore an imbalance is the reduction of the number of features that the algorithm will be trained on. In feature selection, only the features that are deemed informative are selected. For an outline of several feature selection techniques in the context of medical sciences, we refer to Remeseiro et al., 2019 [20]. The original set of features can also be transformed to a new set of features. This can, for example, be done with principal component analysis (PCA), where we could say that the features are “reordered”; an equal number of features are obtained—the “principal components”—that explain variance in the data in a decreasing order. Feature selection can then occur on principal components instead of the original features. The additional benefit of PCA is that it is a solution to the problem of multi-collinearity, in which features are mutually correlated. As a result, two variables might contain similar information, while the resulting principal components from PCA are uncorrelated [21].
3.2. Specific Pitfalls for Medical Data
- Study data versus real-world data. Although the standardization of conditions and minimizing missing values are in general considered good practice, for example, when collecting data as part of a research study, it might limit the use of models in daily clinical routine that are known to be contaminated with, e.g., measurement errors, non-standardized test intervals, and missing values. When an algorithm encounters such inconsistencies during training, it could be expected to perform better on out-of-sample data. Although well-curated study data still dominate the field of prognostic modelling, efforts are underway to expand the use of real-world data [27,28];
- Single-center versus multi-center data. This argument is similar to the former; data from different clinical centers might be different due to discrepancies in testing equipment (e.g., MRI scanner), testing protocols, and patient characteristics. Introducing this heterogeneity already during the training phase might increase generalizability;
- Multiple visits of the same patient. Finally, when using multiple visits of a patient as separate observations in a dataset, one should always remain vigilant that the visits do not get intermingled between train, validation, and test datasets. Since visits are often highly comparable, the performance on an unseen test dataset could be biased, performing better than would be the case when adopting a truly independent test dataset. This could be categorized under the hazard called “leakage”, in which information of the test dataset leaks in the training dataset. To prevent this from occurring, Seccia et al., 2020 applied a correctional method called “leave one group out” (LOGO) [27]. With this method, they withdrew all visits of one subject from the training set and used them as a test set, after which the procedure was repeated for all subjects. This hinders models to recognize patients within a dataset. Other methods were, for example, discussed in Tacchella et al., 2018 [29] and Yperman et al., 2020 [28].
4. Designing an ML Study for Cognitive Prognosis
4.1. Which Outcome to Predict?
4.2. Which Features to Take into Account?
4.3. On Which Time-Frame Should Predictions Be Made?
4.4. Which Machine Learning Algorithm to Use?
4.5. How to Assess a Machine Learning Model?
4.6. How Should Authors Report the Performance of Their Machine Learning Model?
4.7. When Is a Model Ready for Clinical Practice?
4.8. Which Data to Use?
5. State-of-the-Art ML-Powered Cognitive Prognostic Models
- Kiiski et al., 2018
- Using the “Elastic Net” [66] as learning algorithm. This is in essence a linear regression approach, but it uses regularization, which is the addition of constraints to the learning process to increase a model’s generalizability. Specifically, it uses a combination of L1 (Lasso) and L2 (Ridge) regularization, which both tend to shrink large feature weights, whereas Lasso additionally tends to remove unimportant features from the model [66]. The low complexity of linear regression combined with regularization might have increased generalizability;
- Using cross-validation (CV), which is a technique that allows the use of data for both training and validation purposes by training multiple models. If no CV were used, only one model would have been created on a part of the data, whereas validation would happen on the remaining data. Since this is a balance between few data for training (risk for a poorly trained model) and few data for validation (risk for a poor evaluation of the model), CV is a useful technique to minimize both risks.
- Lopez-Soley et al., 2021
6. ML Trends and Opportunities for Prognostic Modelling in MS
6.1. Alternative Approaches for Prognostication
6.2. Simulation of Treatment Response
6.3. Solutions to Scarcity of Longitudinal Data
7. Conclusions
8. Key Messages
- Machine learning is capable of handling multimodal data and could predict disease course on an individual level;
- The literature on cognitive prognosis using machine learning in MS is scarce. Future studies on machine learning for prognosis in MS should not overlook cognitive deterioration;
- Recommendations for the design of studies on machine learning for cognitive prognosis are proposed;
- Researchers should aim to share as many results as possible to allow benchmarking, solid interpretation, and comparison in the field, for example, by sharing raw predictions;
- Several trends in machine learning could overcome current roadblocks in ML-powered prognostic modelling in MS, such as scarcity of longitudinal data.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Winquist, R.J.; Kwong, A.; Ramachandran, R.; Jain, J. The complex etiology of multiple sclerosis. Biochem. Pharmacol. 2007, 74, 1321–1329. [Google Scholar] [CrossRef]
- Brown, F.S.; Glasmacher, S.A.; Kearns, P.K.A.; MacDougall, N.; Hunt, D.; Connick, P.; Chandran, S. Systematic review of prediction models in relapsing remitting multiple sclerosis. PLoS ONE 2020, 15, e0233575. [Google Scholar] [CrossRef] [PubMed]
- Weinstock-Guttman, B.; Sormani, M.P.; Repovic, P. Predicting Long-term Disability in Multiple Sclerosis: A Narrative Review of Current Evidence and Future Directions. Int. J. MS Care 2021. Available online: https://meridian.allenpress.com/ijmsc/article/doi/10.7224/1537-2073.2020-114/471428/Predicting-Long-term-Disability-in-Multiple (accessed on 8 December 2021). [CrossRef]
- Seccia, R.; Romano, S.; Salvetti, M.; Crisanti, A.; Palagi, L.; Grassi, F. Machine Learning Use for Prognostic Purposes in Multiple Sclerosis. Life 2021, 11, 122. [Google Scholar] [CrossRef] [PubMed]
- Moazami, F.; Lefevre-Utile, A.; Papaloukas, C.; Soumelis, V. Machine Learning Approaches in Study of Multiple Sclerosis Disease Through Magnetic Resonance Images. Front. Immunol. 2021, 12, 3205. [Google Scholar] [CrossRef]
- Lejbkowicz, I.; Caspi, O.; Miller, A. Participatory medicine and patient empowerment towards personalized healthcare in multiple sclerosis. Expert Rev. Neurother. 2012, 12, 343–352. [Google Scholar] [CrossRef]
- Reich, D.S.; Lucchinetti, C.F.; Calabresi, P.A. Multiple Sclerosis. N. Engl. J. Med. 2018, 378, 169–180. [Google Scholar] [CrossRef]
- Kister, I.; Bacon, T.E.; Chamot, E.; Salter, A.R.; Cutter, G.R.; Kalina, J.T.; Herbert, J. Natural History of Multiple Sclerosis Symptoms. Int. J. MS Care 2013, 15, 146. [Google Scholar] [CrossRef] [Green Version]
- Ziemssen, T.; Akgün, K.; Brück, W. Molecular biomarkers in multiple sclerosis. J. Neuroinflamm. 2019, 16, 272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macías Islas, M.; Ciampi, E. Assessment and Impact of Cognitive Impairment in Multiple Sclerosis: An Overview. Biomedicines 2019, 7, 22. [Google Scholar] [CrossRef] [Green Version]
- Clemens, L.; Langdon, D. How does cognition relate to employment in multiple sclerosis? A systematic review. Mult. Scler. Relat. Disord. 2018, 26, 183–191. [Google Scholar] [CrossRef]
- Kavaliunas, A.; Karrenbauer, V.D.; Gyllensten, H.; Manouchehrinia, A.; Glaser, A.; Olsson, T.; Alexanderson, K.; Hillert, J. Cognitive function is a major determinant of income among multiple sclerosis patients in Sweden acting independently from physical disability. Mult. Scler. 2019, 25, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Definition of Machine Learning. Oxford University Press. Available online: https://www.lexico.com/definition/machine_learning (accessed on 20 October 2021).
- Lecun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Polikar, R. Ensemble Machine Learning; Zhang, C., Ma, Y., Eds.; Springer: Boston, MA, USA, 2012; ISBN 978-1-4419-9325-0. [Google Scholar]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning; Springer Series in Statistics; Springer: New York, NY, USA, 2009; ISBN 978-0-387-84857-0. [Google Scholar]
- Alibakshi, A. Strategies to develop robust neural network models: Prediction of flash point as a case study. Anal. Chim. Acta. 2018, 1026, 69–76. [Google Scholar] [CrossRef] [PubMed]
- DeVries, T.; Taylor, G.W. Dataset Augmentation in Feature Space. arXiv 2017, arXiv:1702.05538. [Google Scholar]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Networks. Commun. ACM 2014, 63, 139–144. [Google Scholar] [CrossRef]
- Remeseiro, B.; Bolon-Canedo, V. A review of feature selection methods in medical applications. Comput. Biol. Med. 2019, 112, 103375. [Google Scholar] [CrossRef]
- Jolliffe, I.T.; Cadima, J. Principal component analysis: A review and recent developments. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2016, 374, 20150202. [Google Scholar] [CrossRef]
- Bejarano, B.; Bianco, M.; Gonzalez-Moron, D.; Sepulcre, J.; Goñi, J.; Arcocha, J.; Soto, O.; Carro, U.D.; Comi, G.; Leocani, L.; et al. Computational classifiers for predicting the short-term course of Multiple sclerosis. BMC Neurol. 2011, 11, 67. [Google Scholar] [CrossRef] [Green Version]
- Yoo, Y.; Tang, L.W.; Brosch, T.; Li, D.K.B.; Metz, L.; Traboulsee, A.; Tam, R. Deep Learning of Brain Lesion Patterns for Predicting Future Disease Activity in Patients with Early Symptoms of Multiple Sclerosis. In Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Berlin/Heidelberg, Germany, 2016; Volume 10008 LNCS, pp. 86–94. ISBN 9783319469751. [Google Scholar]
- Zhao, Y.; Healy, B.C.; Rotstein, D.; Guttmann, C.R.G.; Bakshi, R.; Weiner, H.L.; Brodley, C.E.; Chitnis, T. Exploration of machine learning techniques in predicting multiple sclerosis disease course. PLoS ONE 2017, 12, e0174866. [Google Scholar] [CrossRef] [Green Version]
- Tousignant, A.; Lemaître, P.; Precup, D.; Arnold, D.L.; Arbel, T. Prediction of Disease Progression in Multiple Sclerosis Patients using Deep Learning Analysis of MRI Data. In Proceedings of the 2nd International Conference on Medical Imaging with Deep Learning, London, UK, 8–10 July 2019; Volume 102. [Google Scholar]
- Buda, M.; Maki, A.; Mazurowski, M.A. A systematic study of the class imbalance problem in convolutional neural networks. Neural Netw. 2018, 106, 249–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seccia, R.; Gammelli, D.; Dominici, F.; Romano, S.; Landi, A.C.; Salvetti, M.; Tacchella, A.; Zaccaria, A.; Crisanti, A.; Grassi, F.; et al. Considering patient clinical history impacts performance of machine learning models in predicting course of multiple sclerosis. PLoS ONE 2020, 15, e0230219. [Google Scholar] [CrossRef] [Green Version]
- Yperman, J.; Becker, T.; Valkenborg, D.; Popescu, V.; Hellings, N.; Van Wijmeersch, B.; Peeters, L.M. Machine learning analysis of motor evoked potential time series to predict disability progression in multiple sclerosis. BMC Neurol. 2020, 20, 105. [Google Scholar] [CrossRef] [Green Version]
- Tacchella, A.; Romano, S.; Ferraldeschi, M.; Salvetti, M.; Zaccaria, A.; Crisanti, A.; Grassi, F. Collaboration between a human group and artificial intelligence can improve prediction of multiple sclerosis course: A proof-of-principle study. F1000Research 2018, 6, 2172. [Google Scholar] [CrossRef]
- Kourou, K.; Exarchos, T.P.; Exarchos, K.P.; Karamouzis, M.V.; Fotiadis, D.I. Machine learning applications in cancer prognosis and prediction. Comput. Struct. Biotechnol. J. 2015, 13, 8–17. [Google Scholar] [CrossRef] [Green Version]
- Sumowski, J.F.; Benedict, R.; Enzinger, C.; Filippi, M.; Geurts, J.J.; Hamalainen, P.; Hulst, H.; Inglese, M.; Leavitt, V.M.; Rocca, M.A.; et al. Cognition in multiple sclerosis: State of the field and priorities for the future. Neurology 2018, 90, 278–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oreja-Guevara, C.; Ayuso Blanco, T.; Brieva Ruiz, L.; Hernández Pérez, M.Á.; Meca-Lallana, V.; Ramió-Torrentà, L. Cognitive Dysfunctions and Assessments in Multiple Sclerosis. Front. Neurol. 2019, 10, 581. [Google Scholar] [CrossRef]
- Ouellette, R.; Bergendal, Å.; Shams, S.; Martola, J.; Mainero, C.; Kristoffersen Wiberg, M.; Fredrikson, S.; Granberg, T. Lesion accumulation is predictive of long-term cognitive decline in multiple sclerosis. Mult. Scler. Relat. Disord. 2018, 21, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Costers, L.; Gielen, J.; Eelen, P.L.; Van Schependom, J.; Laton, J.; Van Remoortel, A.; Vanzeir, E.; Van Wijmeersch, B.; Seeldrayers, P.; Haelewyck, M.C.; et al. Does including the full CVLT-II and BVMT-R improve BICAMS? Evidence from a Belgian (Dutch) validation study. Mult. Scler. Relat. Disord. 2017, 18, 33–40. [Google Scholar] [CrossRef] [Green Version]
- Eijlers, A.J.C.; van Geest, Q.; Dekker, I.; Steenwijk, M.D.; Meijer, K.A.; Hulst, H.E.; Barkhof, F.; Uitdehaag, B.M.J.; Schoonheim, M.M.; Geurts, J.J.G. Predicting cognitive decline in multiple sclerosis: A 5-year follow-up study. Brain 2018, 141, 2605–2618. [Google Scholar] [CrossRef]
- Filippi, M.; Preziosa, P.; Copetti, M.; Riccitelli, G.; Horsfield, M.A.; Martinelli, V.; Comi, G.; Rocca, M.A. Gray matter damage predicts the accumulation of disability 13 years later in MS. Neurology 2013, 81, 1759–1767. [Google Scholar] [CrossRef]
- Colato, E.; Stutters, J.; Tur, C.; Narayanan, S.; Arnold, D.L.; Gandini Wheeler-Kingshott, C.A.M.; Barkhof, F.; Ciccarelli, O.; Chard, D.T.; Eshaghi, A. Predicting disability progression and cognitive worsening in multiple sclerosis using patterns of grey matter volumes. J. Neurol. Neurosurg. Psychiatry 2021, 92, 995–1006. [Google Scholar] [CrossRef] [PubMed]
- Portaccio, E.; Goretti, B.; Zipoli, V.; Iudice, A.; Pina, D.D.; Malentacchi, G.M.; Sabatini, S.; Annunziata, P.; Falcini, M.; Mazzoni, M.; et al. Reliability, practice effects, and change indices for Raos brief repeatable battery. Mult. Scler. 2010, 16, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Cacciaguerra, L.; Pagani, E.; Mesaros, S.; Dackovic, J.; Dujmovic-Basuroski, I.; Drulovic, J.; Valsasina, P.; Filippi, M.; Rocca, M.A. Dynamic volumetric changes of hippocampal subfields in clinically isolated syndrome patients: A 2-year MRI study. Mult. Scler. J. 2019, 25, 1232–1242. [Google Scholar] [CrossRef] [PubMed]
- Beier, M.; Amtmann, D.; Ehde, D.M. Beyond depression: Predictors of self-reported cognitive function in adults living with MS. Rehabil. Psychol. 2015, 60, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Degenhardt, A.; Ramagopalan, S.V.; Scalfari, A.; Ebers, G.C. Clinical prognostic factors in multiple sclerosis: A natural history review. Nat. Rev. Neurol. 2009, 5, 672–682. [Google Scholar] [CrossRef] [PubMed]
- Louapre, C.; Bodini, B.; Lubetzki, C.; Freeman, L.; Stankoff, B. Imaging markers of multiple sclerosis prognosis. Curr. Opin. Neurol. 2017, 30, 231–236. [Google Scholar] [CrossRef]
- Kearney, H.; Miller, D.H.; Ciccarelli, O. Spinal cord MRI in multiple sclerosis—diagnostic, prognostic and clinical value. Nat. Rev. Neurol. 2015, 11, 327–338. [Google Scholar] [CrossRef]
- Davda, N.; Tallantyre, E.; Robertson, N.P. Early MRI predictors of prognosis in multiple sclerosis. J. Neurol. 2019, 266, 3171–3173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leocani, L.; Rocca, M.A.; Comi, G. MRI and neurophysiological measures to predict course, disability and treatment response in multiple sclerosis. Curr. Opin. Neurol. 2016, 29, 243–253. [Google Scholar] [CrossRef]
- Dekker, I.; Eijlers, A.J.C.; Popescu, V.; Balk, L.J.; Vrenken, H.; Wattjes, M.P.; Uitdehaag, B.M.J.; Killestein, J.; Geurts, J.J.G.; Barkhof, F.; et al. Predicting clinical progression in multiple sclerosis after 6 and 12 years. Eur. J. Neurol. 2019, 26, 893–902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuchs, T.A.; Wojcik, C.; Wilding, G.E.; Pol, J.; Dwyer, M.G.; Weinstock-Guttman, B.; Zivadinov, R.; Benedict, R.H. Trait Conscientiousness predicts rate of longitudinal SDMT decline in multiple sclerosis. Mult. Scler. J. 2020, 26, 245–252. [Google Scholar] [CrossRef]
- Hildesheim, F.E.; Benedict, R.H.B.; Zivadinov, R.; Dwyer, M.G.; Fuchs, T.; Jakimovski, D.; Weinstock-Guttman, B.; Bergsland, N. Nucleus basalis of Meynert damage and cognition in patients with multiple sclerosis. J. Neurol. 2021, 268, 4796–4808. [Google Scholar] [CrossRef]
- Bsteh, G.; Hegen, H.; Teuchner, B.; Amprosi, M.; Berek, K.; Ladstätter, F.; Wurth, S.; Auer, M.; Di Pauli, F.; Deisenhammer, F.; et al. Peripapillary retinal nerve fibre layer as measured by optical coherence tomography is a prognostic biomarker not only for physical but also for cognitive disability progression in multiple sclerosis. Mult. Scler. J. 2019, 25, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Gold, S.M.; Raji, A.; Huitinga, I.; Wiedemann, K.; Schulz, K.-H.; Heesen, C. Hypothalamo–pituitary–adrenal axis activity predicts disease progression in multiple sclerosis. J. Neuroimmunol. 2005, 165, 186–191. [Google Scholar] [CrossRef]
- Nauta, I.M.; Kulik, S.D.; Breedt, L.C.; Eijlers, A.J.; Strijbis, E.M.; Bertens, D.; Tewarie, P.; Hillebrand, A.; Stam, C.J.; Uitdehaag, B.M.; et al. Functional brain network organization measured with magnetoencephalography predicts cognitive decline in multiple sclerosis. Mult. Scler. J. 2021, 27, 1727–1737. [Google Scholar] [CrossRef]
- Brichetto, G.; Bragadin, M.M.; Fiorini, S.; Battaglia, M.A.; Konrad, G.; Ponzio, M.; Pedullà, L.; Verri, A.; Barla, A.; Tacchino, A. The hidden information in patient-reported outcomes and clinician-assessed outcomes: Multiple sclerosis as a proof of concept of a machine learning approach. Neurol. Sci. 2020, 41, 459–462. [Google Scholar] [CrossRef] [Green Version]
- De Groot, V.; Beckerman, H.; Uitdehaag, B.M.; Hintzen, R.Q.; Minneboo, A.; Heymans, M.W.; Lankhorst, G.J.; Polman, C.H.; Bouter, L.M. Physical and Cognitive Functioning After 3 Years Can Be Predicted Using Information From the Diagnostic Process in Recently Diagnosed Multiple Sclerosis. Arch. Phys. Med. Rehabil. 2009, 90, 1478–1488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sidey-Gibbons, J.A.M.; Sidey-Gibbons, C.J. Machine learning in medicine: A practical introduction. BMC Med. Res. Methodol. 2019, 19, 64. [Google Scholar] [CrossRef] [Green Version]
- Kuceyeski, A.; Monohan, E.; Morris, E.; Fujimoto, K.; Vargas, W.; Gauthier, S.A. Baseline biomarkers of connectome disruption and atrophy predict future processing speed in early multiple sclerosis. NeuroImage Clin. 2018, 19, 417–424. [Google Scholar] [CrossRef]
- Kiiski, H.; Jollans, L.; Donnchadha, S.Ó.; Nolan, H.; Lonergan, R.; Kelly, S.; O’Brien, M.C.; Kinsella, K.; Bramham, J.; Burke, T.; et al. Machine Learning EEG to Predict Cognitive Functioning and Processing Speed Over a 2-Year Period in Multiple Sclerosis Patients and Controls. Brain Topogr. 2018, 31, 346–363. [Google Scholar] [CrossRef]
- Schulz, K.F. CONSORT 2010 Statement: Updated Guidelines for Reporting Parallel Group Randomized Trials. Ann. Intern. Med. 2010, 152, 726. [Google Scholar] [CrossRef] [Green Version]
- Moons, K.G.M.; Altman, D.G.; Reitsma, J.B.; Ioannidis, J.P.A.; Macaskill, P.; Steyerberg, E.W.; Vickers, A.J.; Ransohoff, D.F.; Collins, G.S. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): Explanation and elaboration. Ann. Intern. Med. 2015, 162, W1–W73. [Google Scholar] [CrossRef] [Green Version]
- Sutton, R.T.; Pincock, D.; Baumgart, D.C.; Sadowski, D.C.; Fedorak, R.N.; Kroeker, K.I. An overview of clinical decision support systems: Benefits, risks, and strategies for success. NPJ Digit. Med. 2020, 3, 17. [Google Scholar] [CrossRef] [Green Version]
- Asan, O.; Bayrak, A.E.; Choudhury, A. Artificial Intelligence and Human Trust in Healthcare: Focus on Clinicians. J. Med. Internet Res. 2020, 22, e15154. [Google Scholar] [CrossRef] [PubMed]
- Romero, K.; Shammi, P.; Feinstein, A. Neurologists’ accuracy in predicting cognitive impairment in multiple sclerosis. Mult. Scler. Relat. Disord. 2015, 4, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Comparison of the Accuracy of the Neurological Prognosis at 6 Months of Traumatic Brain Injury Between Junior and Senior Doctors—Full Text View—ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/NCT04810039 (accessed on 2 November 2021).
- Gunning, D.; Stefik, M.; Choi, J.; Miller, T.; Stumpf, S.; Yang, G.-Z. XAI—Explainable artificial intelligence. Sci. Robot. 2019, 4, eaay7120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez-Soley, E.; Martinez-Heras, E.; Andorra, M.; Solanes, A.; Radua, J.; Montejo, C.; Alba-Arbalat, S.; Sola-Valls, N.; Pulido-Valdeolivas, I.; Sepulveda, M.; et al. Dynamics and Predictors of Cognitive Impairment along the Disease Course in Multiple Sclerosis. J. Pers. Med. 2021, 11, 1107. [Google Scholar] [CrossRef]
- Memarian, N.; Kim, S.; Dewar, S.; Engel, J., Jr.; Staba, R.J. Multimodal data and machine learning for surgery outcome prediction in complicated cases of mesial temporal lobe epilepsy. Comput. Biol. Med. 2015, 64, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, H.; Hastie, T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Ser. B (Stat. Methodol.) 2005, 67, 301–320. [Google Scholar] [CrossRef] [Green Version]
- Voigt, I.; Inojosa, H.; Dillenseger, A.; Haase, R.; Akgün, K.; Ziemssen, T. Digital Twins for Multiple Sclerosis. Front. Immunol. 2021, 12, 1556. [Google Scholar] [CrossRef]
- Pruenza, C.; Solano, M.T.; Diaz, J.; Arroyo-Gonzalez, R.; Izquierdo, G. Model for Prediction of Progression in Multiple Sclerosis. Int. J. Interact. Multimed. Artif. Intell. 2019, 5, 48–53. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Li, F.-F. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef] [Green Version]
- Nanni, L.; Interlenghi, M.; Brahnam, S.; Salvatore, C.; Papa, S.; Nemni, R.; Castiglioni, I.; Initiative, T.A.D.N. Comparison of Transfer Learning and Conventional Machine Learning Applied to Structural Brain MRI for the Early Diagnosis and Prognosis of Alzheimer’s Disease. Front. Neurol. 2020, 11, 576194. [Google Scholar] [CrossRef] [PubMed]
- Van Panhuis, W.G.; Paul, P.; Emerson, C.; Grefenstette, J.; Wilder, R.; Herbst, A.J.; Heymann, D.; Burke, D.S. A systematic review of barriers to data sharing in public health. BMC Public Health 2014, 14, 1144. [Google Scholar] [CrossRef] [Green Version]
- Brisimi, T.S.; Chen, R.; Mela, T.; Olshevsky, A.; Paschalidis, I.C.; Shi, W. Federated learning of predictive models from federated Electronic Health Records. Int. J. Med. Inform. 2018, 112, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Aledhari, M.; Razzak, R.; Parizi, R.M.; Saeed, F. Federated Learning: A Survey on Enabling Technologies, Protocols, and Applications. IEEE Access Pract. Innov. Open Solut. 2020, 8, 140699–140725. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.S.; Lee, A.Y. Clinical applications of continual learning machine learning. Lancet Digit. Health 2020, 2, e279–e281. [Google Scholar] [CrossRef]

| Method | Description | Visualization |
|---|---|---|
| Logistic Regression | Logistic regression identifies the optimal sigmoid curve between the two labels to be predicted, yielding a probability of belonging to either of the two groups. In the illustration: the probability that a person will worsen or stabilize over time. |  |
| Decision Tree | A decision tree is a sequence of decisions that are made on certain criteria. The last leaves of the tree indicate one of the class labels that are to be predicted. |  |
| Random Forest | This is an example of “ensemble learning”, meaning that learning, and thus the resulting model, relies on multiple learning strategies, aiming to average the error out [15]. In this case, a random forest consists of multiple decision trees, mitigating the bias introduced by relying on one single decision tree. The ultimate prediction of a random forest classifier is the majority vote of the predictions of the individual decision trees in the random forest. |  |
| SVM | In case of two features, a support vector machine (SVM) tries to find a line or a curve that separates the two classes of interest. It does so by maximizing the distance between the line and the data-points on both sides of the line, thus maximally separating both classes. |  |
| ANN | An artificial neural network (ANN) was inspired by the neural network of the brain and consists of nodes (weights) and edges that connect the nodes. Input data in either raw form or a feature representation enters the ANN on the left (input layer) and gets modified by the ANN in the hidden layers using the nodes’ weights learned during the training phase, so that the input is optimally reshaped, or “mapped”, to the endpoint that needs to be predicted on the right (output layer). |  |
| Linear Regression | Linear regression is a technique in which the weight of every input feature is learned, which is multiplied with their respective feature and summed together with the so-called “bias” (also a learned weight but not associated to a feature, i.e., a constant), yielding a prediction that minimizes the error with the ground-truth. In the 2D case, this is the line that minimizes the sum of the squared vertical distances of individual points to the regression line. The learned weights in this case are the slope (β1) and intercept (β0, bias) of the line. |  |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Denissen, S.; Chén, O.Y.; De Mey, J.; De Vos, M.; Van Schependom, J.; Sima, D.M.; Nagels, G. Towards Multimodal Machine Learning Prediction of Individual Cognitive Evolution in Multiple Sclerosis. J. Pers. Med. 2021, 11, 1349. https://doi.org/10.3390/jpm11121349
Denissen S, Chén OY, De Mey J, De Vos M, Van Schependom J, Sima DM, Nagels G. Towards Multimodal Machine Learning Prediction of Individual Cognitive Evolution in Multiple Sclerosis. Journal of Personalized Medicine. 2021; 11(12):1349. https://doi.org/10.3390/jpm11121349
Chicago/Turabian StyleDenissen, Stijn, Oliver Y. Chén, Johan De Mey, Maarten De Vos, Jeroen Van Schependom, Diana Maria Sima, and Guy Nagels. 2021. "Towards Multimodal Machine Learning Prediction of Individual Cognitive Evolution in Multiple Sclerosis" Journal of Personalized Medicine 11, no. 12: 1349. https://doi.org/10.3390/jpm11121349






