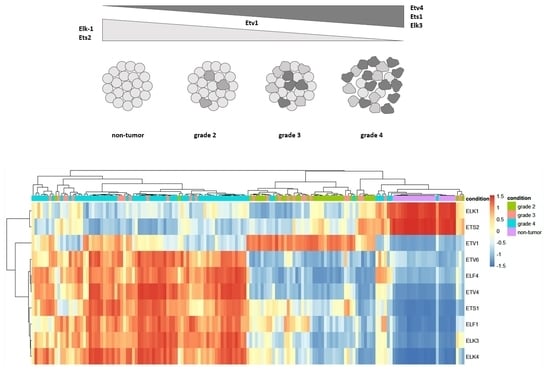
Gene Regulatory Network of ETS Domain Transcription Factors in Different Stages of Glioma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Data Processing and Differentially Gene Expression Analysis
2.3. Gene Regulatory Network Construction
2.4. Functional Enrichment Analysis
3. Results
3.1. Identification of Differentially Expressed Genes in Gliomas
3.2. Transcriptional Gene Regulatory Network Construction
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, Z.; Zhang, Q. Genome-wide identification and evolutionary analysis of the animal specific ETS transcription factor family. Evol. Bioinf. 2009, 5, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Hollenhorst, P.; McIntosh, L.P.; Graves, B.J. Genomic and biochemical insights into the specificity of ETS transcription factors. Annu. Rev. Biochem. 2011, 80, 437–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boros, J.; O’Donnell, A.; Donaldson, I.J.; Kasza, A.; Zeef, L.; Sharrocks, A.D. Overlapping promoter targeting by Elk-1 and other divergent ETS-domain transcription factor family members. Nucleic Acids Res. 2009, 37, 7368–7380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kar, A.; Gutierrez-Hartmann, A. Molecular mechanisms of ETS transcription factor mediated tumorigenesis. Crit. Rev. Biochem. Mol. Biol 2013, 48, 522–543. [Google Scholar] [CrossRef] [Green Version]
- Yuan, C.C.; Dunn, K.J.; Papas, T.S.; Blair, D.G. Properties of a murine retroviral recombinant of avian acute leukemia virus E26: A murine fibroblast assay for v-ets function. J. Virol. 1989, 63, 205–215. [Google Scholar] [CrossRef] [Green Version]
- Shore, P.; Whitmarsh, A.J.; Bhaskaran, R.; Davis, R.J.; Waltho, J.P.; Sharrocks, A.D. Determinants of DNA-binding specificity of ETS-domain transcription factors. Mol. Cell. Biol. 1996, 16, 3338–3349. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.-H.; Whitmarsh, A.J.; Davis, R.J.; Sharrocks, A.D. Differential targeting of MAP kinases to the ETS-domain transcription factor Elk-1. EMBO J. 1998, 17, 1740–1749. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.-H.; Shore, P.; Willingham, N.; Lakey, J.H.; Sharrocks, A.D. The mechanism of phosphorylation-inducible activation of the ETS domain transcription factor Elk-1. EMBO J. 1999, 18, 5666–5674. [Google Scholar] [CrossRef] [Green Version]
- Hsu, T.; Trojanowska, M.; Watson, D.K. Ets proteins in biological control and cancer. J Cell. Biochem. 2004, 91, 896–903. [Google Scholar] [CrossRef] [Green Version]
- Furlan, A.; Vercamer, C.; Heliot, L.; Wernert, N.; Desbiens, X.; Pourtier, A. Ets-1 drives breast cancer cell angiogenic potential and interactions between breast cancer and endothelial cells. Int. J. Oncol. 2019, 54, 29–40. [Google Scholar] [CrossRef] [Green Version]
- Verschoor, M.L.; Verschoor, C.P.; Singh, G. Ets-1 global gene expression profile reveals associations with metabolism and oxidative stress in ovarian and breast cancers. Cancer Metab. 2013, 1, 17. [Google Scholar] [CrossRef] [Green Version]
- Fry, E.A.; Inoue, K. Aberrant expression of ETS1 and ETS2 proteins in cancer. Cancer Rep. Rev. 2018, 2. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.F.; Sul, J.-Y.; Kim, M.-S.; Klein-Szanto, A.J.; Schochet, T.; Rustgi, A.; Eberwine, J.H. Elk-1 phosphorylated at threonine-417 is present in diverse cancers and correlates with differentiation grade of colonic adenocarcinoma. Hum. Pathol. 2013, 44, 766–776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pardy, L.; Rosati, R.; Soave, C.; Huang, Y.; Kim, S.; Ratnam, M. The ternary complex factor protein ELK-1 is an independent prognosticator of disease recurrence in prostate cancer. Prostate 2019, 80, 198–208. [Google Scholar] [CrossRef]
- Ahmad, A.; Hayat, A. Expression of oncogenes ELK1 and ELK3 in cancer. Annal. Colorec. Cancer Res. 2019, 1, 1–6. [Google Scholar]
- Yue, C.-H.; Huang, C.-Y.; Tsai, J.-H.; Hsu, C.-W.; Hsieh, Y.-H.; Liu, J.-Y. MZF-1/Elk-1 complex binds to protein kinase Calpha promoter and is involved in hepatocellular carcinoma. PLoS ONE 2015, 10, e0127420. [Google Scholar] [CrossRef] [Green Version]
- Kherrouche, Z.; Monte, D.; Werkmeister, E.; Stoven, L.; de Launoit, Y.; Cortot, A.B.; Tulasne, D.; Chotteau-Lelievre, A. PEA3 transcription factors are dowmstream effectors of Met signaling involved in migration and invasiveness of Met-addicted tumor cells. Mol. Oncol. 2015, 9, 1852–1867. [Google Scholar] [CrossRef]
- Mesci, A.; Taeb, S.; Huang, X.; Jairath, R.; Sivaloganathan, D.; Liu, S.K. PEA3 expression promotes the invasive and metastatic potential of colorectal carcinoma. World J. Gastroenterol. 2014, 20, 17376–17387. [Google Scholar] [CrossRef]
- Qi, T.; Qu, Q.; Li, G.; Wang, J.; Zhu, H.; Yang, Z.; Sun, Y.; Lu, Q.; Qu, J. Function and regulation of the PEA3 subfamily of ETS transcription factors in cancer. Am. J. Cancer Res. 2020, 10, 3083–3105. [Google Scholar]
- Olar, A.; Sulman, E.P. Molecular markers in low grade glioma—toward tumor reclassification. Semin. Radiat. Oncol. 2015, 25, 155–163. [Google Scholar] [CrossRef] [Green Version]
- Wood, M.D.; Halfpenny, A.M.; Moore, S.R. Applications of molecular neurooncology- a review of diffuse glioma integrated diagnosis and emerging molecular entities. Diag. Pathol. 2019, 14, 29. [Google Scholar] [CrossRef] [Green Version]
- Mehta, S.; Lo Cascio, C. Developmentally regulated signaling pathways in glioma invasion. Cell. Mol. Life Sci. 2018, 75, 385–402. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Gomez, G.A.; Zhao, Y.; Yang, Y.; Cao, D.; Lu, J.; Yang, H.; Lin, S. ETV2 mediates endothelial transdifferentiation of glioblastoma. Signal Transduc. Tar. Ther. 2018, 3, 4. [Google Scholar] [CrossRef]
- Haber, M.A.; Iranmahboob, A.; Thomas, C.; Liu, M.; Najjar, A.; Zagzag, D. ERG is a novel and reliable marker for endothelial cells in central nervous system tumors. Clin. Neuropathol. 2015, 34, 117–127. [Google Scholar] [CrossRef] [Green Version]
- Vyazunova, I.; Maklakova, V.I.; Berman, S.; De, I.; Steffen, M.D.; Hong, W.; Lincoln, H.; Morrissy, A.S.; Taylor, M.D.; Akagi, K.; et al. Sleeping beauty mouse models identify candidate genes involved in gliomagenesis. PLoS ONE 2014, 9, e113489. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Gu, S.; Bi, Y.; Qi, X.; Yan, Y.; Lou, M. Transcription factor PU.1 is involved in the progression of glioma. Oncol. Lett. 2018, 15, 3753–3759. [Google Scholar] [CrossRef] [Green Version]
- Pan, Y.; Duron, C.; Bush, E.C.; Ma, Y.; Sims, P.A.; Gutmann, D.H.; Radunskaya, A.; Hardin, J. Graph complexity analysis identifies an ETV5 tumor-specific network in human and murine low-grade glioma. PLoS ONE 2018, 13, e0190001. [Google Scholar] [CrossRef]
- Barthel, F.P.; Wesseling, P.; Verhaak, R.G.W. Reconstructing the molecular life history of gliomas. Acta Neuropathol. 2018, 135, 649–670. murine low-grade glioma. PLoS ONE 2018, 13, e0190001. [Google Scholar] [CrossRef] [Green Version]
- Torres, A.; Alshalalfa, M.; Tomlins, S.A.; Erho, N.; Gibb, E.A.; Chelliserry, J.; Lim, L.; Lam, L.L.C.; Faraj, S.F.; Bezerra, S.M.; et al. Comprehensive determination of prostate tumor ETS gene status in clinical samples using the CLIA decipher assay. J. Mol. Diag. 2017, 19, 475–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.; Hui, A.-M.; Su, Q.; Vortmeyer, A.; Kotliarov, Y.; Pastorino, S.; Passaniti, A.; Menon, J.; Walling, J.; Bailey, R.; et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell 2006, 9, 287–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, S.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Blighe, K.; Rana, S.; Lewis, M. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling. 2020. Available online: https://github.com/kevinblighe/EnhancedVolcano (accessed on 14 December 2020).
- Campbell, T.M.; Castro, M.A.A.; de Santiago, I.; Fletcher, M.N.C.; Halim, S.; Prathalingam, R.; Ponder, B.A.J.; Meyer, K.B. FGFR2 risk SNPs confer breast cancer risk by augmenting oestrogen responsiveness. Carcinogenesis 2016, 37, 741–750. [Google Scholar] [CrossRef] [Green Version]
- Castro, M.A.A.; de Santiago, I.; Campbell, T.M.; Vaughn, C.; Hickey, T.E.; Ross, E.; Tilley, W.D.; Markowetz, F.; Ponder, B.A.J.; Meyer, K.B. Regulators of genetic risk of breast cancer identified by integrative network analysis. Nat. Genet. 2016, 48, 12–21. [Google Scholar] [CrossRef] [Green Version]
- Fletcher, M.N.C.; Castro, M.A.A.; Wang, X.; de Santiago, I.; O’Reilly, M.; Chin, S.-F.; Rueda, O.M.; Caldas, C.; Ponder, B.A.J.; Markowetz, F.; et al. Master regulators of FGFR2 signalling and breast cancer risk. Nat. Commun. 2013, 4, 2464. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.; Han, Y.; He, Q. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Xu, H.; Zhao, G.; Zhang, Y.; Jiang, H.; Wang, W.; Zhao, D.; Yu, H.; Qi, L. Long non-coding RNA PAXIP1-AS1 facilitates cell invasion and angiogenesis of glioma by recruiting transcription factor ETS1 to upregulate KIF14 expression. J. Exp. Clin. Cancer Res. 2019, 38, 486. [Google Scholar] [CrossRef] [Green Version]
- Cam, M.; Charan, M.; Welker, A.M.; Dravid, P.; Studebaker, A.W.; Leonard, J.R.; Pierson, C.R.; Nakano, I.; Beattie, C.E.; Hwang, E.I.; et al. DeltaNp73/ETS2 complex drives glioblastoma pathogenesis-targeting downstream mediators by rebastinib prolongs survival in preclinical models of glioblastoma. Neuro-oncology 2020, 22, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Giese, A. Glioma invasion-pattern of dissemination by mechanisms of invasion and surgical intervention, pattern of gene expression and its regulatory control by tumor suppressor p53 and proto-oncogene ETS-1. Acta Neurochir. Suppl. 2003, 88, 153–162. [Google Scholar] [PubMed]
- Mut, M.; Lule, S.; Demir, O.; Kurnaz, I.A.; Vural, I. Both mitogen-activated protein kinase (MAPK)/extracellular signal regulated kinases (ERK) ½ and phosphatidylinositide-3-OH kinase (PI3K)/Akt pathways regulate activation of E-twenty-six (ETS)-like transcription factor-1 (Elk-1) in U138 glioblastoma cells. Int. J. Biochem. Cell. Biol. 2012, 44, 302–310. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yuan, H.; Sun, C.; Xu, L.; Chen, Y.; Zhu, Q.; Zhao, H.; Huang, Q.; Dong, J.; Lan, Q. GATA2 promotes glioma progression through EGFR/ERK/Elk-1 pathway. Med. Oncol. 2015, 32, 87. [Google Scholar] [CrossRef]
- Savasan Sogut, M.; Venugopal, C.; Kandemir, B.; Dag, U.; Mahendram, S.; Singh, S.; Gulfidan, G.; Arga, K.Y.; Yilmaz, B.; Aksan Kurnaz, I. ETS-domainn transcription factor Elk-1 in brain tumors and CD133+ brain tumor initiating cells. J. Per. Med. 2021, 11, 125. [Google Scholar] [CrossRef]
- Cheng, M.; Zeng, Y.; Zhang, T.; Xu, M.; Li, Z.; Wu, Y. Transcription factor ELF1 activates MEIS1 transcription factor and then regulates the GFI1/FBW7 axis to promote the development of glioma. Mol. Ther. Nucleic Acids 2020, 23, 418–430. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yang, C.; Liu, X.; Zheng, J.; Xue, Y.; Ruan, X.; Shen, S.; Wang, D.; Li, Z.; Cai, H.; et al. An upstream open reading frame regulates vasculogenic mimicry of glioma via ZNRD1-AS1/miR-499a-5p/ELF1/EMI1 pathway. Cell. Mol. Med. 2020, 24, 6120–6136. [Google Scholar] [CrossRef]
- Padul, V.; Epari, S.; Moiyadi, A.; Shetty, P.; Shirsat, N.V. ETV/PEA3 family transcription factor-encoding genes are overexpressed in CIC-mutant oligodendrogliomas. Genes Chrom. Cancer 2015, 54, 725–733. [Google Scholar] [CrossRef] [PubMed]
- Gatalica, Z.; Xiu, J.; Swensen, J.; Vranic, S. Molecular characterization of cancers with MTRK gene fusions. Mod. Pathol. 2019, 32, 147–153. [Google Scholar] [CrossRef]
- Busse, T.M.; Roth, J.J.; Wilmoth, D.; Wainright, L.; Tooke, L.; Biegel, J.A. Copy number alterations determined by single nucleotide polymorphism array testing in the clinical laboratory are indicative of gene fusions in pediatric cancer patients. Genes Chrom. Cancer 2017, 56, 730–749. [Google Scholar] [CrossRef]
- Kjær, C.; Barzaghi, G.; Bak, L.K.; Goetze, J.P.; Yde, C.W.; Woldbye, D.; Pinborg, L.H.; Jensen, L.J. Transcriptome analysis in patients with temporal lobe epilepsy. Brain 2019, 142, e55. [Google Scholar] [CrossRef]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Babal, Y.K.; Kandemir, B.; Kurnaz, I.A. Gene Regulatory Network of ETS Domain Transcription Factors in Different Stages of Glioma. J. Pers. Med. 2021, 11, 138. https://doi.org/10.3390/jpm11020138
Babal YK, Kandemir B, Kurnaz IA. Gene Regulatory Network of ETS Domain Transcription Factors in Different Stages of Glioma. Journal of Personalized Medicine. 2021; 11(2):138. https://doi.org/10.3390/jpm11020138
Chicago/Turabian StyleBabal, Yigit Koray, Basak Kandemir, and Isil Aksan Kurnaz. 2021. "Gene Regulatory Network of ETS Domain Transcription Factors in Different Stages of Glioma" Journal of Personalized Medicine 11, no. 2: 138. https://doi.org/10.3390/jpm11020138
APA StyleBabal, Y. K., Kandemir, B., & Kurnaz, I. A. (2021). Gene Regulatory Network of ETS Domain Transcription Factors in Different Stages of Glioma. Journal of Personalized Medicine, 11(2), 138. https://doi.org/10.3390/jpm11020138