Deep Learning Can Differentiate IDH-Mutant from IDH-Wild GBM
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Histopathological Analysis
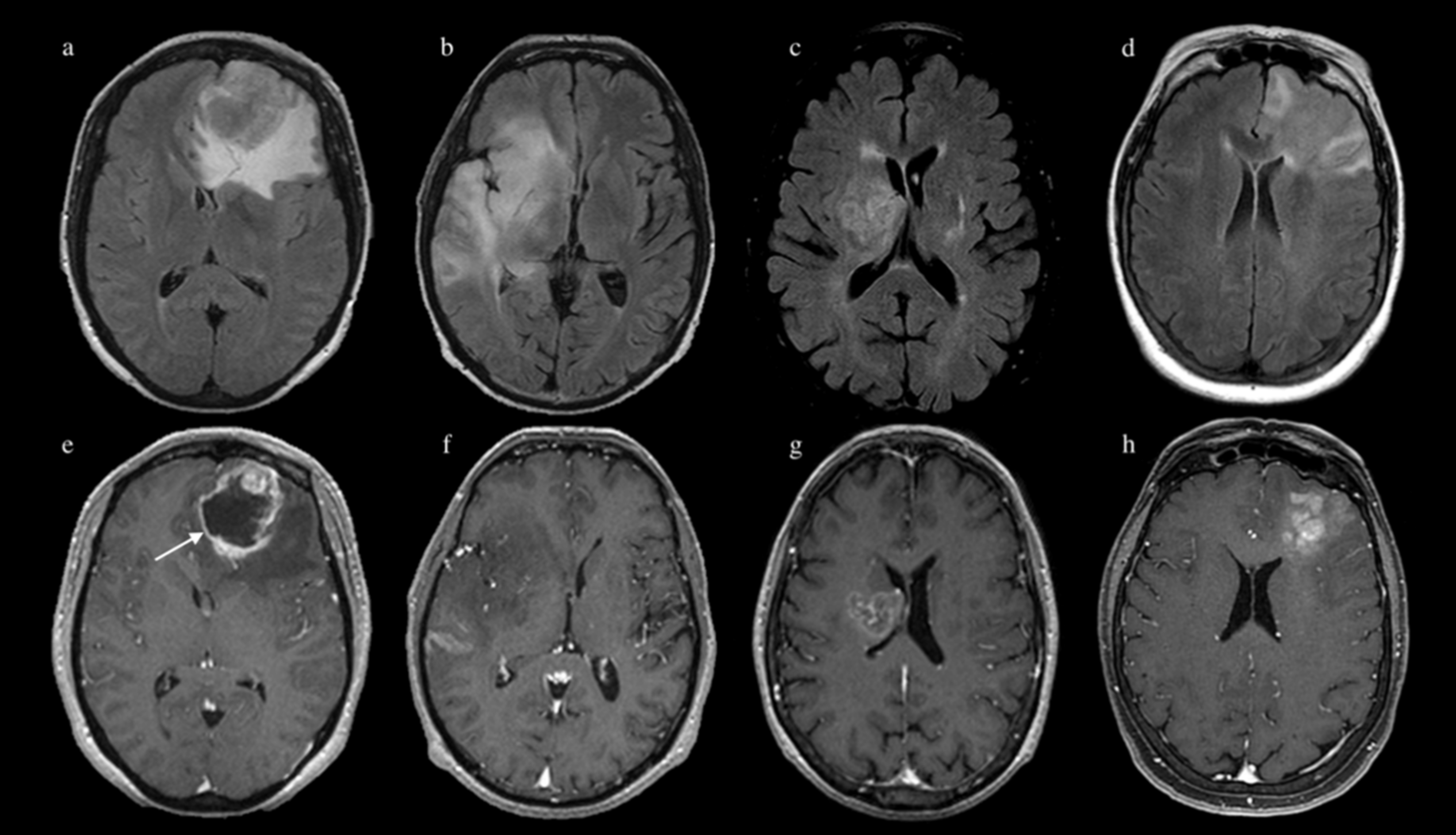
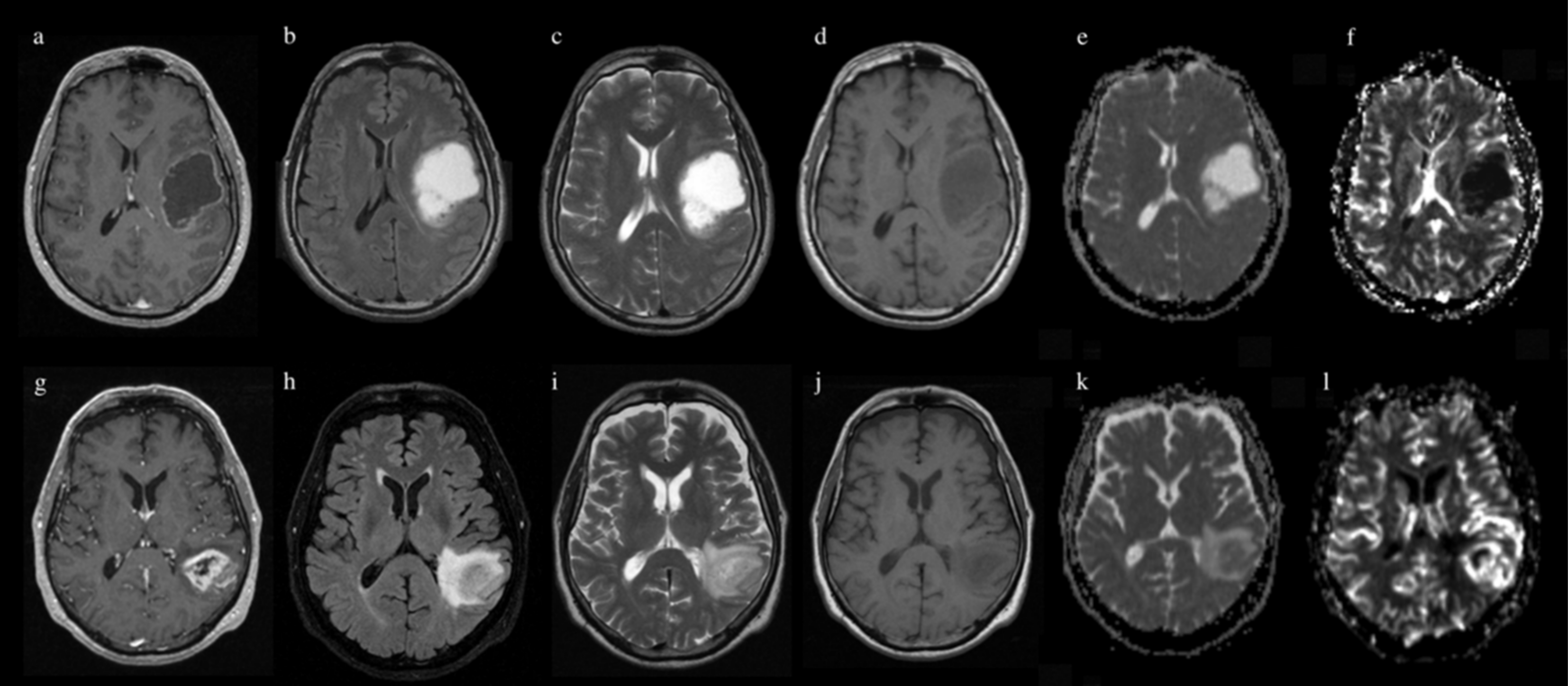
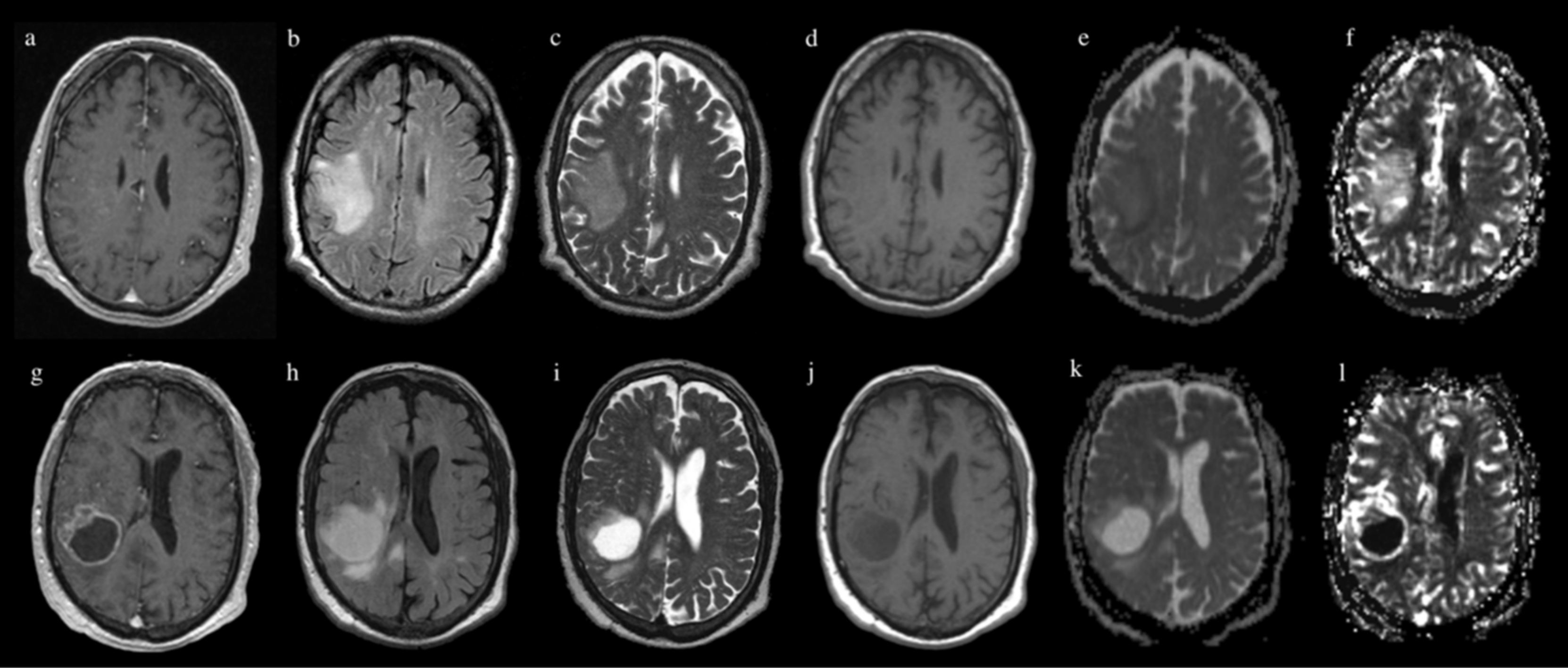
2.3. MR Image Acquisition
2.4. Image Processing
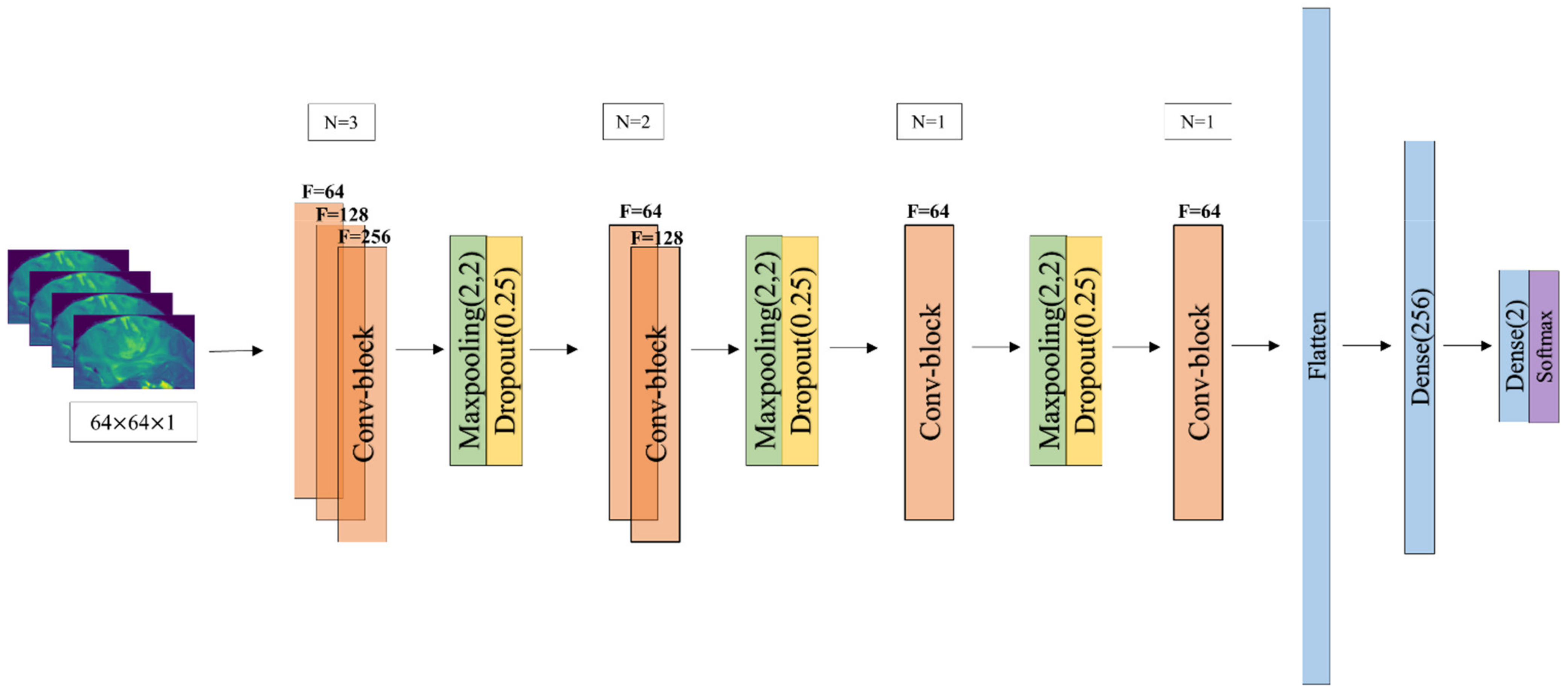
2.5. CNN Architecture
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Molinaro, A.M.; Taylor, J.W.; Wiencke, J.K.; Wrensch, M.R. Genetic and Molecular Epidemiology of Adult Diffuse Glioma. Nat. Rev. Neurol. 2019, 15, 405–417. [Google Scholar] [CrossRef] [PubMed]
- Tamimi, A.F.; Juweid, M. Epidemiology and Outcome of Glioblastoma. In Glioblastoma; De Vleeschouwer, S., Ed.; Codon Publications: Brisbane, Australia, 2017. [Google Scholar]
- Wang, J.; Bettegowda, C. Genomic Discoveries in Adult Astrocytoma. Curr. Opin. Genet. Dev. 2015, 30, 17–24. [Google Scholar] [CrossRef]
- Han, S.; Liu, Y.; Cai, S.J.; Qian, M.; Ding, J.; Larion, M.; Gilbert, M.R.; Yang, C. IDH Mutation in Glioma: Molecular Mechanisms and Potential Therapeutic Targets. Br. J. Cancer 2020, 122, 1580–1589. [Google Scholar] [CrossRef]
- Johnson, D.R.; Guerin, J.B.; Giannini, C.; Morris, J.M.; Eckel, L.J.; Kaufmann, T.J. 2016 Updates to the WHO Brain Tumor Classification System: What the Radiologist Needs to Know. Radiographics 2017, 37, 2164–2180. [Google Scholar] [CrossRef]
- Nobusawa, S.; Watanabe, T.; Kleihues, P.; Ohgaki, H. IDH1 Mutations as Molecular Signature and Predictive Factor of Secondary Glioblastomas. Clin. Cancer Res. 2009, 15, 6002–6007. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. Mutations in Gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Dang, L.; White, D.W.; Gross, S.; Bennett, B.D.; Bittinger, M.A.; Driggers, E.M.; Fantin, V.R.; Jang, H.G.; Jin, S.; Keenan, M.C.; et al. Cancer-Associated IDH1 Mutations Produce 2-Hydroxyglutarate. Nature 2009, 462, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, C.; Hentschel, B.; Wick, W.; Capper, D.; Felsberg, J.; Simon, M.; Westphal, M.; Schackert, G.; Meyermann, R.; Pietsch, T.; et al. Patients with IDH1 Wild Type Anaplastic Astrocytomas Exhibit Worse Prognosis than IDH1-Mutated Glioblastomas, and IDH1 Mutation Status Accounts for the Unfavorable Prognostic Effect of Higher Age: Implications for Classification of Gliomas. Acta Neuropathol. 2010, 120, 707–718. [Google Scholar] [CrossRef]
- Louis, D.N.; Wesseling, P.; Aldape, K.; Brat, D.J.; Capper, D.; Cree, I.A.; Eberhart, C.; Figarella-Branger, D.; Fouladi, M.; Fuller, G.N.; et al. CIMPACT-NOW Update 6: New Entity and Diagnostic Principle Recommendations of the CIMPACT-Utrecht Meeting on Future CNS Tumor Classification and Grading. Brain Pathol. 2020, 30, 844–856. [Google Scholar] [CrossRef] [PubMed]
- Raab, S.; Grzybicki, D.; Janosky, J. Clinical Impact and Frequency of Anatomic Pathology Errors in Cancer Diagnoses. Cancer 2005, 104, 2205–2213. [Google Scholar] [CrossRef] [PubMed]
- Loponte, S.; Lovisa, S.; Deem, A.K.; Carugo, A.; Viale, A. The Many Facets of Tumor Heterogeneity: Is Metabolism Lagging Behind? Cancers 2019, 11, 1574. [Google Scholar] [CrossRef] [PubMed]
- Cryan, J.B.; Haidar, S.; Ramkissoon, L.A.; Bi, W.L.; Knoff, D.S.; Schultz, N.; Abedalthagafi, M.; Brown, L.; Wen, P.Y.; Reardon, D.A.; et al. Clinical Multiplexed Exome Sequencing Distinguishes Adult Oligodendroglial Neoplasms from Astrocytic and Mixed Lineage Gliomas. Oncotarget 2014, 5, 8083–8092. [Google Scholar] [CrossRef] [PubMed]
- Hegi, M.E.; Diserens, A.-C.; Gorlia, T.; Hamou, M.-F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT Gene Silencing and Benefit from Temozolomide in Glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Gutman, D.A.; Dunn, W.D.; Grossmann, P.; Cooper, L.A.D.; Holder, C.A.; Ligon, K.L.; Alexander, B.M.; Aerts, H.J.W.L. Somatic Mutations Associated with MRI-Derived Volumetric Features in Glioblastoma. Neuroradiology 2015, 57, 1227–1237. [Google Scholar] [CrossRef] [PubMed]
- Fathi Kazerooni, A.; Bakas, S.; Saligheh Rad, H.; Davatzikos, C. Imaging Signatures of Glioblastoma Molecular Characteristics: A Radiogenomics Review. J. Magn. Reson. Imaging 2020, 52, 54–69. [Google Scholar] [CrossRef]
- Chow, D.; Chang, P.; Weinberg, B.D.; Bota, D.A.; Grinband, J.; Filippi, C.G. Imaging Genetic Heterogeneity in Glioblastoma and Other Glial Tumors: Review of Current Methods and Future Directions. Am. J. Roentgenol. 2018, 210, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Smits, M.; van den Bent, M.J. Imaging Correlates of Adult Glioma Genotypes. Radiology 2017, 284, 316–331. [Google Scholar] [CrossRef] [PubMed]
- Shaver, M.M.; Kohanteb, P.A.; Chiou, C.; Bardis, M.D.; Chantaduly, C.; Bota, D.; Filippi, C.G.; Weinberg, B.; Grinband, J.; Chow, D.S.; et al. Optimizing Neuro-Oncology Imaging: A Review of Deep Learning Approaches for Glioma Imaging. Cancers 2019, 11, 829. [Google Scholar] [CrossRef] [PubMed]
- Ostergaard, L.; Weisskoff, R.M.; Chesler, D.A.; Gyldensted, C.; Rosen, B.R. High Resolution Measurement of Cerebral Blood Flow Using Intravascular Tracer Bolus Passages. Part I: Mathematical Approach and Statistical Analysis. Magn. Reson. Med. 1996, 36, 715–725. [Google Scholar] [CrossRef] [PubMed]
- Boxerman, J.L.; Schmainda, K.M.; Weisskoff, R.M. For Contrast Agent Extravasation Significantly Correlate with Glioma Tumor Grade, Whereas. Ajnr. Am. J. Neuroradiol. 2006, 27, 859–867. [Google Scholar] [PubMed]
- Hao, J.; Kim, Y.; Kim, T.K.; Kang, M. PASNet: Pathway-Associated Sparse Deep Neural Network for Prognosis Prediction from High-Throughput Data. BMC Bioinform. 2018, 19, 1–13. [Google Scholar] [CrossRef]
- Nie, D.; Lu, J.; Zhang, H.; Adeli, E.; Wang, J.; Yu, Z.; Liu, L.Y.; Wang, Q.; Wu, J.; Shen, D. Multi-Channel 3D Deep Feature Learning for Survival Time Prediction of Brain Tumor Patients Using Multi-Modal Neuroimages. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef]
- Han, L.; Kamdar, M.R. MRI to MGMT: Predicting Methylation Status in Glioblastoma Patients Using Convolutional Recurrent Neural Networks. Pac. Symp. Biocomput. 2018, 23, 331–342. [Google Scholar] [CrossRef]
- Chang, K.; Bai, H.X.; Zhou, H.; Su, C.; Bi, W.L.; Agbodza, E.; Kavouridis, V.K.; Senders, J.T.; Boaro, A.; Beers, A.; et al. Residual Convolutional Neural Network for the Determination of IDH Status in Low- and High-Grade Gliomas from Mr Imaging. Clin. Cancer Res. 2018, 24, 1073–1081. [Google Scholar] [CrossRef]
- Liang, S.; Zhang, R.; Liang, D.; Song, T.; Ai, T.; Xia, C.; Xia, L.; Wang, Y. Multimodal 3D Densenet for IDH Genotype Prediction in Gliomas. Genes 2018, 9, 382. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A Survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Pasquini, L.; Napolitano, A.; Lucignani, M.; Tagliente, E.; Rossi-espagnet, M.C.; Vidiri, A.; Ranazzi, G.; Stoppacciaro, A.; Romano, A.; di Napoli, A.; et al. Comparison of Machine Learning Classifiers to Predict Patient Survival and Genetics of GBM: Towards a Standardized Model for Clinical Implementation. arXiv 2021, arXiv:2102.06526. [Google Scholar]
- Suter, Y.; Jungo, A.; Rebsamen, M.; Knecht, U.; Herrmann, E.; Wiest, R.; Reyes, M. Deep Learning Versus Classical Regression for Brain Tumor Patient Survival Prediction. arXiv 2018, arXiv:1811.04907. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2015, arXiv:1412.6980. [Google Scholar]
- Chang, P.; Grinband, J.; Weinberg, B.D.; Bardis, M.; Khy, M.; Cadena, G.; Su, M.Y.; Cha, S.; Filippi, C.G.; Bota, D.; et al. Deep-Learning Convolutional Neural Networks Accurately Classify Genetic Mutations in Gliomas. Am. J. Neuroradiol. 2018, 39, 1201–1207. [Google Scholar] [CrossRef]
- Lu, C.; Ward, P.S.; Kapoor, G.S.; Rohle, D.; Turcan, S.; Abdel-Wahab, O.; Edwards, C.R.; Khanin, R.; Figueroa, M.E.; Melnick, A.; et al. IDH Mutation Impairs Histone Demethylation and Results in a Block to Cell Differentiation. Nature 2012, 483, 474–478. [Google Scholar] [CrossRef]
- Barajas, R.F.; Hodgson, J.G.; Chang, J.S.; Vandenberg, S.R.; Yeh, R.F.; Parsa, A.T.; McDermott, M.W.; Berger, M.S.; Dillon, W.P.; Cha, S. Glioblastoma Multiforme Regional Genetic and Cellular Expression Patterns: Influence on Anatomic and Physiologic MR Imaging. Radiology 2010, 254, 564–576. [Google Scholar] [CrossRef]
- Barajas, R.F.; Phillips, J.J.; Parvataneni, R.; Molinaro, A.; Essock-burns, E.; Bourne, G.; Parsa, A.T.; Aghi, M.K.; Mcdermott, M.W.; Berger, M.S.; et al. Regional Variation in Histopathologic Features. Neuro Oncol. 2012, 14, 942–954. [Google Scholar] [CrossRef]
- Hsieh, K.L.C.; Chen, C.Y.; Lo, C.M. Radiomic Model for Predicting Mutations in the Isocitrate Dehydrogenase Gene in Glioblastomas. Oncotarget 2017, 8, 45888–45897. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Chang, K.; Ramkissoon, S.; Tanguturi, S.; Bi, W.L.; Reardon, D.A.; Ligon, K.L.; Alexander, B.M.; Wen, P.Y.; Huang, R.Y. Multimodal MRI Features Predict Isocitrate Dehydrogenase Genotype in High-Grade Gliomas. Neuro Oncol. 2017, 19, 109–117. [Google Scholar] [CrossRef]
- Parmar, C.; Grossmann, P.; Bussink, J.; Lambin, P.; Aerts, H.J.W.L. Machine Learning Methods for Quantitative Radiomic Biomarkers. Sci. Rep. 2015, 5, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Larue, R.T.H.M.; Defraene, G.; de Ruysscher, D.; Lambin, P.; van Elmpt, W. Quantitative Radiomics Studies for Tissue Characterization: A Review of Technology and Methodological Procedures. Br. J. Radiol. 2017, 90, 20160665. [Google Scholar] [CrossRef] [PubMed]
- Jang, K.; Russo, C.; di Ieva, A. Radiomics in Gliomas: Clinical Implications of Computational Modeling and Fractal-Based Analysis. Neuroradiology 2020, 62, 771–790. [Google Scholar] [CrossRef] [PubMed]
- Zeiler, M.; Fergus, R. Visualizing and Understanding Convolutional Networks. arXiv 2013, arXiv:1311.2901. [Google Scholar]
- Kickingereder, P.; Sahm, F.; Radbruch, A.; Wick, W.; Heiland, S.; von Deimling, A.; Bendszus, M.; Wiestler, B. IDH Mutation Status Is Associated with a Distinct Hypoxia/Angiogenesis Transcriptome Signature Which Is Non-Invasively Predictable with RCBV Imaging in Human Glioma. Sci. Rep. 2015, 5, 1–9. [Google Scholar] [CrossRef]
- Wu, H.; Tong, H.; Du, X.; Guo, H.; Ma, Q.; Zhang, Y.; Zhou, X.; Liu, H.; Wang, S.; Fang, J.; et al. Vascular Habitat Analysis Based on Dynamic Susceptibility Contrast Perfusion MRI Predicts IDH Mutation Status and Prognosis in High-Grade Gliomas. Eur. Radiol. 2020, 30, 3254–3265. [Google Scholar] [CrossRef] [PubMed]
- Law, M.; Yang, S.; Babb, J.S.; Knopp, E.A.; Golfinos, J.G.; Zagzag, D.; Johnson, G. Comparison of Cerebral Blood Volume and Vascular Permeability from Dynamic Susceptibility Contrast-Enhanced Perfusion MR Imaging with Glioma Grade. Am. J. Neuroradiol. 2004, 25, 746–755. [Google Scholar] [PubMed]
- Romano, A.; Pasquini, L.; di Napoli, A.; Tavanti, F.; Boellis, A.; Rossi Espagnet, M.C.; Minniti, G.; Bozzao, A. Prediction of Survival in Patients Affected by Glioblastoma: Histogram Analysis of Perfusion MRI. J. Neuro-Oncol. 2018, 139, 455–460. [Google Scholar] [CrossRef]
- Yalaza, C.; Ak, H.; Cagli, M.S.; Ozgiray, E.; Atay, S.; Aydin, H.H. R132H Mutation in IDH1 Gene Is Associated with Increased Tumor HIF1-Alpha and Serum VEGF Levels in Primary Glioblastoma Multiforme. Ann. Clin. Lab. Sci. 2017, 47, 362–364. [Google Scholar]
- Chow, D.S.; Horenstein, C.I.; Canoll, P.; Lignelli, A.; Hillman, E.M.C.; Filippi, C.G.; Grinband, J. Glioblastoma Induces Vascular Dysregulation in Nonenhancing Peritumoral Regions in Humans. Am. J. Roentgenol. 2016, 206, 1073–1081. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gadda, D.; Mazzoni, L.N.; Pasquini, L.; Busoni, S.; Simonelli, P.; Giordano, G.P. Relationship between Apparent Diffusion Coefficients and MR Spectroscopy Findings in High-Grade Gliomas. J. Neuroimaging 2017. [Google Scholar] [CrossRef]
- La Violette, P.S.; Mickevicius, N.J.; Cochran, E.J.; Rand, S.D.; Connelly, J.; Bovi, J.A.; Malkin, M.G.; Mueller, W.M.; Schmainda, K.M. Precise Ex Vivo Histological Validation of Heightened Cellularity and Diffusion-Restricted Necrosis in Regions of Dark Apparent Diffusion Coefficient in 7 Cases of High-Grade Glioma. Neuro Oncol. 2014, 16, 1599–1606. [Google Scholar] [CrossRef] [PubMed]
- Romano, A.; Calabria, L.F.; Tavanti, F.; Minniti, G.; Rossi-Espagnet, M.C.; Coppola, V.; Pugliese, S.; Guida, D.; Francione, G.; Colonnese, C.; et al. Apparent Diffusion Coefficient Obtained by Magnetic Resonance Imaging as a Prognostic Marker in Glioblastomas: Correlation with MGMT Promoter Methylation Status. Eur. Radiol. 2013, 23, 513–520. [Google Scholar] [CrossRef] [PubMed]


| Sequence | Subjects Intersection | Train/Test (Split = 0.2) | AugTrain/Test | Train:0/1 | Test:0/1 |
|---|---|---|---|---|---|
| T1 | 83 | 65/17 | 106/17 | 53/53 | 5/12 |
| T2 | 84 | 66/17 | 108/17 | 54/54 | 5/14 |
| FLAIR | 96 | 76/19 | 124/19 | 62/62 | 5/14 |
| MPRAGE | 97 | 76/20 | 128/20 | 64/64 | 5/15 |
| rCBV | 66 | 52/14 | 90/14 | 45/45 | 5/9 |
| ADC | 93 | 73/19 | 124/19 | 62/62 | 5/14 |
| Sequence | Test Acc | Test Loss | SN | SP | AUC |
|---|---|---|---|---|---|
| T1 | 0.77 ± 0.11 | 1.4± 09 | 0.36 ± 0.29 | 0.95 ± 0.07 | 0.71 ± 0.23 |
| T2 | 0.67 ± 0.12 | 2.41 ± 0.77 | 0.48 ± 0.16 | 0.75 ± 0.12 | 0.63 ± 0.07 |
| FLAIR | 0.77 ± 0.11 | 1.98 ± 1.06 | 0.28 ± 0.27 | 0.95 ± 0.03 | 0.74 ± 0.13 |
| MPRAGE | 0.66 ± 0.1 | 2.55 ± 0.93 | 0.43 ± 0.24 | 0.74 ± 0.17 | 0.62 ± 0.15 |
| rCBV | 0.83 ± 0.07 | 0.64 ± 0.23 | 0.76 ± 0.08 | 0.86 ± 0.08 | 0.86 ± 0.05 |
| ADC | 0.56 ± 0.11 | 2.53 ± 0.28 | 0.14 ± 0.07 | 0.73 ± 0.16 | 0.45 ± 0.04 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pasquini, L.; Napolitano, A.; Tagliente, E.; Dellepiane, F.; Lucignani, M.; Vidiri, A.; Ranazzi, G.; Stoppacciaro, A.; Moltoni, G.; Nicolai, M.; et al. Deep Learning Can Differentiate IDH-Mutant from IDH-Wild GBM. J. Pers. Med. 2021, 11, 290. https://doi.org/10.3390/jpm11040290
Pasquini L, Napolitano A, Tagliente E, Dellepiane F, Lucignani M, Vidiri A, Ranazzi G, Stoppacciaro A, Moltoni G, Nicolai M, et al. Deep Learning Can Differentiate IDH-Mutant from IDH-Wild GBM. Journal of Personalized Medicine. 2021; 11(4):290. https://doi.org/10.3390/jpm11040290
Chicago/Turabian StylePasquini, Luca, Antonio Napolitano, Emanuela Tagliente, Francesco Dellepiane, Martina Lucignani, Antonello Vidiri, Giulio Ranazzi, Antonella Stoppacciaro, Giulia Moltoni, Matteo Nicolai, and et al. 2021. "Deep Learning Can Differentiate IDH-Mutant from IDH-Wild GBM" Journal of Personalized Medicine 11, no. 4: 290. https://doi.org/10.3390/jpm11040290
APA StylePasquini, L., Napolitano, A., Tagliente, E., Dellepiane, F., Lucignani, M., Vidiri, A., Ranazzi, G., Stoppacciaro, A., Moltoni, G., Nicolai, M., Romano, A., Di Napoli, A., & Bozzao, A. (2021). Deep Learning Can Differentiate IDH-Mutant from IDH-Wild GBM. Journal of Personalized Medicine, 11(4), 290. https://doi.org/10.3390/jpm11040290








