Hypoxia Transcriptomic Modifications Induced by Proton Irradiation in U87 Glioblastoma Multiforme Cell Line
Abstract
:1. Introduction
2. Materials and Methods
2.1. GSI Hypoxic Chambers
2.2. Cell Culture Preparation and Proton Irradiation Set-Up
2.3. Clonogenic Assay
2.4. Whole-Genome cDNA Microarray Expression Analysis
3. Results
3.1. Survival Curves
3.2. Overview of cDNA Microarray Gene Expression Analyses under PT/Hypoxia Conditions
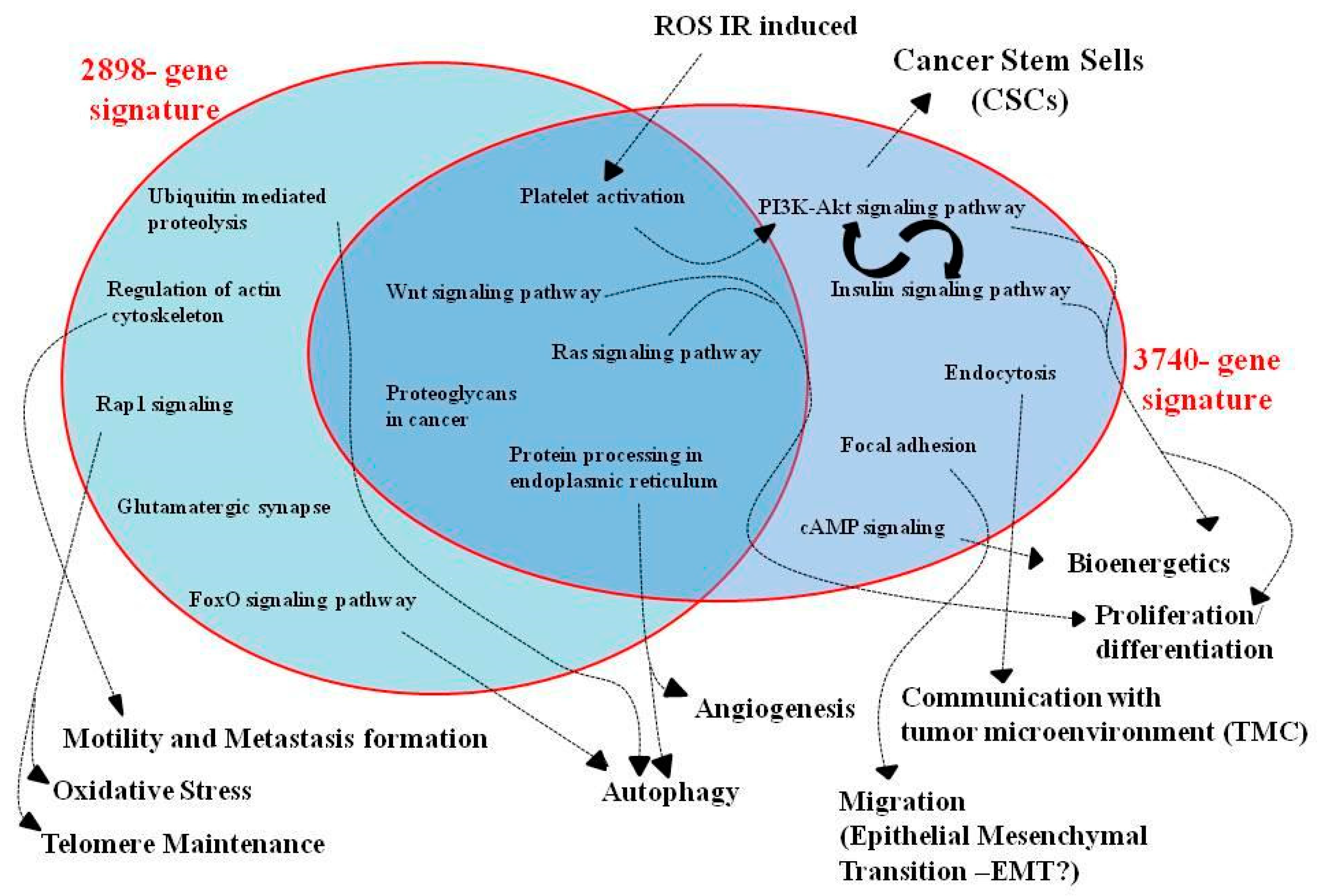
3.3. Pathway Analysis of GEP Lists under Combined PT/Hypoxia Conditions
3.4. Commonly Deregulated Genes and Pathways among PT-Treated Samples under Normoxia vs. Hypoxia Condition
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mudassar, F.; Shen, H.; O’Neill, G.; Hau, E. Targeting tumor hypoxia and mitochondrial metabolism with anti-parasitic drugs to improve radiation response in high-grade gliomas. J. Exp. Clin. Cancer Res. 2020, 39, 208. [Google Scholar] [CrossRef]
- Montemurro, N. Glioblastoma Multiforme and Genetic Mutations: The Issue Is Not Over Yet. An Overview of the Current Literature. J. Neurol. Surg. A Cent. Eur. Neurosurg. 2020, 81, 64–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Yan, Q.; Liao, B.; Zhao, L.; Xiong, S.; Wang, J.; Zou, D.; Pan, J.; Wu, L.; Deng, Y.; et al. The HIF1α/HIF2α-miR210-3p network regulates glioblastoma cell proliferation, dedifferentiation and chemoresistance through EGF under hypoxic conditions. Cell Death Dis. 2020, 11, 992. [Google Scholar] [CrossRef] [PubMed]
- Brahimi-Horn, M.; Chiche, J.; Pouysségur, J. Hypoxia and cancer. J. Mol. Med. 2007, 85, 1301–1307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baumann, R.; Depping, R.; Delaperriere, M.; Dunst, J. Targeting hypoxia to overcome radiation resistance in head & neck cancers: Real challenge or clinical fairytale? Expert Rev. Anticancer Ther. 2016, 16, 751–758. [Google Scholar] [PubMed]
- William, R.W.; Michael, P.H. Targeting hypoxia in cancer therapy. Nat. Rev. Cancer 2011, 11, 393. [Google Scholar]
- Tinganelli, W.; Durante, M.; Hirayama, R. Kill-painting of hypoxic tumours in charged particle therapy. Sci. Rep. 2015, 5, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Torrisi, F.; Minafra, L.; Cammarata, F.P.; Savoca, G.; Calvaruso, M.; Vicario, N.; Maccari, L.; Pérès, E.A.; Özçelik, H.; Bernaudin, M.; et al. SRC Tyrosine Kinase Inhibitor and X-rays Combined Effect on Glioblastoma Cell Lines. Int. J. Mol. Sci. 2020, 21, 3917. [Google Scholar] [CrossRef]
- Wenzl, T.; Wilkens, J.J. Theoretical analysis of the dose dependence of the oxygen enhancement ratio and its relevance for clinical applications. Radiat. Oncol. 2011, 6, 171. [Google Scholar] [CrossRef] [Green Version]
- Cabrera, A.R.; Kirkpatrick, J.P.; Fiveash, J.B.; Shih, H.A.; Koay, E.J.; Lutz, S.; Petit, J.; Chao, S.T.; Brown, P.D.; Vogelbaum, M.; et al. Radiation therapy for glioblastoma: Executive summary of an American Society for Radiation Oncology Evidence-Based Clinical Practice Guideline. Pract. Radiat. Oncol. 2016, 6, 217–225. [Google Scholar] [CrossRef] [Green Version]
- Cammarata, F.P.; Torrisi, F.; Forte, G.I.; Minafra, L.; Bravatà, V.; Pisciotta, P.; Savoca, G.; Calvaruso, M.; Petringa, G.; Cirrone, G.A.P.; et al. Proton Therapy and Src Family Kinase Inhibitor Combined Treatments on U87 Human Glioblastoma Multiforme Cell Line. Int. J. Mol. Sci. 2019, 20, 4745. [Google Scholar] [CrossRef] [Green Version]
- Saeed, A.M.; Khairnar, R.; Sharma, A.M.; Larson, G.L.; Tsai, H.K.; Wang, C.J.; Halasz, L.M.; Chinnaiyan, P.; Vargas, C.E.; Mishra, M.V. Clinical Outcomes in Patients with Recurrent Glioblastoma Treated with Proton Beam Therapy Reirradiation: Analysis of the Multi-Institutional Proton Collaborative Group Registry. Adv. Radiat. Oncol. 2020, 5, 978–983. [Google Scholar] [CrossRef]
- Combs, S.E.; Schmid, T.E.; Vaupel, P.; Multhoff, G. Stress Response Leading to Resistance in Glioblastoma-The Need for Innovative Radiotherapy (iRT) Concepts. Cancers 2016, 8, 15. [Google Scholar] [CrossRef] [Green Version]
- Mahmoudi, K.; Bouras, A.; Bozec, D.; Ivkov, R.; Hadjipanayis, C. Magnetic hyperthermia therapy for the treatment of glioblastoma: A review of the therapy’s history, efficacy and application in humans. Int. J. Hyperth. 2018, 34, 1316–1328. [Google Scholar] [CrossRef] [Green Version]
- Mohan, R.; Liu, A.Y.; Brown, P.D.; Mahajan, A.; Dinh, J.; Chung, C.; McAvoy, S.; McAleer, M.F.; Lin, S.H.; Li, J.; et al. Proton Therapy Reduces the Likelihood of High-Grade Radiation-Induced Lymphopenia in Glioblastoma Patients: Phase II Randomized Study of Protons vs. Photons. Neuro Oncol. 2020, 23, 284–294. [Google Scholar] [CrossRef] [PubMed]
- Tinganelli, W.; Ma, N.Y.; Von Neubeck, C.; Maier, A.; Schicker, C.; Kraft-Weyrather, W.; Durante, M. Influence of acute hypoxia and radiation quality on cell survival. J. Radiat. Res. 2013, 54, i23–i30. [Google Scholar] [CrossRef]
- Petringa, G.; Romano, F.; Manti, L.; Pandola, L.; Attili, A.; Cammarata, F.; Cuttone, G.; Forte, G.; Manganaro, L.; Pipek, J.; et al. Radiobiological quantities in proton-therapy: Estimation and validation using Geant4-based Monte Carlo simulations. Phys. Med. 2019, 58, 72–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bravatà, V.; Cammarata, F.P.; Minafra, L.; Musso, R.; Pucci, G.; Spada, M.; Fazio, I.; Russo, G.; Forte, G.I. Gene Expression Profiles Induced by High-dose Ionizing Radiation in MDA-MB-231 Triple-negative Breast Cancer Cell Line. Cancer Genom. Proteom. 2019, 16, 257–266. [Google Scholar] [CrossRef] [Green Version]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Kimberly, A.; Marshall, K.; Phillippy, K.H.; Sherman, P.M.; et al. NCI GEO: Archive for functional genomics data sets-update. Nucleic. Acids. Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becker, K.G.; Hosack, D.A.; Dennis, G.; Lempicki, R.A.; Bright, T.J.; Cheadle, C.; Engel, J. PubMatrix: A tool for multiplex literature mining. BMC Bioinform. 2003, 10, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bravatà, V.; Minafra, L.; Cammarata, F.P.; Pisciotta, P.; Lamia, D.; Marchese, V.; Petringa, G.; Manti, L.; Cirrone, G.A.; Gilardi, M.C.; et al. Gene expression profiling of breast cancer cell lines treated with proton and electron radiations. Br. J. Radiol. 2018, 91, 20170934. [Google Scholar] [CrossRef]
- Trone, J.C.; Vallard, A.; Sotton, S.; Ben Mrad, M.; Jmour, O.; Magné, N.; Pommier, B.; Laporte, S.; Ollier, E. Survival after hypofractionation in glioblastoma: A systematic review and meta-analysis. Radiat. Oncol. 2020, 15, 145. [Google Scholar] [CrossRef]
- Minafra, L.; Bravatà, V.; Cammarata, F.P.; Russo, G.; Gilardi, M.C.; Forte, G.I. Radiation Gene-expression Signatures in Primary Breast Cancer Cells. Anticancer Res. 2018, 38, 2707–2715. [Google Scholar]
- Di Maggio, F.M.; Minafra, L.; Forte, G.I.; Cammarata, F.P.; Lio, D.; Messa, C.; Gilardi, M.C.; Bravatà, V. Portrait of inflammatory response to ionizing radiation treatment. J. Inflamm. 2015, 12, 14. [Google Scholar] [CrossRef] [Green Version]
- Veeraraghavan, J.; Natarajan, M.; Aravindan, S.; Herman, T.S.; Aravindan, N. Radiation-triggered tumor necrosis factor (TNF) alpha-NFkappaB cross-signaling favors survival advantage in human neuroblastoma cells. J. Biol. Chem. 2011, 286, 21588–21600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jhaveri, K.; Teplinsky, E.; Silvera, D.; Valeta-Magara, A.; Arju, R.; Giashuddin, S.; Sarfraz, Y.; Alexander, M.; Darvishian, F.; Levine, P.H.; et al. Hyperactivated mTOR and JAK2/STAT3 Pathways: Molecular Drivers and Potential Therapeutic Targets of Inflammatory and Invasive Ductal Breast Cancers After Neoadjuvant Chemotherapy. Clin. Breast Cancer 2016, 16, 113–122. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.L.; Sun, Y.; Wu, S. Gamma-irradiation induces matrix metalloproteinase II expression in a p53-dependent manner. Mol. Carcinog. 2000, 27, 252–258. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Sun, R.; Zou, J.; Ying, Y.; Luo, Z. Dual Roles of the AMP-Activated Protein Kinase Pathway in Angiogenesis. Cells 2019, 8, 752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marques, B.L.; Carvalho, G.A.; Freitas, E.M.M.; Chiareli, R.A.; Barbosa, T.G.; Di Araújo, A.G.P.; Nogueira, Y.L.; Ribeiro, R.I.; Parreira, R.C.; Vieira, M.S.; et al. The role of neurogenesis in neurorepair after ischemic stroke. Semin. Cell Dev. Biol. 2019, 95, 98–110. [Google Scholar] [CrossRef]
- Huang, W.J.; Chen, W.W.; Zhang, X. Glioblastoma multiforme: Effect of hypoxia and hypoxia inducible factors on therapeutic approaches. Oncol. Lett. 2016, 12, 2283–2288. [Google Scholar] [CrossRef] [PubMed]
- Bravatà, V.; Cammarata, F.P.; Minafra, L.; Pisciotta, P.; Scazzone, C.; Manti, L.; Savoca, G.; Petringa, G.; Cirrone, G.A.P.; Cuttone, G.; et al. Proton-irradiated breast cells: Molecular points of view. J. Radiat. Res. 2019, 60, 451–465. [Google Scholar] [CrossRef] [PubMed]
- Cammarata, F.P.; Forte, G.I.; Broggi, G.; Bravatà, V.; Minafra, L.; Pisciotta, P.; Calvaruso, M.; Tringali, R.; Tomasello, B.; Torrisi, F.; et al. Molecular Investigation on a Triple Negative Breast Cancer Xenograft Model Exposed to Proton Beams. Int. J. Mol. Sci. 2020, 21, 6337. [Google Scholar] [CrossRef] [PubMed]
- Green, Y.S.; Sargis, T.; Reichert, E.C.; Rudasi, E.; Fuja, D.; Jonasch, E.; Koh, M.Y. Hypoxia-Associated Factor (HAF) Mediates Neurofibromin Ubiquitination and Degradation Leading to Ras-ERK Pathway Activation in Hypoxia. Mol. Cancer Res. 2019, 17, 1220–1232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, K.; Fu, P.; Juerchott, K.; Motaln, H.; Selbig, J.; Lah, T.; Tonn, J.C.; Schichor, C. The expression of Wnt-inhibitor DKK1 (Dickkopf 1) is determined by intercellular crosstalk and hypoxia in human malignant gliomas. J. Cancer Res. Clin. Oncol. 2014, 140, 1261–1270. [Google Scholar] [CrossRef]
- Maity, A.; Pore, N.; Lee, J.; Solomon, D.; O’Rourke, D.M. Epidermal growth factor receptor transcriptionally up-regulates vascular endothelial growth factor expression in human glioblastoma cells via a pathway involving phosphatidylinositol 3’-kinase and distinct from that induced by hypoxia. Cancer Res. 2000, 60, 5879–5886. [Google Scholar]
- Vallée, A.; Lecarpentier, Y.; Guillevin, R.; Vallée, J.N. Interactions between TGF-β1, canonical WNT/β-catenin pathway and PPAR γ in radiation-induced fibrosis. Oncotarget 2017, 8, 90579–90604. [Google Scholar] [CrossRef] [Green Version]
- Martínez, P.; Gómez-López, G.; Pisano, D.G.; Flores, J.M.; Blasco, M.A. A genetic interaction between RAP1 and telomerase reveals an unanticipated role for RAP1 in telomere maintenance. Aging. Cell 2016, 15, 1113–1125. [Google Scholar] [CrossRef]
- Swanson, M.J.; Baribault, M.E.; Israel, J.N.; Bae, N.S. Telomere protein RAP1 levels are affected by cellular aging and oxidative stress. Biomed. Rep. 2016, 5, 181–187. [Google Scholar] [CrossRef] [Green Version]
- Sayyah, J.; Bartakova, A.; Nogal, N.; Quilliam, L.A.; Stupack, D.G.; Brown, J.H. The Ras-related protein, Rap1A, mediates thrombin-stimulated, integrin-dependent glioblastoma cell proliferation and tumor growth. J. Biol. Chem. 2014, 289, 17689–17698. [Google Scholar] [CrossRef] [Green Version]
- Ji, J.; Ding, K.; Luo, T.; Xu, R.; Zhang, X.; Huang, B.; Chen, A.; Zhang, D.; Miletic, H.; Bjerkvig, R.; et al. PMEPA1 isoform a drives progression of glioblastoma by promoting protein degradation of the Hippo pathway kinase LATS1. Oncogene 2020, 39, 1125–1139. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Wei, Y.; Zhang, L.; Yee, P.P.; Johnson, M.; Zhang, X.; Gulley, M.; Atkinson, J.M.; Trebak, M.; Wang, H.G.; et al. Induction of store-operated calcium entry (SOCE) suppresses glioblastoma growth by inhibiting the Hippo pathway transcriptional coactivators YAP/TAZ. Oncogene 2019, 38, 120–139. [Google Scholar] [CrossRef]
- Zhang, L.; Cheng, F.; Wei, Y.; Zhang, L.; Guo, D.; Wang, B.; Li, W. Inhibition of TAZ contributes radiation-induced senescence and growth arrest in glioma cells. Oncogene 2019, 38, 2788–2799. [Google Scholar] [CrossRef] [PubMed]
- Wade, A.; Robinson, A.E.; Engler, J.R.; Petritsch, C.; James, C.D.; Phillips, J.J. Proteoglycans and their roles in brain cancer. FEBS J. 2013, 280, 2399–2417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kardideh, B.; Samimi, Z.; Norooznezhad, F.; Kiani, S.; Mansouri, K. Autophagy, cancer and angiogenesis: Where is the link? Cell Biosci. 2019, 9, 65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Li, J.; Guo, X.; Pan, H.; Zhou, Q. Absence of HTATIP2 Expression in A549 Lung Adenocarcinoma Cells Promotes Tumor Plasticity in Response to Hypoxic Stress. Cancers 2020, 12, 1538. [Google Scholar] [CrossRef] [PubMed]
- Firat, E.; Niedermann, G. FoxO proteins or loss of functional p53 maintain stemness of glioblastoma stem cells and survival after ionizing radiation plus PI3K/mTOR inhibition. Oncotarget 2016, 7, 54883–54896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chi, Y.; Shi, C.; Zhao, Y.; Guo, C. Forkhead box O (FOXO) 3 modulates hypoxia-induced autophagy through AMPK signalling pathway in cardiomyocytes. Biosci. Rep. 2016, 36, e00345. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Wang, J.; Jin, T.; Zhou, Y.; Chen, Q. FoxG1 facilitates proliferation and inhibits differentiation by downregulating FoxO/Smad signaling in glioblastoma. Biochem. Biophys. Res. Commun. 2018, 504, 46–53. [Google Scholar] [CrossRef]
- Carruthers, R.D.; Ahmed, S.U.; Ramachandran, S.; Strathdee, K.; Kurian, K.M.; Hedley, A.; Gomez-Roman, N.; Kalna, G.; Neilson, M.; Gilmour, L.; et al. Replication Stress Drives Constitutive Activation of the DNA Damage Response and Radioresistance in Glioblastoma Stem-like Cells. Cancer Res. 2018, 78, 5060–5071. [Google Scholar] [CrossRef] [Green Version]
- Altaner, C. Glioblastoma and stem cells. Neoplasma 2008, 55, 369–374. [Google Scholar]
- Neftel, C.; Laffy, J.; Filbin, M.G.; Hara, T.; Shore, M.E.; Rahme, G.J.; Richman, A.R.; Silverbush, D.; Shaw, M.L.; Hebert, C.M.; et al. An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell 2019, 178, 835–849. [Google Scholar] [CrossRef]
- Xu, F.; Zhang, J.; Hu, G.; Liu, L.; Liang, W. Hypoxia and TGF-β1 induced PLOD2 expression improve the migration and invasion of cervical cancer cells by promoting epithelial-to-mesenchymal transition (EMT) and focal adhesion formation. Cancer Cell Int. 2017, 17, 54. [Google Scholar] [CrossRef]
- Chen, E.Y.; Mazure, N.M.; Cooper, J.A.; Giaccia, A.J. Hypoxia activates a platelet-derived growth factor receptor/phosphatidylinositol 3-kinase/Akt pathway that results in glycogen synthase kinase-3 inactivation. Cancer Res. 2001, 61, 2429–2433. [Google Scholar] [PubMed]
- Liu, R.; Chen, Y.; Liu, G.; Li, C.; Song, Y.; Cao, Z.; Li, W.; Hu, J.; Lu, C.; Liu, Y. PI3K/AKT pathway as a key link modulates the multidrug resistance of cancers. Cell Death Dis. 2020, 11, 797. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Sun, S.; Ho, A.S.; Kiang, K.M.; Zhang, X.Q.; Xu, F.F.; Leung, G.K. Hyperoxia resensitizes chemoresistant glioblastoma cells to temozolomide through unfolded protein response. Anticancer Res. 2014, 34, 2957–2966. [Google Scholar] [PubMed]
- Lai, H.H.; Li, J.N.; Wang, M.Y.; Huang, H.Y.; Croce, C.M.; Sun, H.L.; Lyu, Y.J.; Kang, J.W.; Chiu, C.F.; Hung, M.C.; et al. HIF-1α promotes autophagic proteolysis of Dicer and enhances tumor metastasis. J. Clin. Invest. 2018, 128, 625–643. [Google Scholar] [CrossRef] [PubMed]
- Fujimura, A.; Michiue, H.; Cheng, Y.; Uneda, A.; Tani, Y.; Nishiki, T.; Ichikawa, T.; Wei, F.Y.; Tomizawa, K.; Matsui, H. Cyclin G2 promotes hypoxia-driven local invasion of glioblastoma by orchestrating cytoskeletal dynamics. Neoplasia 2013, 15, 1272–1281. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.W.; Ryu, Y.K.; Ji, Y.H.; Kang, J.H.; Moon, E.Y. Hypoxia/reoxygenation-experienced cancer cell migration and metastasis are regulated by Rap1- and Rac1-GTPase activation via the expression of thymosin beta-4. Oncotarget 2015, 6, 9820–9833. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Wang, J.J.; Fu, X.L.; Guang, R.; To, S.T. Advances in the targeting of HIF-1α and future therapeutic strategies for glioblastoma multiforme. Oncol. Rep. 2017, 37, 657–670. [Google Scholar] [CrossRef] [Green Version]
- Kim, I.G.; Lee, J.H.; Kim, S.Y.; Hwang, H.M.; Kim, T.R.; Cho, E.W. Hypoxia-inducible transgelin 2 selects epithelial-to-mesenchymal transition and γ-radiation-resistant subtypes by focal adhesion kinase-associated insulin-like growth factor 1 receptor activation in non-small-cell lung cancer cells. Cancer Sci. 2018, 109, 3519–3531. [Google Scholar] [CrossRef]
- Chhipa, R.R.; Fan, Q.; Anderson, J.; Muraleedharan, R.; Huang, Y.; Ciraolo, G.; Chen, X.; Waclaw, R.; Chow, L.M.; Khuchua, Z.; et al. AMP kinase promotes glioblastoma bioenergetics and tumour growth. Nat. Cell Biol. 2018, 20, 823–835. [Google Scholar] [CrossRef] [PubMed]
- Meyer, R.G.; Küpper, J.H.; Kandolf, R.; Rodemann, H.P. Early growth response-1 gene (Egr-1) promoter induction by ionizing radiation in U87 malignant glioma cells in vitro. Eur. J. Biochem. 2002, 269, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Sun, T.; Hu, J.; Zhang, R.; Rao, Y.; Wang, S.; Bigner, D.D.; Chen, R.; McLendon, R.E.; Friedman, A.H.; et al. miR-33a promotes glioma-initiating cell self-renewal via PKA and NOTCH pathways. J. Clin. Invest. 2014, 124, 4489–4502. [Google Scholar] [CrossRef] [PubMed]
- Daniel, P.M.; Filiz, G.; Mantamadiotis, T. Sensitivity of GBM cells to cAMP agonist-mediated apoptosis correlates with CD44 expression and agonist resistance with MAPK signaling. Cell Death Dis. 2016, 7, e2494. [Google Scholar] [CrossRef]
- Bullen, J.W.; Tchernyshyov, I.; Holewinski, R.J.; DeVine, L.; Wu, F.; Venkatraman, V.; Kass, D.L.; Cole, R.N.; Van Eyk, J.; Semenza, G.L. Protein kinase A-dependent phosphorylation stimulates the transcriptional activity of hypoxia-inducible factor 1. Sci. Signal 2016, 9, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torrisi, F.; Vicario, N.; Spitale, F.M.; Cammarata, F.P.; Minafra, L.; Salvatorelli, L.; Russo, G.; Cuttone, G.; Valable, S.; Gulino, R.; et al. The role of hypoxia and SRC tyrosine kinase in glioblastoma invasiveness and radioresistance. Cancers 2020, 12, 2860. [Google Scholar] [CrossRef]



| Genes Differentially Expressed (>2-fold) | |||
| Configuration | Number of Genes | Downregulated | Upregulated |
| U87_2Gy_ Hyp | 3275 | 773 | 2502 |
| U87_10Gy_ Hyp | 4232 | 1605 | 2627 |
| Genes Differentially Expressed (>5-fold) | |||
| Configuration | Number of Genes | Downregulated | Upregulated |
| U87_2Gy_ Hyp | 207 | 1 | 206 |
| U87_10Gy_ Hyp | 293 | 119 | 174 |
| Pathways | Gene Count | p Value | Genes |
|---|---|---|---|
| Proteoglycans in cancer | 47 | 9.80 × 108 | CAV2, FGFR1, LUM, PPP1R12C, SDC4, MMP2, PDCD4, ITGB1, IQGAP1, PXN, TGFB2, CTNNB1, PTK2, KRAS, ANK2, GAB1, PPP1R12A, PRKACB, THBS1, WNT6, PIK3R1, AKT2, FN1, TWIST1, PIK3R2, ACTB, ROCK1, ROCK2, MAP2K2, MET, ITGA2, ARHGEF12, PPP1CC, FLNC, PPP1CB, STAT3, ITPR1, FLNA, PRKCB, FZD6, PTPN11, CCND1, CBLB, MAPK12, ITGA5, VEGFA, HBEGF |
| Pathways in cancer | 74 | 5.22 × 109 | GNA13, FGF5, FGF7, PTGS2, PGF, STAT5B, NFKB2, MMP2, TGFB2, CTNNB1, EDNRA, CUL2, CASP8, RALB, RARB, PRKACB, WNT6, AKT2, CTBP1, BCR, ROCK1, PTGER4, ROCK2, FADD, RB1, ARHGEF12, DAPK3, CDK2, CTNNA2, PRKCB, CCND1, EP300, GNB2, GNAQ, GNB1, LPAR6, VEGFA, FGFR1, XIAP, GNAI1, PML, BCL2L1, ITGB1, TPM3, PTK2, KRAS, RUNX1, AXIN2, PIK3R1, FN1, APC, PIK3R2, CEBPA, DVL3, EPAS1, MAP2K2, MET, SMAD4, ITGA2, STAT3, COL4A6, DVL1, FZD6, CBLB, CDKN1B, ADCY9, ITGA6, ETS1, BAX, RASSF1, GSK3B, JAK1, ABL1, CRK |
| Hippo signaling pathway | 37 | 6.31 × 109 | YWHAZ, SOX2, BMPR2, LATS1, CTNNB1, TGFB2, DLG4, LIMD1, YAP1, AXIN2, WNT6, APC, ACTB, DVL3, PARD6B, NF2, SMAD4, PPP1CC, SNAI2, YWHAE, PPP1CB, TP73, CTNNA2, FZD6, DVL1, CCND1, YWHAG, YWHAH, CCND3, ID2, CSNK1E, CCND2, BBC3, GSK3B, RASSF1, PARD6G, BMP8B |
| Focal adhesion | 43 | 6.55 × 109 | CAV2, TLN1, XIAP, PGF, PPP1R12C, ARHGAP35, ITGB1, PXN, CTNNB1, MYL9, PTK2, PAK2, COL6A3, PPP1R12A, COL6A2, COL6A1, SHC1, THBS1, RAPGEF1, PIK3R1, PIK3R2, FN1, AKT2, ACTB, ROCK1, ROCK2, MET, ITGA2, FLNC, PPP1CC, PPP1CB, FLNA, COL4A6, PRKCB, CCND1, CCND3, ITGA6, ITGA5, CCND2, ITGA8, GSK3B, VEGFA, CRK |
| Signaling pathways regulating pluripotency of stem cells | 32 | 1.24 × 1011 | BMI1, FGFR1, FGFR4, ONECUT1, IL6ST, SOX2, BMPR2, REST, CTNNB1, ACVR1C, PCGF5, KRAS, SKIL, AXIN2, WNT6, PIK3R1, PIK3R2, APC, AKT2, DVL3, MAP2K2, SMAD4, LIFR, STAT3, FZD6, DVL1, ID2, RIF1, MAPK12, GSK3B, JAK1, KAT6A |
| FoxO signaling pathway | 29 | 7.03 × 1011 | STK11, PRKAG2, BNIP3, CCNG2, TGFB2, KRAS, PRKAA2, INSR, PIK3R1, PIK3R2, AKT2, IRS2, SGK2, MAP2K2, SMAD4, GRM1, IRS1, CDK2, STAT3, SOD2, CCND1, PLK4, CDKN1B, EP300, MAPK12, CSNK1E, CCND2, SETD7, GADD45B |
| p53 signaling pathway | 18 | 8.34 × 1011 | ZMAT3, RRM2B, CCNG1, CCNG2, SESN1, CDK2, TP73, CCND1, CCND3, BBC3, CCND2, BAX, RRM2, CASP8, SIAH1, MDM4, THBS1, GADD45B |
| Rap1 signaling pathway | 39 | 0.001 | FGFR1, FGF5, TLN1, FGFR4, FGF7, GNAI1, PGF, EFNA3, CTNND1, ITGB1, CTNNB1, PFN2, KRAS, RALB, RAPGEF4, RAPGEF2, THBS1, RAPGEF1, INSR, PIK3R1, PIK3R2, AKT2, ACTB, PARD6B, GNAO1, MAP2K2, MET, GRIN2A, SIPA1L3, PRKCB, DOCK4, ADCY9, MAPK12, GNAQ, KRIT1, VEGFA, PARD6G, CRK, CALM1 |
| Cell cycle | 25 | 0.004 | FZR1, YWHAZ, E2F4, E2F5, CDC14B, SMAD4, PRKDC, RB1, YWHAE, WEE1, CDK2, TGFB2, CCND1, YWHAG, RAD21, YWHAH, EP300, CDKN1B, CCND3, CCND2, GSK3B, ANAPC7, ABL1, GADD45B, STAG2 |
| Phosphatidylinositol signaling system | 21 | 0.005 | IMPAD1, IMPA1, PIK3C2A, SYNJ1, PI4K2B, PIP5K1A, ITPR1, PRKCB, DGKA, MTM1, MTMR14, PIKFYVE, PLCD3, INPP5E, PIP4K2A, MTMR6, IPMK, PIK3R1, INPP5A, CALM1, PIK3R2 |
| Ras signaling pathway | 39 | 0.006 | FGFR1, FGF5, FGFR4, FGF7, PGF, EFNA3, ARF6, BCL2L1, KRAS, REL, PAK2, GAB1, RALB, SHC1, PRKACB, INSR, PIK3R1, RASA2, PIK3R2, AKT2, PLA2G16, MAP2K2, NF1, MET, GRIN2A, PRKCB, PTPN11, PLA2G4A, KSR2, GNB2, GNB1, ETS1, ETS2, RASSF1, VEGFA, RAB5A, PLA2G2A, ABL1, CALM1 |
| Wnt signaling pathway | 26 | 0.009 | PPP3R1, CTNNB1, CSNK2A1, PRKACB, WNT6, NFATC2, AXIN2, FOSL1, APC, CSNK1A1, TBL1XR1, DVL3, CTBP1, ROCK2, SMAD4, FZD6, DVL1, PRKCB, CCND1, EP300, CCND3, CSNK1E, CCND2, SFRP2, GSK3B, SIAH1 |
| Inositol phosphate metabolism | 16 | 0.01 | MINPP1, IMPAD1, IMPA1, PIK3C2A, SYNJ1, PI4K2B, PIP5K1A, MTM1, MTMR14, PIKFYVE, PLCD3, INPP5E, PIP4K2A, MTMR6, IPMK, INPP5A |
| PI3K-Akt signaling pathway | 53 | 0.01 | FGF5, FGF7, PGF, PPP2R5A, EFNA3, PKN3, INSR, GHR, AKT2, SGK2, PKN2, IRS1, CDK2, IFNAR2, CCND1, CCND3, GNB2, LPAR6, CCND2, GNB1, VEGFA, FGFR1, YWHAZ, FGFR4, STK11, BCL2L1, ITGB1, ATF2, PTK2, KRAS, COL6A3, COL6A2, COL6A1, PRKAA2, THBS1, PIK3R1, FN1, PIK3R2, MAP2K2, CREB1, MET, ITGA2, YWHAE, COL4A6, YWHAG, YWHAH, CDKN1B, EIF4E, ITGA6, ITGA5, ITGA8, GSK3B, JAK1 |
| VEGF signaling pathway | 14 | 0.01 | PTGS2, MAP2K2, PPP3R1, PXN, PRKCB, PLA2G4A, PTK2, KRAS, MAPK12, VEGFA, NFATC2, PIK3R1, AKT2, PIK3R2 |
| Pathways | Gene Count | p Values | Genes |
|---|---|---|---|
| Endocytosis | 53 | 1.43 × 1011 | HRAS, CHMP3, RAB5B, CAPZA2, EPS15L1, PIP5K1A, MVB12A, PIP5KL1, VPS4B, DNAJC6, AGAP3, PLD1, HLA-A, HLA-C, HLA-B, HLA-E, LDLRAP1, ACAP3, ACAP2, RAB5A, MDM2, PDCD6IP, SNX12, VPS26B, SH3GL1, CAV2, WASH1, STAM2, ASAP2, PML, ASAP1, HSPA1A, CYTH2, ARF6, CHMP2B, SH3GLB2, RAB11B, RAB11A, NEDD4L, HSPA8, EHD4, GIT1, PARD6B, RAB8A, VTA1, EPS15, AP2A2, AP2A1, HGS, SMURF2, PARD6G, ARAP2, DNM2 |
| Hippo signaling pathway | 37 | 3.38 × 1010 | YWHAZ, APC2, SOX2, BMPR2, LATS1, CTNNB1, TGFB2, CTGF, DLG4, YAP1, WNT6, PPP2R2C, APC, ACTB, DVL3, PARD6B, NF2, SMAD4, WWTR1, PPP1CC, PPP1CB, TP73, STK3, CTNNA2, DVL1, AMH, CCND1, YWHAH, CCND3, CSNK1E, CCND2, BBC3, RASSF1, PARD6G, BMP8B, BMPR1A, PPP2R2A |
| Proteoglycans in cancer | 45 | 3.99 × 1010 | CAV2, FGFR1, HRAS, GRB2, LUM, PPP1R12C, ELK1, RPS6KB2, SDC4, MMP2, PDCD4, IQGAP1, PXN, TGFB2, CTNNB1, CTTN, KRAS, ANK2, GAB1, PPP1R12A, PRKACB, WNT6, PIK3R1, AKT2, TWIST1, PIK3R2, ACTB, ROCK2, MAP2K2, MET, ITGA2, ARHGEF12, PPP1CC, FLNC, PPP1CB, STAT3, FLNA, EIF4B, CCND1, CDKN1A, SDC1, MAPK12, ARAF, HBEGF, MDM2 |
| FoxO signaling pathway | 33 | 8.84 × 1010 | HRAS, GRB2, STK11, PRKAG2, CCNG2, TGFB2, PRMT1, KRAS, PRKAA2, INSR, PIK3R1, PIK3R2, AKT2, SGK1, MAP2K2, SMAD4, PCK2, IRS1, CDK2, STAT3, SOD2, CCND1, CDKN1A, PLK4, CDKN1B, PLK2, MAPK12, CSNK1E, CCND2, ARAF, MDM2, GADD45B, GADD45A |
| AMPK signaling pathway | 31 | 9.67 × 1010 | CAB39L, PFKFB3, STK11, PPP2R5A, LEPR, PPP2R5D, PRKAG2, RPS6KB2, CAMKK1, AKT1S1, FASN, RAB11B, PRKAA2, INSR, PPP2R2C, PIK3R1, PIK3R2, AKT2, RAB2A, RAB8A, PFKL, CREB3, SCD, ADIPOR1, CREB5, EEF2, ACACB, PCK2, IRS1, CCND1, PPP2R2A |
| p53 signaling pathway | 18 | 0.001 | ZMAT3, RRM2B, CCNG2, SESN1, CDK2, TP73, CCND1, CDKN1A, CCND3, BBC3, CCND2, RRM2, BAX, CASP8, MDM2, SIAH1, GADD45B, GADD45A |
| Focal adhesion | 39 | 0.004 | CAV2, TLN1, HRAS, GRB2, PGF, PPP1R12C, ELK1, ARHGAP35, PXN, CTNNB1, MYL9, BCL2, PPP1R12A, COL6A2, SHC1, RAPGEF1, PIK3R1, PIK3R2, AKT2, ACTB, TNXB, ROCK2, MET, ITGA2, BAD, FLNC, PPP1CC, PPP1CB, FLNA, COL4A6, COL4A5, VEGFB, CCND1, LAMA3, CCND3, ITGA6, CCND2, LAMC2, CRK |
| PI3K-Akt signaling pathway | 59 | 0.004 | HRAS, PGF, FGF14, EFNA1, PPP2R5A, PPP2R5D, EFNA3, RPS6KB2, PKN3, MLST8, GNG3, INSR, AKT2, SGK1, PKN2, PKN1, IRS1, CDK2, VEGFB, IFNAR2, CCND1, GNB2, CCND3, CCND2, MDM2, LAMC2, PPP2R2A, FGFR1, YWHAZ, GRB2, STK11, BCL2L1, CDC37, KRAS, BCL2, COL6A2, PRKAA2, PPP2R2C, PIK3R1, PIK3R2, TNXB, CREB3, MAP2K2, MET, ITGA2, NR4A1, CREB5, BAD, PCK2, COL4A6, COL4A5, EIF4B, CDKN1A, ATF4, LAMA3, YWHAH, CDKN1B, EIF4E, ITGA6 |
| Neurotrophin signaling pathway | 24 | 0.01 | IRAK1, HRAS, MAP2K2, GRB2, NFKBIB, BAD, IRS1, TP73, ATF4, KRAS, PSEN1, MAPK12, BCL2, BAX, GAB1, PSEN2, SHC1, SH2B1, RAPGEF1, CRK, ARHGDIA, PIK3R1, AKT2, PIK3R2 |
| Signaling pathways regulating pluripotency of stem cells | 27 | 0.01 | BMI1, FGFR1, HRAS, APC2, GRB2, IL6ST, SOX2, BMPR2, CTNNB1, ACVR1C, KRAS, WNT6, PIK3R1, PIK3R2, APC, AKT2, DVL3, TBX3, MAP2K2, OTX1, SMAD4, LIFR, STAT3, DVL1, RIF1, MAPK12, BMPR1A |
| mTOR signaling pathway | 14 | 0.01 | CAB39L, STK11, RPS6KB2, IRS1, RRAGB, EIF4B, AKT1S1, EIF4E, ULK3, MLST8, PRKAA2, PIK3R1, AKT2, PIK3R2 |
| Wnt signaling pathway | 26 | 0.02 | APC2, PPP3R1, PPP3R2, CTNNB1, PLCB3, PRKACB, SOX17, WNT6, NFATC2, NFATC3, FOSL1, APC, CSNK1A1, TBL1XR1, DVL3, CTBP1, ROCK2, SMAD4, DVL1, CCND1, CCND3, PSEN1, CSNK1E, CCND2, SFRP2, SIAH1 |
| Rap1 signaling pathway | 36 | 0.02 | FGFR1, TLN1, HRAS, GNAI1, ADORA2A, PGF, FGF14, EFNA1, EFNA3, CTNND1, ITGAM, CTNNB1, PLCB3, PFN2, KRAS, RAPGEF4, RAPGEF2, RAPGEF1, INSR, PIK3R1, PIK3R2, AKT2, ACTB, PARD6B, MAP2K2, GRIN1, MET, SIPA1L3, RGS14, DOCK4, VEGFB, PRKD2, MAPK12, KRIT1, PARD6G, CRK |
| TNF signaling pathway | 21 | 0.02 | CEBPB, CREB3, PTGS2, CXCL3, CXCL2, FADD, CREB5, JUNB, VCAM1, CASP10, FOS, TNFRSF1B, ATF4, RPS6KA4, MAPK12, PGAM5, CASP8, TNFAIP3, PIK3R1, AKT2, PIK3R2 |
| Cell cycle | 23 | 0.03 | ANAPC2, FZR1, YWHAZ, E2F4, E2F5, DBF4, SMAD4, TTK, PRKDC, WEE1, CDK2, TGFB2, CCND1, CDKN1A, YWHAH, CDKN1B, CCND3, CCND2, MDM2, ANAPC7, GADD45B, GADD45A, STAG2 |
| Pathways | Gene Count | p Value | Genes |
|---|---|---|---|
| Hippo signaling pathway | 33 | 2.22 × 109 | YWHAZ, SOX2, BMPR2, LATS1, CTNNB1, TGFB2, WNT3, SERPINE1, DLG4, YAP1, WNT6, PPP2R2C, APC, ACTB, DVL3, PARD6B, NF2, SMAD4, PPP1CC, PPP1CB, TP73, CTNNA2, DVL1, CCND1, YWHAH, RASSF6, CCND3, CSNK1E, CCND2, BBC3, RASSF1, PARD6G, BMP8B |
| Proteoglycans in cancer | 37 | 2.64 × 1011 | CAV2, LUM, PPP1R12C, SDC4, MMP2, PDCD4, PXN, IQGAP1, CTNNB1, TGFB2, KRAS, WNT3, ANK2, GAB1, PPP1R12A, MSN, PRKACB, PIK3R3, WNT6, PIK3R1, TWIST1, PIK3R2, AKT2, ACTB, MAP2K2, ROCK2, MET, ITGA2, FLNC, ARHGEF12, PPP1CC, PPP1CB, STAT3, FLNA, CCND1, MAPK12, HBEGF |
| Endocytosis | 40 | 1.42 × 1012 | FGFR2, CAV2, CHMP3, WASH1, CAPZA2, STAM2, ASAP2, PIP5K1B, PML, ASAP1, HSPA1A, ARF6, PIP5K1A, AMPH, CHMP2B, SH3GLB2, RAB11B, DNAJC6, RAB11A, NEDD4L, AGAP3, EHD4, GIT1, PARD6B, VTA1, HLA-A, HLA-C, HLA-B, HLA-E, RAB11FIP4, EPS15, ACAP3, AP2A1, ACAP2, RAB5A, SMURF2, PARD6G, PDCD6IP, ARAP2, SH3GL1 |
| p53 signaling pathway | 17 | 1.85 × 1012 | ZMAT3, RRM2B, CCNG2, SESN1, CDK2, TP73, CCND1, CCND3, BBC3, CCND2, SERPINB5, RRM2, BAX, CASP8, SERPINE1, SIAH1, GADD45B |
| Focal adhesion | 34 | 5.50 × 1011 | CAV2, TLN1, XIAP, TLN2, PGF, PPP1R12C, ARHGAP35, PXN, CTNNB1, MYL9, PPP1R12A, COL6A2, SHC1, PIK3R3, RAPGEF1, PIK3R1, PIK3R2, AKT2, ACTB, ROCK2, MYLK3, MET, ITGA2, FLNC, PPP1CC, PPP1CB, COL4A6, FLNA, CCND1, CCND3, ITGA6, CCND2, COL24A1, CRK |
| Signaling pathways regulating pluripotency of stem cells | 25 | 0.001 | FGFR2, BMI1, IL6ST, SOX2, BMPR2, CTNNB1, ACVR1C, WNT3, KRAS, WNT6, PIK3R3, PIK3R1, PIK3R2, APC, AKT2, DVL3, MAP2K2, OTX1, SMAD4, LIFR, STAT3, DVL1, RIF1, MAPK12, JAK3 |
| FoxO signaling pathway | 24 | 0.001 | STK11, MAP2K2, PRKAG2, SMAD4, CCNG2, IRS1, CDK2, STAT3, SOD2, TGFB2, CCND1, PLK4, KRAS, CDKN1B, MAPK12, CSNK1E, CCND2, PRKAA2, PIK3R3, GADD45B, INSR, PIK3R1, AKT2, PIK3R2 |
| Wnt signaling pathway | 23 | 0.004 | CSNK1A1, DVL3, TBL1XR1, CTBP1, ROCK2, SMAD4, PPP3R1, DKK4, DVL1, CTNNB1, CCND1, WNT3, SOST, CCND3, CSNK1E, SFRP2, CCND2, SIAH1, PRKACB, WNT6, NFATC2, FOSL1, APC |
| Rap1 signaling pathway | 31 | 0.005 | FGFR2, FGF5, TLN1, TLN2, PGF, GNAI1, EFNA3, CTNNB1, PFN2, KRAS, GRIN2B, RAPGEF4, RAPGEF2, PIK3R3, INSR, RAPGEF1, PIK3R1, PIK3R2, AKT2, ACTB, FYB, PARD6B, MAGI1, MAP2K2, MET, SIPA1L3, DOCK4, MAPK12, KRIT1, PARD6G, CRK |
| VEGF signaling pathway | 11 | 0.04 | KRAS, MAPK12, PTGS2, MAP2K2, PPP3R1, NFATC2, PIK3R3, PIK3R1, PXN, AKT2, PIK3R2 |
| PubMatrix | Hypoxia | GBM | RT | PT | Cancer | IR | Cell Death | Cell Cycle | Hif1-Alpha |
|---|---|---|---|---|---|---|---|---|---|
| Ubiquitin-mediated proteolysis | 85 | 10 | 11 | 0 | 1043 | 54 | 346 | 802 | 1 |
| Regulation of actin cytoskeleton | 106 | 65 | 12 | 3 | 2466 | 32 | 610 | 1180 | 1 |
| Rap1 signaling pathway | 10 | 5 | 3 | 0 | 322 | 3 | 74 | 92 | 1 |
| Protein processing in endoplasmic reticulum | 184 | 27 | 17 | 4 | 1676 | 27 | 1640 | 506 | 0 |
| Proteoglycans in cancer | 195 | 284 | 339 | 11 | 81 | 1158 | 1299 | 4 | |
| Platelet activation | 584 | 136 | 151 | 101 | 5387 | 268 | 1651 | 1857 | 2 |
| Ras signaling pathway | 266 | 200 | 201 | 13 | 11,296 | 201 | 2907 | 3446 | 7 |
| Glutamatergic synapse | 48 | 3 | 1 | 1 | 95 | 1 | 144 | 50 | 0 |
| Wnt signaling pathway | 310 | 209 | 191 | 11 | 10,182 | 70 | 2563 | 2294 | 4 |
| FoxO signaling pathway | 53 | 8 | 3 | 0 | 491 | 9 | 329 | 265 | 5 |
| PubMatrix | Hypoxia | GBM | RT | PT | Cancer | IR | Cell Death | Cell Cycle | Hif1-Alpha |
|---|---|---|---|---|---|---|---|---|---|
| PI3K-Akt signaling pathway | 1063 | 427 | 368 | 8 | 12,427 | 191 | 6798 | 3323 | 26 |
| Insulin signaling pathway | 591 | 89 | 82 | 8 | 6338 | 100 | 3505 | 2647 | 16 |
| Platelet activation | 584 | 136 | 151 | 101 | 5387 | 268 | 1651 | 1857 | 2 |
| Endocytosis | 384 | 245 | 274 | 107 | 13,851 | 391 | 6505 | 3903 | 2 |
| Wnt signaling pathway | 310 | 209 | 191 | 11 | 10,182 | 70 | 2563 | 2294 | 4 |
| Ras signaling pathway | 266 | 200 | 201 | 13 | 11,296 | 201 | 2907 | 3446 | 7 |
| Focal adhesion | 261 | 255 | 127 | 3 | 7394 | 91 | 1858 | 1874 | 3 |
| cAMP signaling pathway | 248 | 47 | 29 | 4 | 2908 | 117 | 1273 | 1490 | 6 |
| Proteoglycans in cancer | 195 | 284 | 339 | 11 | 13,155 | 81 | 1158 | 1299 | 4 |
| Protein processing in endoplasmic reticulum | 184 | 27 | 17 | 4 | 1676 | 27 | 1640 | 506 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bravatà, V.; Tinganelli, W.; Cammarata, F.P.; Minafra, L.; Calvaruso, M.; Sokol, O.; Petringa, G.; Cirrone, G.A.P.; Scifoni, E.; Forte, G.I.; et al. Hypoxia Transcriptomic Modifications Induced by Proton Irradiation in U87 Glioblastoma Multiforme Cell Line. J. Pers. Med. 2021, 11, 308. https://doi.org/10.3390/jpm11040308
Bravatà V, Tinganelli W, Cammarata FP, Minafra L, Calvaruso M, Sokol O, Petringa G, Cirrone GAP, Scifoni E, Forte GI, et al. Hypoxia Transcriptomic Modifications Induced by Proton Irradiation in U87 Glioblastoma Multiforme Cell Line. Journal of Personalized Medicine. 2021; 11(4):308. https://doi.org/10.3390/jpm11040308
Chicago/Turabian StyleBravatà, Valentina, Walter Tinganelli, Francesco P. Cammarata, Luigi Minafra, Marco Calvaruso, Olga Sokol, Giada Petringa, Giuseppe A.P. Cirrone, Emanuele Scifoni, Giusi I. Forte, and et al. 2021. "Hypoxia Transcriptomic Modifications Induced by Proton Irradiation in U87 Glioblastoma Multiforme Cell Line" Journal of Personalized Medicine 11, no. 4: 308. https://doi.org/10.3390/jpm11040308
APA StyleBravatà, V., Tinganelli, W., Cammarata, F. P., Minafra, L., Calvaruso, M., Sokol, O., Petringa, G., Cirrone, G. A. P., Scifoni, E., Forte, G. I., & Russo, G. (2021). Hypoxia Transcriptomic Modifications Induced by Proton Irradiation in U87 Glioblastoma Multiforme Cell Line. Journal of Personalized Medicine, 11(4), 308. https://doi.org/10.3390/jpm11040308















