Therapeutic Value of Single Nucleotide Polymorphisms on the Efficacy of New Therapies in Patients with Multiple Sclerosis
Abstract
1. Introduction
2. Pharmacogenetics of New Therapies in MS
2.1. Dimethyl Fumarate
2.1.1. Zinc Finger MIZ-Type Containing 1; ZMIZ1
2.1.2. NADPH Oxidase 3; NOX3
2.2. Teriflunomide
2.2.1. ATP-Binding Cassette, Subfamily G, Member 2; ABCG2
2.2.2. Dihydroorotate Dehydrogenase; DHODH
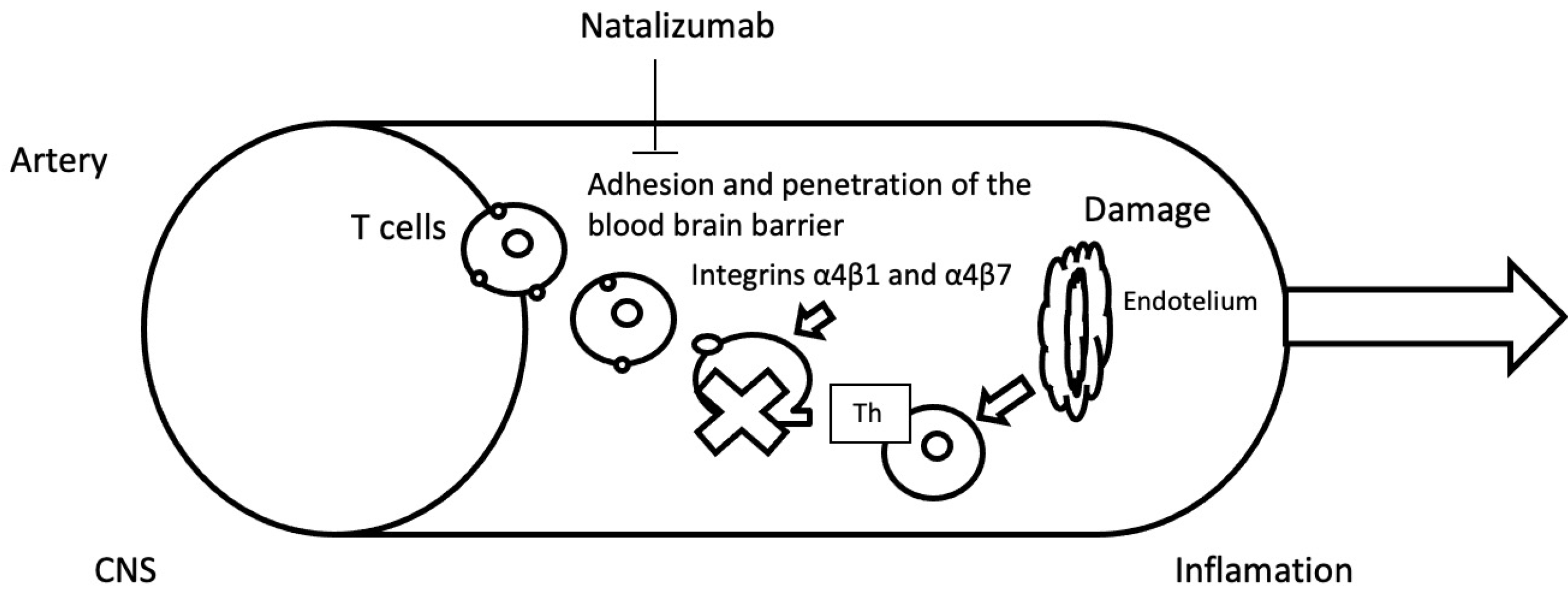
2.3. Natalizumab
2.3.1. Glutathione S-Transferase Pi 1; GSTP1
2.3.2. Integrin Subunit Alpha 4; ITGA4
2.3.3. NAD(P)H Quinone Dehydrogenase 1; NQO1
2.3.4. AKT Serine/Threonine Kinase 1; AKT1
2.3.5. Glycoprotein VI Platelet; GP6
2.4. Fingolimod
Zinc Finger MIZ-Type Containing 1; ZMIZ1
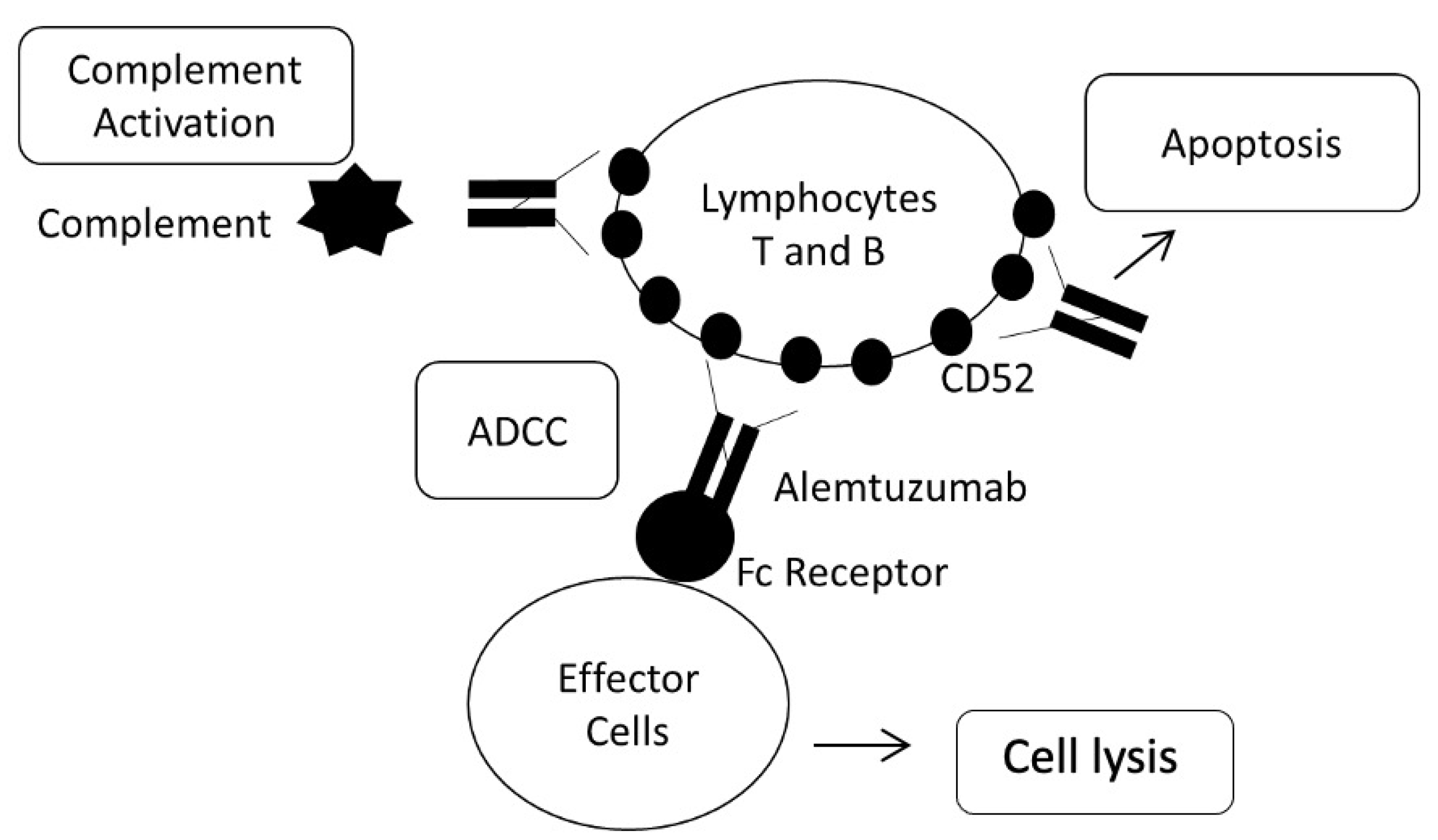
2.5. Alemtuzumab
2.5.1. Fc Fragment of IgG Receptors; FCGR2A and FCGR3A
2.5.2. CD52 Molecule; CD52
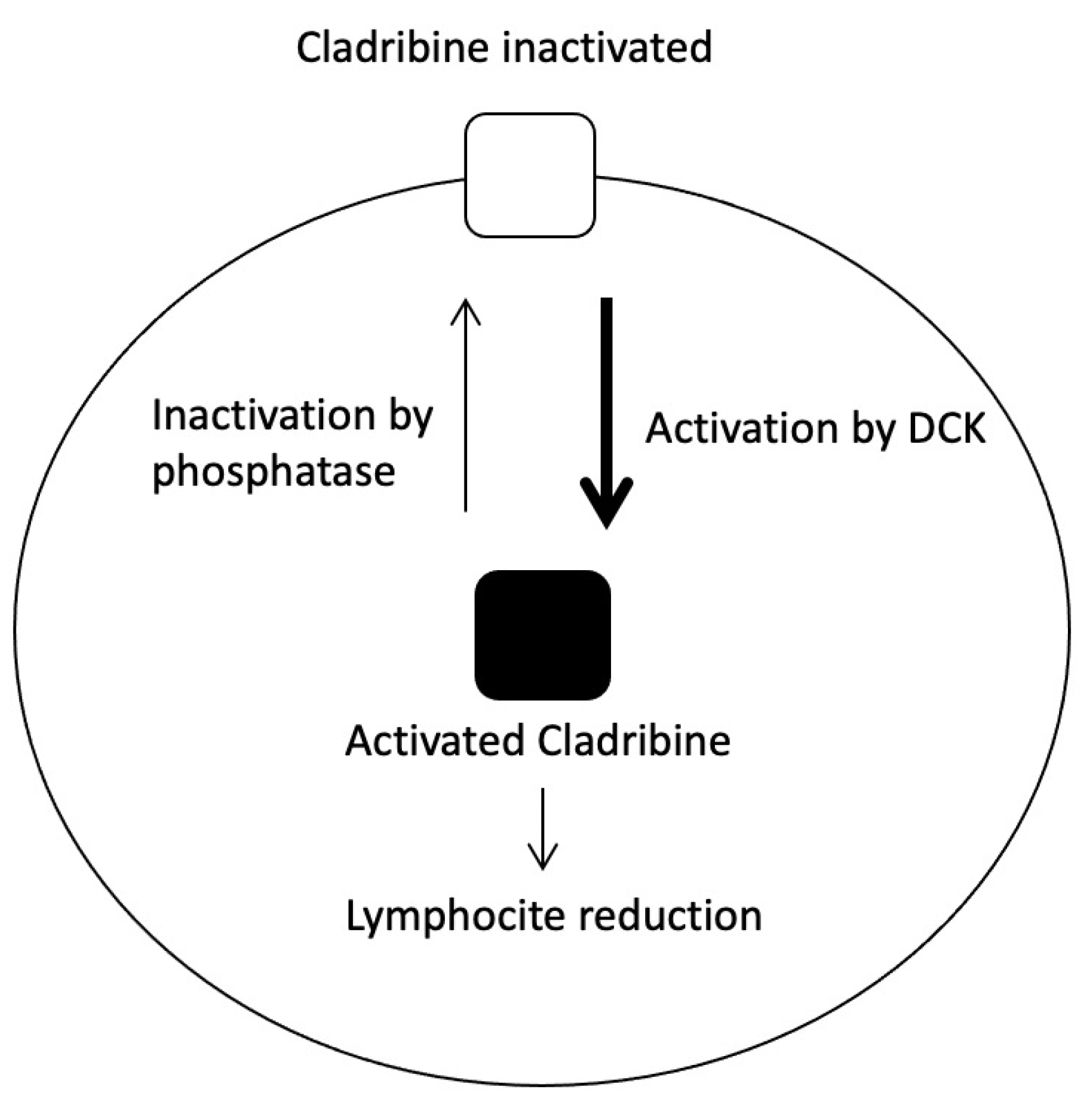
2.6. Cladribine
2.6.1. Ribonucleotide Reductase, M1 and M2 Subunits; RRM1 and RRM2
2.6.2. Adenosine Deaminase; ADA
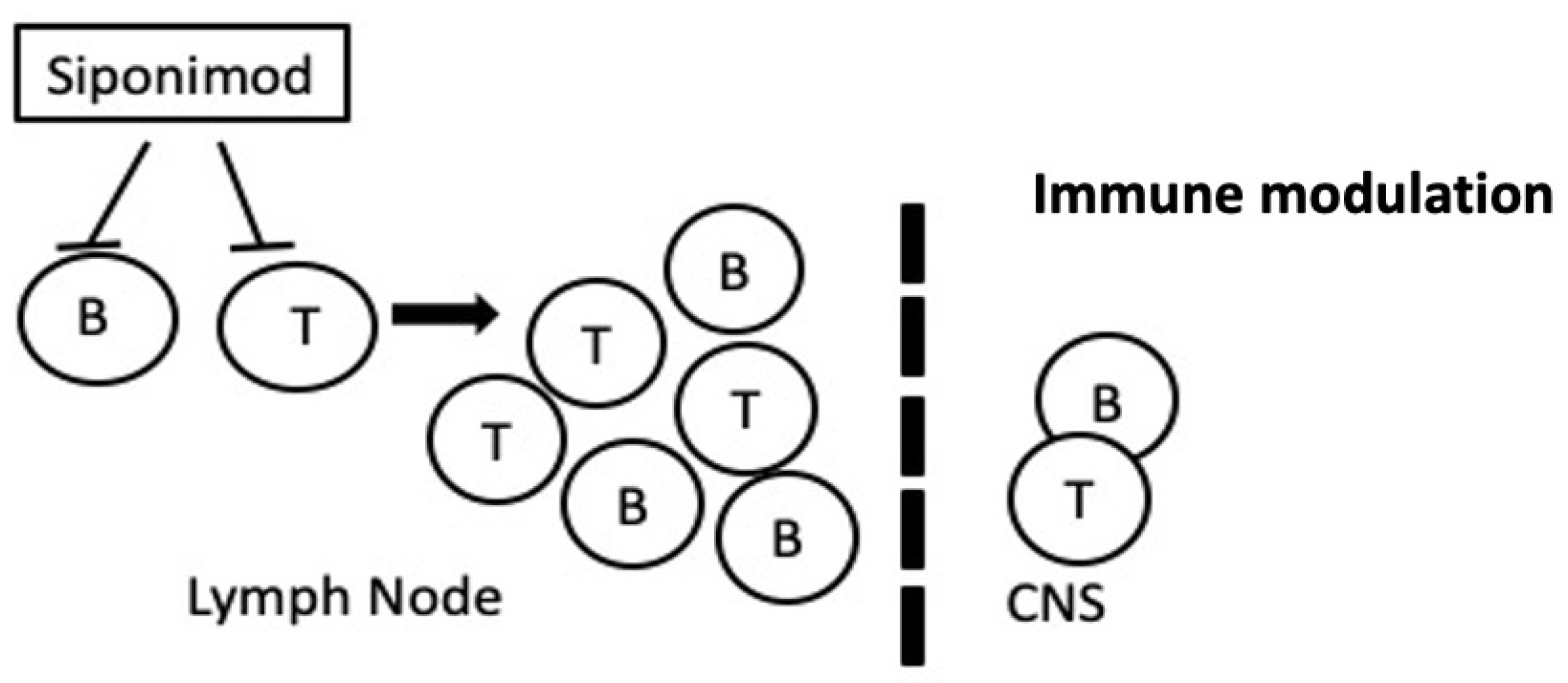
2.7. Siponimod
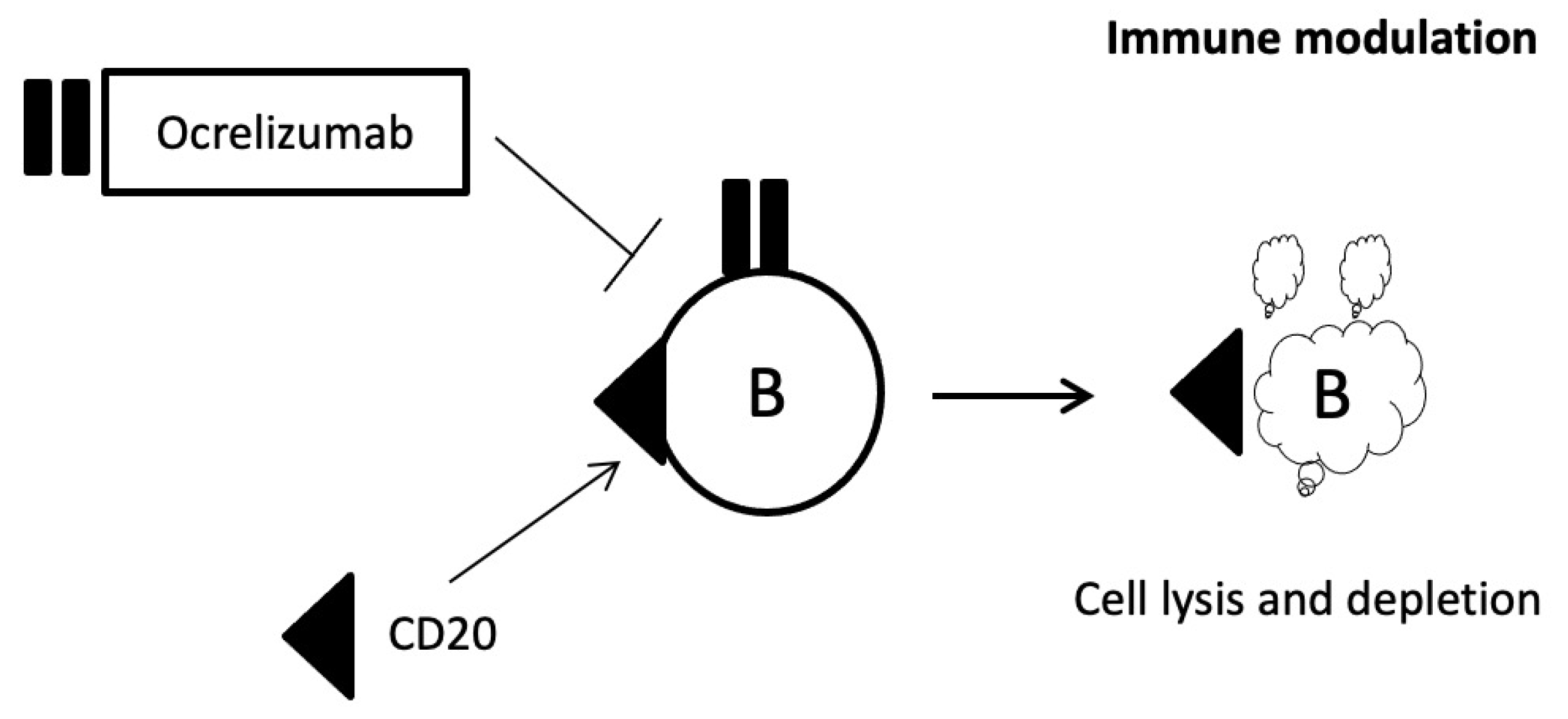
2.8. Ocrelizumab
3. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pravica, V.; Popadic, D.; Savic, E.; Markovic, M.; Drulovic, J.; Mostarica-Stojkovic, M. Single nucleotide polymorphisms in multiple sclerosis: Disease susceptibility and treatment response biomarkers. Immunol. Res. 2012, 52, 42–52. [Google Scholar] [CrossRef]
- Browne, P.; Chandraratna, D.; Angood, C.; Tremlett, H.; Baker, C.; Taylor, B.V.; Thompson, A.J. Atlas of Multiple Sclerosis 2013: A growing global problem with widespread inequity. Neurology 2014, 83, 1022–1024. [Google Scholar] [CrossRef]
- Hauser, S.L.; Oksenberg, J.R. The neurobiology of multiple sclerosis: Genes, inflammation, and neurodegeneration. Neuron 2006, 52, 61–76. [Google Scholar] [CrossRef]
- McDonald, W.I. The mystery of the origin of multiple sclerosis. J. Neurol. Neurosurg. Psychiatry 1986, 49, 113–123. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ebers, G.C. McAlpine’s multiple sclerosis. JAMA Neurol. 1991, 48, 1214. [Google Scholar] [CrossRef]
- Freal, J.E.; Kraft, G.H.; Coryell, J.K. Symptomatic fatigue in multiple sclerosis. Arch. Phys. Med. Rehabil. 1984, 65, 135–138. [Google Scholar]
- Krupp, L.B.; Alvarez, L.A.; LaRocca, N.G.; Scheinberg, L.C. Fatigue in multiple sclerosis. Arch. Neurol. 1988, 45, 435–437. [Google Scholar] [CrossRef]
- Compston, A.; Coles, A. Multiple sclerosis. Lancet 2008, 372, 1502–1517. [Google Scholar] [CrossRef]
- Lucchinetti, C.; Bruck, W.; Parisi, J.; Scheithauer, B.; Rodriguez, M.; Lassmann, H. Heterogeneity of multiple sclerosis lesions: Implications for the pathogenesis of demyelination. Ann. Neurol. 2000, 47, 707–717. [Google Scholar] [CrossRef]
- Boster, A.L.; Ford, C.C.; Neudorfer, O.; Gilgun-Sherki, Y. Glatiramer acetate: Long-term safety and efficacy in relapsing-remitting multiple sclerosis. Expert Rev. Neurother. 2015, 15, 575–586. [Google Scholar] [CrossRef]
- Rojas, J.I.; Pappolla, A.; Patrucco, L.; Cristiano, E.; Sanchez, F. Do clinical trials for new disease modifying treatments include real world patients with multiple sclerosis? Mult. Scler. Relat. Disord. 2020, 39, 101931. [Google Scholar] [CrossRef] [PubMed]
- Ta, S.O. Recent advances in the treatment for multiple sclerosis; Current new drugs specific for multiple sclerosis. Noro Psikiyatr. Ars. 2018, 55 (Suppl. S1), S15–S20. [Google Scholar]
- Sorensen, P.S.; Fox, R.J.; Comi, G. The window of opportunity for treatment of progressive multiple sclerosis. Curr. Opin. Neurol. 2020, 33, 262–270. [Google Scholar] [CrossRef]
- Gold, R.; Kappos, L.; Arnold, D.L.; Bar-Or, A.; Giovannoni, G.; Selmaj, K.; Tornatore, C.; Sweetser, M.T.; Yang, M.; Sheikh, S.I.; et al. Placebo-controlled phase 3 study of oral BG-12 for relapsing multiple sclerosis. N. Engl. J. Med. 2012, 367, 1098–1107. [Google Scholar] [CrossRef]
- Fox, R.J.; Miller, D.H.; Phillips, J.T.; Hutchinson, M.; Havrdova, E.; Kita, M.; Yang, M.; Raghupathi, K.; Novas, M.; Sweetser, M.T.; et al. Placebo-controlled phase 3 study of oral BG-12 or glatiramer in multiple sclerosis. N. Engl. J. Med. 2012, 367, 1087–1097. [Google Scholar] [CrossRef]
- O’Connor, P.; Wolinsky, J.S.; Confavreux, C.; Comi, G.; Kappos, L.; Olsson, T.P.; Benzerdjeb, H.; Truffinet, P.; Wang, L.; Miller, A.; et al. Randomized trial of oral teriflunomide for relapsing multiple sclerosis. N. Engl. J. Med. 2011, 365, 1293–1303. [Google Scholar] [CrossRef] [PubMed]
- Polman, C.H.; O’Connor, P.W.; Havrdova, E.; Hutchinson, M.; Kappos, L.; Miller, D.H.; Phillips, J.T.; Lublin, F.D.; Giovannoni, G.; Wajgt, A.; et al. A randomized, placebo-controlled trial of natalizumab for relapsing multiple sclerosis. N. Engl. J. Med. 2006, 354, 899–910. [Google Scholar] [CrossRef]
- Cohen, J.A.; Barkhof, F.; Comi, G.; Hartung, H.P.; Khatri, B.O.; Montalban, X.; Pelletier, J.; Capra, R.; Gallo, P.; Izquierdo, G.; et al. Oral fingolimod or intramuscular interferon for relapsing multiple sclerosis. N. Engl. J. Med. 2010, 362, 402–415. [Google Scholar] [CrossRef]
- Kappos, L.; Bar-Or, A.; Cree, B.A.C.; Fox, R.J.; Giovannoni, G.; Gold, R.; Vermersch, P.; Arnold, D.L.; Arnould, S.; Scherz, T.; et al. Siponimod versus placebo in secondary progressive multiple sclerosis (EXPAND): A double-blind, randomised, phase 3 study. Lancet 2018, 391, 1263–1273. [Google Scholar] [CrossRef]
- Havrdova, E.; Arnold, D.L.; Cohen, J.A.; Hartung, H.P.; Fox, E.J.; Giovannoni, G.; Schippling, S.; Selmaj, K.W.; Traboulsee, A.; Compston, D.A.S.; et al. Alemtuzumab CARE-MS I 5-year follow-up: Durable efficacy in the absence of continuous MS therapy. Neurology 2017, 89, 1107–1116. [Google Scholar] [CrossRef]
- Tuohy, O.; Costelloe, L.; Hill-Cawthorne, G.; Bjornson, I.; Harding, K.; Robertson, N.; May, K.; Button, T.; Azzopardi, L.; Kousin-Ezewu, O.; et al. Alemtuzumab treatment of multiple sclerosis: Long-term safety and efficacy. J. Neurol. Neurosurg. Psychiatry 2015, 86, 208–215. [Google Scholar] [CrossRef]
- Kappos, L.; Radue, E.W.; O’Connor, P.; Polman, C.; Hohlfeld, R.; Calabresi, P.; Selmaj, K.; Agoropoulou, C.; Leyk, M.; Zhang-Auberson, L.; et al. A placebo-controlled trial of oral fingolimod in relapsing multiple sclerosis. N. Engl. J. Med. 2010, 362, 387–401. [Google Scholar] [CrossRef]
- Giovannoni, G.; Soelberg Sorensen, P.; Cook, S.; Rammohan, K.W.; Rieckmann, P.; Comi, G.; Dangond, F.; Hicking, C.; Vermersch, P. Efficacy of Cladribine Tablets in high disease activity subgroups of patients with relapsing multiple sclerosis: A post hoc analysis of the CLARITY study. Mult. Scler. 2019, 25, 819–827. [Google Scholar] [CrossRef]
- Rose, J.W.; Giovannoni, G.; Wiendl, H.; Gold, R.; Havrdova, E.; Kappos, L.; Selmaj, K.W.; Zhao, J.; Riester, K.; Tsao, L.C.; et al. Consistent efficacy of daclizumab beta across patient demographic and disease activity subgroups in patients with relapsing-remitting multiple sclerosis. Mult. Scler. Relat. Disord. 2017, 17, 32–40. [Google Scholar] [CrossRef]
- Wolinsky, J.S.; Engmann, N.J.; Pei, J.; Pradhan, A.; Markowitz, C.; Fox, E.J. An exploratory analysis of the efficacy of ocrelizumab in patients with multiple sclerosis with increased disability. Mult. Scler. J. Exp. Transl. Clin. 2020, 6, 2055217320911939. [Google Scholar] [CrossRef] [PubMed]
- Comabella, M.; Vandenbroeck, K. Pharmacogenomics and multiple sclerosis: Moving toward individualized medicine. Curr. Neurol. Neurosci. Rep. 2011, 11, 484–491. [Google Scholar] [CrossRef] [PubMed]
- Gopal, S.; Mikulskis, A.; Gold, R.; Fox, R.J.; Dawson, K.T.; Amaravadi, L. Evidence of activation of the Nrf2 pathway in multiple sclerosis patients treated with delayed-release dimethyl fumarate in the Phase 3 DEFINE and CONFIRM studies. Mult. Scler. 2017, 23, 1875–1883. [Google Scholar] [CrossRef] [PubMed]
- Van Horssen, J.; Drexhage, J.A.; Flor, T.; Gerritsen, W.; van der Valk, P.; de Vries, H.E. Nrf2 and DJ1 are consistently upregulated in inflammatory multiple sclerosis lesions. Free Radic. Biol. Med. 2010, 49, 1283–1289. [Google Scholar] [CrossRef]
- Biotti, D.; Ciron, J. First-line therapy in relapsing remitting multiple sclerosis. Rev. Neurol (Paris) 2018, 174, 419–428. [Google Scholar] [CrossRef]
- Sharma, M.; Li, X.; Wang, Y.; Zarnegar, M.; Huang, C.Y.; Palvimo, J.J.; Lim, B.; Sun, Z. hZimp10 is an androgen receptor co-activator and forms a complex with SUMO-1 at replication foci. EMBO J. 2003, 22, 6101–6114. [Google Scholar] [CrossRef]
- Fewings, N.L.; Gatt, P.N.; McKay, F.C.; Parnell, G.P.; Schibeci, S.D.; Edwards, J.; Basuki, M.A.; Goldinger, A.; Fabis-Pedrini, M.J.; Kermode, A.G.; et al. The autoimmune risk gene ZMIZ1 is a vitamin D responsive marker of a molecular phenotype of multiple sclerosis. J. Autoimmun. 2017, 78, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Paffenholz, R.; Bergstrom, R.A.; Pasutto, F.; Wabnitz, P.; Munroe, R.J.; Jagla, W.; Heinzmann, U.; Marquardt, A.; Bareiss, A.; Laufs, J.; et al. Vestibular defects in head-tilt mice result from mutations in Nox3, encoding an NADPH oxidase. Genes Dev. 2004, 18, 486–491. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, H.; Hikage, M.; Miyashita, H.; Fukumoto, M. NADPH oxidase subunit, gp91(phox) homologue, preferentially expressed in human colon epithelial cells. Gene 2000, 254, 237–243. [Google Scholar] [CrossRef]
- Accetta, R.; Damiano, S.; Morano, A.; Mondola, P.; Paterno, R.; Avvedimento, E.V.; Santillo, M. Reactive Oxygen Species Derived from NOX3 and NOX5 drive differentiation of human oligodendrocytes. Front. Cell. Neurosci. 2016, 10, 146. [Google Scholar] [CrossRef] [PubMed]
- Carlstrom, K.E.; Ewing, E.; Granqvist, M.; Gyllenberg, A.; Aeinehband, S.; Enoksson, S.L.; Checa, A.; Badam, T.V.S.; Huang, J.; Gomez-Cabrero, D.; et al. Therapeutic efficacy of dimethyl fumarate in relapsing-remitting multiple sclerosis associates with ROS pathway in monocytes. Nat. Commun. 2019, 10, 3081. [Google Scholar] [CrossRef] [PubMed]
- Bruneau, J.M.; Yea, C.M.; Spinella-Jaegle, S.; Fudali, C.; Woodward, K.; Robson, P.A.; Sautes, C.; Westwood, R.; Kuo, E.A.; Williamson, R.A.; et al. Purification of human dihydro-orotate dehydrogenase and its inhibition by A77 1726, the active metabolite of leflunomide. Biochem. J. 1998, 336 Pt 2, 299–303. [Google Scholar] [CrossRef]
- Cherwinski, H.M.; Cohn, R.G.; Cheung, P.; Webster, D.J.; Xu, Y.Z.; Caulfield, J.P.; Young, J.M.; Nakano, G.; Ransom, J.T. The immunosuppressant leflunomide inhibits lymphocyte proliferation by inhibiting pyrimidine biosynthesis. J. Pharmacol. Exp. Ther. 1995, 275, 1043–1049. [Google Scholar]
- Ruckemann, K.; Fairbanks, L.D.; Carrey, E.A.; Hawrylowicz, C.M.; Richards, D.F.; Kirschbaum, B.; Simmonds, H.A. Leflunomide inhibits pyrimidine de novo synthesis in mitogen-stimulated T-lymphocytes from healthy humans. J. Biol. Chem. 1998, 273, 21682–21691. [Google Scholar] [CrossRef]
- Loffler, M.; Klein, A.; Hayek-Ouassini, M.; Knecht, W.; Konrad, L. Dihydroorotate dehydrogenase mRNA and protein expression analysis in normal and drug-resistant cells. Nucleosides Nucleotides Nucleic Acids 2004, 23, 1281–1285. [Google Scholar] [CrossRef]
- Gold, R.; Wolinsky, J.S. Pathophysiology of multiple sclerosis and the place of teriflunomide. Acta Neurol. Scand. 2011, 124, 75–84. [Google Scholar] [CrossRef]
- Munier-Lehmann, H.; Vidalain, P.O.; Tangy, F.; Janin, Y.L. On dihydroorotate dehydrogenases and their inhibitors and uses. J. Med. Chem. 2013, 56, 3148–3167. [Google Scholar] [CrossRef]
- Allikmets, R.; Schriml, L.M.; Hutchinson, A.; Romano-Spica, V.; Dean, M. A human placenta-specific ATP-binding cassette gene (ABCP) on chromosome 4q22 that is involved in multidrug resistance. Cancer Res. 1998, 58, 5337–5339. [Google Scholar] [PubMed]
- Kim, K.A.; Joo, H.J.; Park, J.Y. Effect of ABCG2 genotypes on the pharmacokinetics of A771726, an active metabolite of prodrug leflunomide, and association of A771726 exposure with serum uric acid level. Eur. J. Clin. Pharmacol. 2011, 67, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Barnes, T.; Parry, P.; Hart, I.; Jones, C.; Minet, M.; Patterson, D. Regional mapping of the gene encoding dihydroorotate dehydrogenase, an enzyme involved in UMP synthesis, electron transport, and superoxide generation, to human chromosome region 16q22. Somat. Cell Mol. Genet. 1993, 19, 405–411. [Google Scholar] [CrossRef]
- Mladenovic, V.; Domljan, Z.; Rozman, B.; Jajic, I.; Mihajlovic, D.; Dordevic, J.; Popovic, M.; Dimitrijevic, M.; Zivkovic, M.; Campion, G.; et al. Safety and effectiveness of leflunomide in the treatment of patients with active rheumatoid arthritis. Results of a randomized, placebo-controlled, phase II study. Arthritis Rheum. 1995, 38, 1595–1603. [Google Scholar] [CrossRef]
- Pawlik, A.; Herczynska, M.; Kurzawski, M.; Safranow, K.; Dziedziejko, V.; Drozdzik, M. The effect of exon (19C > A) dihydroorotate dehydrogenase gene polymorphism on rheumatoid arthritis treatment with leflunomide. Pharmacogenomics 2009, 10, 303–309. [Google Scholar] [CrossRef]
- Steinman, L. Blocking adhesion molecules as therapy for multiple sclerosis: Natalizumab. Nat. Rev. Drug Discov. 2005, 4, 510–518. [Google Scholar] [CrossRef]
- Martinez-Forero, I.; Pelaez, A.; Villoslada, P. Pharmacogenomics of multiple sclerosis: In search for a personalized therapy. Expert Opin. Pharmacother. 2008, 9, 3053–3067. [Google Scholar] [CrossRef]
- Alexoudi, A.; Zachaki, S.; Stavropoulou, C.; Gavrili, S.; Spiliopoulou, C.; Papadodima, S.; Karageorgiou, C.E.; Sambani, C. Possible Implication of GSTP1 and NQO1 Polymorphisms on Natalizumab Response in Multiple Sclerosis. Ann. Clin. Lab. Sci. 2016, 46, 586–591. [Google Scholar]
- Sousa, L.; de Sa, J.; Sa, M.J.; Cerqueira, J.J.; Martins-Silva, A.; Portugal Experience with Natalizumab Study, G. The efficacy and safety of natalizumab for the treatment of multiple sclerosis in Portugal: A retrospective study. Rev. Neurol. 2014, 59, 399–406. [Google Scholar]
- Board, P.G.; Coggan, M.; Woodcock, D.M. The human Pi class glutathione transferase sequence at 12q13-q14 is a reverse-transcribed pseudogene. Genomics 1992, 14, 470–473. [Google Scholar] [CrossRef]
- Ketterer, B.; Coles, B.; Meyer, D.J. The role of glutathione in detoxication. Environ. Health Perspect. 1983, 49, 59–69. [Google Scholar] [CrossRef]
- Zhang, Z.H.; Vekemans, S.; Aly, M.S.; Jaspers, M.; Marynen, P.; Cassiman, J.J. The gene for the alpha 4 subunit of the VLA-4 integrin maps to chromosome 2Q31-32. Blood 1991, 78, 2396–2399. [Google Scholar] [CrossRef]
- Berlin, C.; Bargatze, R.F.; Campbell, J.J.; von Andrian, U.H.; Szabo, M.C.; Hasslen, S.R.; Nelson, R.D.; Berg, E.L.; Erlandsen, S.L.; Butcher, E.C. Alpha 4 integrins mediate lymphocyte attachment and rolling under physiologic flow. Cell 1995, 80, 413–422. [Google Scholar] [CrossRef]
- Miller, D.H.; Khan, O.A.; Sheremata, W.A.; Blumhardt, L.D.; Rice, G.P.; Libonati, M.A.; Willmer-Hulme, A.J.; Dalton, C.M.; Miszkiel, K.A.; O’Connor, P.W.; et al. A controlled trial of natalizumab for relapsing multiple sclerosis. N. Engl. J. Med. 2003, 348, 15–23. [Google Scholar] [CrossRef]
- Durmanova, V.; Shawkatova, I.; Javor, J.; Parnicka, Z.; Copikova-Cundrakova, D.; Turcani, P.; Lisa, I.; Gajdosechova, B.; Buc, M.; Bucova, M. VLA4 gene polymorphism and susceptibility to multiple sclerosis in Slovaks. Folia Biol. 2015, 61, 8–13. [Google Scholar]
- Siegel, D.; Gustafson, D.L.; Dehn, D.L.; Han, J.Y.; Boonchoong, P.; Berliner, L.J.; Ross, D. NAD(P)H:quinone oxidoreductase 1: Role as a superoxide scavenger. Mol. Pharmacol. 2004, 65, 1238–1247. [Google Scholar] [CrossRef]
- Haider, L.; Fischer, M.T.; Frischer, J.M.; Bauer, J.; Hoftberger, R.; Botond, G.; Esterbauer, H.; Binder, C.J.; Witztum, J.L.; Lassmann, H. Oxidative damage in multiple sclerosis lesions. Brain J. Neurol. 2011, 134 Pt 7, 1914–1924. [Google Scholar] [CrossRef]
- Staal, S.P.; Huebner, K.; Croce, C.M.; Parsa, N.Z.; Testa, J.R. The AKT1 proto-oncogene maps to human chromosome 14, band q32. Genomics 1988, 2, 96–98. [Google Scholar] [CrossRef]
- Rossi, S.; Motta, C.; Studer, V.; Monteleone, F.; De Chiara, V.; Buttari, F.; Barbieri, F.; Bernardi, G.; Battistini, L.; Cutter, G.; et al. A genetic variant of the anti-apoptotic protein Akt predicts natalizumab-induced lymphocytosis and post-natalizumab multiple sclerosis reactivation. Mult. Scler. 2013, 19, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Jandrot-Perrus, M.; Busfield, S.; Lagrue, A.H.; Xiong, X.; Debili, N.; Chickering, T.; Le Couedic, J.P.; Goodearl, A.; Dussault, B.; Fraser, C.; et al. Cloning, characterization, and functional studies of human and mouse glycoprotein VI: A platelet-specific collagen receptor from the immunoglobulin superfamily. Blood 2000, 96, 1798–1807. [Google Scholar] [CrossRef]
- Bender, M.; Hofmann, S.; Stegner, D.; Chalaris, A.; Bosl, M.; Braun, A.; Scheller, J.; Rose-John, S.; Nieswandt, B. Differentially regulated GPVI ectodomain shedding by multiple platelet-expressed proteinases. Blood 2010, 116, 3347–3355. [Google Scholar] [CrossRef] [PubMed]
- Marshall, O.; Lu, H.; Brisset, J.C.; Xu, F.; Liu, P.; Herbert, J.; Grossman, R.I.; Ge, Y. Impaired cerebrovascular reactivity in multiple sclerosis. JAMA Neurol 2014, 71, 1275–1281. [Google Scholar] [CrossRef] [PubMed]
- Malak Al-Mojel, R.A. Texy Kannankeril, Mohammed Dashti and Rabeah Al-Temaimi, GP6 rs2304166 polymorphism is associated with response to natalizumab in multiple sclerosis patients. BMC 2019, 4, 1–6. [Google Scholar]
- Brinkmann, V.; Davis, M.D.; Heise, C.E.; Albert, R.; Cottens, S.; Hof, R.; Bruns, C.; Prieschl, E.; Baumruker, T.; Hiestand, P.; et al. The immune modulator FTY720 targets sphingosine 1-phosphate receptors. J. Biol. Chem. 2002, 277, 21453–21457. [Google Scholar] [CrossRef] [PubMed]
- Mandala, S.; Hajdu, R.; Bergstrom, J.; Quackenbush, E.; Xie, J.; Milligan, J.; Thornton, R.; Shei, G.J.; Card, D.; Keohane, C.; et al. Alteration of lymphocyte trafficking by sphingosine-1-phosphate receptor agonists. Science 2002, 296, 346–349. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, C.; Dev, K.K. The structure and function of the S1P1 receptor. Trends Pharmacol. Sci. 2013, 34, 401–412. [Google Scholar] [CrossRef]
- Fujiwara, M.; Anstadt, E.J.; Khanna, K.M.; Clark, R.B. Cbl-b-deficient mice express alterations in trafficking-related molecules but retain sensitivity to the multiple sclerosis therapeutic agent, FTY720. Clin. Immunol. 2015, 158, 103–113. [Google Scholar] [CrossRef]
- Esposito, F.; Ferre, L.; Clarelli, F.; Rocca, M.A.; Sferruzza, G.; Storelli, L.; Radaelli, M.; Sangalli, F.; Moiola, L.; Colombo, B.; et al. Effectiveness and baseline factors associated to fingolimod response in a real-world study on multiple sclerosis patients. J. Neurol. 2018, 265, 896–905. [Google Scholar] [CrossRef]
- Arnold, D.L.; Banwell, B.; Bar-Or, A.; Ghezzi, A.; Greenberg, B.M.; Waubant, E.; Giovannoni, G.; Wolinsky, J.S.; Gartner, J.; Rostasy, K.; et al. Effect of fingolimod on MRI outcomes in patients with paediatric-onset multiple sclerosis: Results from the phase 3 PARADIGMS study. J. Neurol. Neurosurg. Psychiatry 2020, 91, 483–492. [Google Scholar] [CrossRef]
- Pinnell, N.; Yan, R.; Cho, H.J.; Keeley, T.; Murai, M.J.; Liu, Y.; Alarcon, A.S.; Qin, J.; Wang, Q.; Kuick, R.; et al. The PIAS-like Coactivator Zmiz1 Is a Direct and Selective Cofactor of Notch1 in T Cell Development and Leukemia. Immunity 2015, 43, 870–883. [Google Scholar] [CrossRef]
- Hu, Q.D.; Ang, B.T.; Karsak, M.; Hu, W.P.; Cui, X.Y.; Duka, T.; Takeda, Y.; Chia, W.; Sankar, N.; Ng, Y.K.; et al. F3/contactin acts as a functional ligand for Notch during oligodendrocyte maturation. Cell 2003, 115, 163–175. [Google Scholar] [CrossRef]
- Bandala-Sanchez, E.; Zhang, Y.; Reinwald, S.; Dromey, J.A.; Lee, B.H.; Qian, J.; Bohmer, R.M.; Harrison, L.C. T cell regulation mediated by interaction of soluble CD52 with the inhibitory receptor Siglec-10. Nat. Immunol. 2013, 14, 741–748. [Google Scholar] [CrossRef]
- Toh, B.H.; Kyaw, T.; Tipping, P.; Bobik, A. Immune regulation by CD52-expressing CD4 T cells. Cell. Mol. Immunol. 2013, 10, 379–382. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.P.; Sancho, J.; Campos-Rivera, J.; Boutin, P.M.; Severy, P.B.; Weeden, T.; Shankara, S.; Roberts, B.L.; Kaplan, J.M. Human peripheral blood mononuclear cells exhibit heterogeneous CD52 expression levels and show differential sensitivity to alemtuzumab mediated cytolysis. PLoS ONE 2012, 7, e39416. [Google Scholar] [CrossRef]
- Rodig, S.J.; Abramson, J.S.; Pinkus, G.S.; Treon, S.P.; Dorfman, D.M.; Dong, H.Y.; Shipp, M.A.; Kutok, J.L. Heterogeneous CD52 expression among hematologic neoplasms: Implications for the use of alemtuzumab (CAMPATH-1H). Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2006, 12, 7174–7179. [Google Scholar] [CrossRef] [PubMed]
- Ginaldi, L.; De Martinis, M.; Matutes, E.; Farahat, N.; Morilla, R.; Dyer, M.J.; Catovsky, D. Levels of expression of CD52 in normal and leukemic B and T cells: Correlation with in vivo therapeutic responses to Campath-1H. Leuk. Res. 1998, 22, 185–191. [Google Scholar] [CrossRef]
- Hale, G. The CD52 antigen and development of the CAMPATH antibodies. Cytotherapy 2001, 3, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Ruck, T.; Bittner, S.; Wiendl, H.; Meuth, S.G. Alemtuzumab in Multiple Sclerosis: Mechanism of action and beyond. Int. J. Mol. Sci. 2015, 16, 16414–16439. [Google Scholar] [CrossRef] [PubMed]
- Rawstron, A.C.; Kennedy, B.; Moreton, P.; Dickinson, A.J.; Cullen, M.J.; Richards, S.J.; Jack, A.S.; Hillmen, P. Early prediction of outcome and response to alemtuzumab therapy in chronic lymphocytic leukemia. Blood 2004, 103, 2027–2031. [Google Scholar] [CrossRef]
- Coles, A.J.; Twyman, C.L.; Arnold, D.L.; Cohen, J.A.; Confavreux, C.; Fox, E.J.; Hartung, H.P.; Havrdova, E.; Selmaj, K.W.; Weiner, H.L.; et al. Alemtuzumab for patients with relapsing multiple sclerosis after disease-modifying therapy: A randomised controlled phase 3 trial. Lancet 2012, 380, 1829–1839. [Google Scholar] [CrossRef]
- Sammartino, L.; Webber, L.M.; Hogarth, P.M.; McKenzie, I.F.; Garson, O.M. Assignment of the gene coding for human FcRII (CD32) to bands q23q24 on chromosome 1. Immunogenetics 1988, 28, 380–381. [Google Scholar] [CrossRef] [PubMed]
- Hibbs, M.L.; Bonadonna, L.; Scott, B.M.; McKenzie, I.F.; Hogarth, P.M. Molecular cloning of a human immunoglobulin G Fc receptor. Proc. Natl. Acad. Sci. USA 1988, 85, 2240–2244. [Google Scholar] [CrossRef]
- Keller, C.W.; Ruck, T.; McHugh, D.; Pfeuffer, S.; Gross, C.C.; Korsukewitz, C.; Melzer, N.; Klotz, L.; Meuth, S.G.; Munz, C.; et al. Impact of FcgammaR variants on the response to alemtuzumab in multiple sclerosis. Ann. Clin. Transl Neurol. 2019, 6, 2586–2594. [Google Scholar] [CrossRef]
- Xia, M.Q.; Tone, M.; Packman, L.; Hale, G.; Waldmann, H. Characterization of the CAMPATH-1 (CDw52) antigen: Biochemical analysis and cDNA cloning reveal an unusually small peptide backbone. Eur. J. Immunol. 1991, 21, 1677–1684. [Google Scholar] [CrossRef]
- Oko, A.; Wyrwicz, L.S.; Glyda, M.; Idasiak-Piechocka, I.; Binczak-Kuleta, A.; Kaczmarczyk, M.; Drozd, A.; Ciechanowicz, A.; Czekalski, S. CD52 gene polymorphism and its potential effect on the response to alemtuzumab in renal transplant recipients. Ann. Acad. Med. Stetin. 2009, 55, 22–26. [Google Scholar] [PubMed]
- Leist, T.P.; Comi, G.; Cree, B.A.; Coyle, P.K.; Freedman, M.S.; Hartung, H.P.; Vermersch, P.; Casset-Semanaz, F.; Scaramozza, M.; on behalf of the oral cladribine for early MS (ORACLE MS) Study Group. Effect of oral cladribine on time to conversion to clinically definite multiple sclerosis in patients with a first demyelinating event (ORACLE MS): A phase 3 randomised trial. Lancet Neurol 2014, 13, 257–267. [Google Scholar] [CrossRef]
- Rice, G.P.; Filippi, M.; Comi, G.; Cladribine MRI Study Group. Cladribine and progressive MS: Clinical and MRI outcomes of a multicenter controlled trial. Neurology 2000, 54, 1145–1155. [Google Scholar] [CrossRef] [PubMed]
- Kopadze, T.; Dobert, M.; Leussink, V.I.; Dehmel, T.; Kieseier, B.C. Cladribine impedes in vitro migration of mononuclear cells: A possible implication for treating multiple sclerosis. Eur. J. Neurol. 2009, 16, 409–412. [Google Scholar] [CrossRef]
- Pavloff, N.; Rivard, D.; Masson, S.; Shen, S.H.; Mes-Masson, A.M. Sequence analysis of the large and small subunits of human ribonucleotide reductase. DNA Seq. J. DNA Seq. Mapp. 1992, 2, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Byrne, J.A.; Smith, P.J. The 11p15.5 ribonucleotide reductase M1 subunit locus is not imprinted in Wilms’ tumour and hepatoblastoma. Hum. Genet. 1993, 91, 275–277. [Google Scholar] [CrossRef]
- Yang-Feng, T.L.; Barton, D.E.; Thelander, L.; Lewis, W.H.; Srinivasan, P.R.; Francke, U. Ribonucleotide reductase M2 subunit sequences mapped to four different chromosomal sites in humans and mice: Functional locus identified by its amplification in hydroxyurea-resistant cell lines. Genomics 1987, 1, 77–86. [Google Scholar] [CrossRef]
- Lotfi, K.; Juliusson, G.; Albertioni, F. Pharmacological basis for cladribine resistance. Leuk. Lymphoma 2003, 44, 1705–1712. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Mitra, A.K.; Pounds, S.; Crews, K.R.; Gandhi, V.; Plunkett, W.; Dolan, M.E.; Hartford, C.; Raimondi, S.; Campana, D.; et al. RRM1 and RRM2 pharmacogenetics: Association with phenotypes in HapMap cell lines and acute myeloid leukemia patients. Pharmacogenomics 2013, 14, 1449–1466. [Google Scholar] [CrossRef] [PubMed]
- Jhanwar, S.C.; Berkvens, T.M.; Breukel, C.; van Ormondt, H.; van der Eb, A.J.; Meera Khan, P. Localization of human adenosine deaminase (ADA) gene sequences to the q12----q13.11 region of chromosome 20 by in situ hybridization. Cytogenet. Cell Genet. 1989, 50, 168–171. [Google Scholar] [CrossRef] [PubMed]
- Polachini, C.R.; Spanevello, R.M.; Casali, E.A.; Zanini, D.; Pereira, L.B.; Martins, C.C.; Baldissareli, J.; Cardoso, A.M.; Duarte, M.F.; da Costa, P.; et al. Alterations in the cholinesterase and adenosine deaminase activities and inflammation biomarker levels in patients with multiple sclerosis. Neuroscience 2014, 266, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Samuraki, M.; Sakai, K.; Odake, Y.; Yoshita, M.; Misaki, K.; Nakada, M.; Yamada, M. Multiple sclerosis showing elevation of adenosine deaminase levels in the cerebrospinal fluid. Mult. Scler. Relat. Disord. 2017, 13, 44–46. [Google Scholar] [CrossRef]
- Laugel, B.; Borlat, F.; Galibert, L.; Vicari, A.; Weissert, R.; Chvatchko, Y.; Bruniquel, D. Cladribine inhibits cytokine secretion by T cells independently of deoxycytidine kinase activity. J. Neuroimmunol. 2011, 240–241, 52–57. [Google Scholar] [CrossRef]
- Stampanoni Bassi, M.; Buttari, F.; Simonelli, I.; Gilio, L.; Furlan, R.; Finardi, A.; Marfia, G.A.; Visconti, A.; Paolillo, A.; Storto, M.; et al. A single nucleotide ADA genetic variant is associated to central inflammation and clinical presentation in MS: Implications for cladribine treatment. Genes 2020, 11, 1152. [Google Scholar] [CrossRef]
- Samjoo, I.A.; Worthington, E.; Haltner, A.; Cameron, C.; Nicholas, R.; Rouyrre, N.; Dahlke, F.; Adlard, N. Matching-adjusted indirect treatment comparison of siponimod and other disease modifying treatments in secondary progressive multiple sclerosis. Curr. Med. Res. Opin. 2020, 36, 1–10. [Google Scholar] [CrossRef]
- Wu, Q.; Mills, E.A.; Wang, Q.; Dowling, C.A.; Fisher, C.; Kirch, B.; Lundy, S.K.; Fox, D.A.; Mao-Draayer, Y.; Group, A.M.S.S. Siponimod enriches regulatory T and B lymphocytes in secondary progressive multiple sclerosis. JCI Insight 2020, 5, e134251. [Google Scholar] [CrossRef] [PubMed]
- Huth, F.; Gardin, A.; Umehara, K.; He, H. Prediction of the impact of cytochrome P450 2C9 genotypes on the drug-drug interaction potential of siponimod with physiologically-based pharmacokinetic modeling: A comprehensive approach for drug label recommendations. Clin. Pharmacol. Ther. 2019, 106, 1113–1124. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Borell, H.; Gardin, A.; Ufer, M.; Huth, F.; Camenisch, G. In vitro studies and in silico predictions of fluconazole and CYP2C9 genetic polymorphism impact on siponimod metabolism and pharmacokinetics. Eur. J. Clin. Pharmacol. 2018, 74, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Hohlfeld, R.; Meinl, E. Ocrelizumab in multiple sclerosis: Markers and mechanisms. Lancet. Neurol. 2017, 16, 259–261. [Google Scholar] [CrossRef]
- Sorensen, P.S.; Blinkenberg, M. The potential role for ocrelizumab in the treatment of multiple sclerosis: Current evidence and future prospects. Ther. Adv. Neurol. Disord. 2016, 9, 44–52. [Google Scholar] [CrossRef]
- Hauser, S.L.; Bar-Or, A.; Comi, G.; Giovannoni, G.; Hartung, H.P.; Hemmer, B.; Lublin, F.; Montalban, X.; Rammohan, K.W.; Selmaj, K.; et al. Ocrelizumab versus Interferon Beta-1a in relapsing multiple sclerosis. N. Engl. J. Med. 2017, 376, 221–234. [Google Scholar] [CrossRef]



| Therapy | Gene | Population | N | Pathology | SNP | p-Value | OR (95% CI) | Genotype Associated | PMID |
|---|---|---|---|---|---|---|---|---|---|
| Dimethyl fumarate | ZMIZ1 | Caucasian | 39 | MS | rs1782645 | 0.031 | - | - | 28063629 |
| NOX3 | Caucasian | 564 | RRMS | rs6919626 | 0.036 | 1.57 | G | 31300673 | |
| Teriflunomide | ABCG2 | Asian | 24 | Healthy Subjects | rs2231142 | 0.004 | - | C | 20972558 |
| DHODH | Caucasian | 147 | Rheumatoid Arthritis | rs3213422 | 0.048 | 1.98 (1.00–3.94) | C | 19207032 | |
| Natalizumab | GSTP1 | Caucasian | 129 | MS | rs1695 | 0.861 | - | A | 27993870 |
| ITGA4 | Caucasian | 117 | MS | rs1143676 | 0.024 | 1.73 (1.07–2.81) | AG | 25958306 | |
| NQO1 | Caucasian | 124 | MS | rs1800566 | 0.068 | C | 27993870 | ||
| AKT1 | Caucasian | 67 | RRMS | rs2498804 | <0.01 | 11.79 (3.72–37.38) | T | 22577119 | |
| GP6 | Asian | 67 | MS | rs2304166 | <0.01 | 22.18 (5.76–95.88) | C | 20644114 | |
| Fingolimod | ZMIZ1 | Caucasian | 39 | MS | rs1782645 | 0.039 | - | - | 29435643 |
| Alemtuzumab | FCGR2A FCGR3A | Caucasian | 85 | RRMS | rs396991 rs1801274 | >0.05 | - | - | 31682087 |
| CD52 | Caucasian | 108 | Kidney transplant | rs107184 rs17645 | - | - | - | 20349607 | |
| Cladribine | RRM1 | Caucasian African ancestry | 90 | Myeloid leukemia | rs11030918 | 0.04 | - | - | 24024897 |
| rs12806698 | 0.0004 | - | - | ||||||
| RRM2 | rs1042927 | 0.030 | - | - | |||||
| rs1138729 | 0.001 | - | - | ||||||
| ADA | Caucasian | 561 | RRMS | rs244072 | 0.011 | - | C | 33007809 | |
| Siponimod | CYP2C9 | - | - | MS | CYP2C9*1/*1 CYP2C9*2/*2 CYP2C9*3/*3 | - | - | CYP2C9*3/*3 | 24024897 |
| Ocrelizumab | - | - | - | - | - | - | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zarzuelo Romero, M.J.; Pérez Ramírez, C.; Carrasco Campos, M.I.; Sánchez Martín, A.; Calleja Hernández, M.Á.; Ramírez Tortosa, M.C.; Jiménez Morales, A. Therapeutic Value of Single Nucleotide Polymorphisms on the Efficacy of New Therapies in Patients with Multiple Sclerosis. J. Pers. Med. 2021, 11, 335. https://doi.org/10.3390/jpm11050335
Zarzuelo Romero MJ, Pérez Ramírez C, Carrasco Campos MI, Sánchez Martín A, Calleja Hernández MÁ, Ramírez Tortosa MC, Jiménez Morales A. Therapeutic Value of Single Nucleotide Polymorphisms on the Efficacy of New Therapies in Patients with Multiple Sclerosis. Journal of Personalized Medicine. 2021; 11(5):335. https://doi.org/10.3390/jpm11050335
Chicago/Turabian StyleZarzuelo Romero, María José, Cristina Pérez Ramírez, María Isabel Carrasco Campos, Almudena Sánchez Martín, Miguel Ángel Calleja Hernández, María Carmen Ramírez Tortosa, and Alberto Jiménez Morales. 2021. "Therapeutic Value of Single Nucleotide Polymorphisms on the Efficacy of New Therapies in Patients with Multiple Sclerosis" Journal of Personalized Medicine 11, no. 5: 335. https://doi.org/10.3390/jpm11050335
APA StyleZarzuelo Romero, M. J., Pérez Ramírez, C., Carrasco Campos, M. I., Sánchez Martín, A., Calleja Hernández, M. Á., Ramírez Tortosa, M. C., & Jiménez Morales, A. (2021). Therapeutic Value of Single Nucleotide Polymorphisms on the Efficacy of New Therapies in Patients with Multiple Sclerosis. Journal of Personalized Medicine, 11(5), 335. https://doi.org/10.3390/jpm11050335







