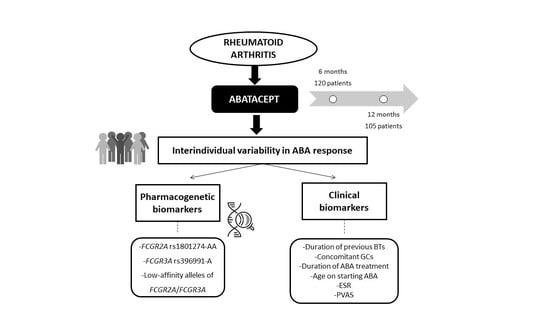
Influence of the FCGR2A rs1801274 and FCGR3A rs396991 Polymorphisms on Response to Abatacept in Patients with Rheumatoid Arthritis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Ethics Statements
2.3. Study Population
2.4. Sociodemographic and Clinical Variables
2.5. Genetic Variables
2.5.1. DNA Isolation
2.5.2. Detection of Gene Polymorphisms
2.6. Response Variables
2.7. Statistical Analysis
3. Results
3.1. Patient Characteristics
3.2. Clinical Effectiveness of ABA
3.3. Distribution of the Genotypes Analyzed
3.4. ABA Response Predictors at 6 Months
3.4.1. EULAR Response
3.4.2. Low Disease Activity (LDA)
3.4.3. Remission
3.5. ABA Response Predictors at 12 Months
3.5.1. EULAR Response
3.5.2. Low Disease Activity (LDA)
3.5.3. Remission
3.6. Association between Low-Affinity FCGR2A/FCGR3A Haplotypes and ABA Response
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABA | abatacept |
| ACPA | anti-cyclic citrullinated peptide antibodies |
| ACR | American College of Rheumatology |
| ADCC | antibody-dependent cellular cytotoxicity |
| Arg | arginine |
| bDMARDs | biologic disease-modifying antirheumatic drugs |
| BT | biological therapy |
| CRP | C-reactive protein |
| csDMARDs | conventional synthetic disease-modifying antirheumatic drugs |
| CTLA-4 | cytotoxic T-lymphocyte-associated antigen 4 |
| DAS28 | 28-joints Disease Activity Score |
| DMARDs | disease-modifying antirheumatic drugs |
| ESR | erythrocyte sedimentation rate |
| EULAR | European League Against Rheumatism |
| Fc | fragment crystallizable |
| FCGR | Fc-gamma receptor |
| GC | glucocorticoid |
| HAQ | Health Assessment Questionnaire score |
| His | histidine |
| HWE | Hardy–Weinberg equilibrium |
| IFX | infliximab |
| IgG1 | human immunoglobulin G1 |
| IV | intravenous |
| LDA | low-activity disease |
| LFN | leflunomide |
| MTX | methotrexate |
| NIJ | number of inflamed joints |
| NK | natural killer |
| NPJ | number of painful joints |
| OR | odds ratio |
| PCR | polymerase chain reaction |
| Phe | phenylalanine |
| PVAS | patient’s visual analogue scale |
| RA | rheumatoid arthritis |
| RF | rheumatoid factor |
| RTX | rituximab |
| SC | subcutaneous |
| SNP | single-nucleotide polymorphism |
| TCZ | tocilizumab |
| TNFi | tumor necrosis factor inhibitor |
| tsDMARDs | targeted synthetic disease-modifying antirheumatic drugs |
| Val | valine |
References
- Blair, H.A.; Deeks, E.D. Abatacept: A Review in Rheumatoid Arthritis. Drugs 2017, 77, 1221–1233. [Google Scholar] [CrossRef]
- Gazeau, P.; Alegria, G.C.; Devauchelle-Pensec, V.; Jamin, C.; Lemerle, J.; Bendaoud, B.; Brooks, W.H.; Saraux, A.; Cornec, D.; Renaudineau, Y. Memory B Cells and Response to Abatacept in Rheumatoid Arthritis. Clin. Rev. Allergy Immunol. 2017, 53, 166–176. [Google Scholar] [CrossRef]
- Lobo, E.D.; Hansen, R.J.; Balthasar, J.P. Antibody Pharmacokinetics and Pharmacodynamics. J. Pharm. Sci. 2004, 93, 2645–2668. [Google Scholar] [CrossRef]
- Montes, A.; Perez-Pampin, E.; Joven, B.; Carreira, P.; Fernandez-Nebro, A.; Ordóñez, M.D.C.; Navarro-Sarabia, F.; Moreira, V.; Vasilopoulos, Y.; Sarafidou, T.; et al. FCGR polymorphisms in the treatment of rheumatoid arthritis with Fc-containing TNF inhibitors. Pharmacogenomics 2015, 16, 333–345. [Google Scholar] [CrossRef] [PubMed]
- Tracey, D.; Klareskog, L.; Sasso, E.H.; Salfeld, J.G.; Tak, P.P. Tumor necrosis factor antagonist mechanisms of action: A comprehensive review. Pharmacol. Ther. 2008, 117, 244–279. [Google Scholar] [CrossRef] [PubMed]
- Nimmerjahn, F.; Ravetch, J.V. Fcγ Receptors: Old Friends and New Family Members. Immunology 2006, 24, 19–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, X.; Meng, G.; Dickinson, B.L.; Li, X.; Mizoguchi, E.; Miao, L.; Wang, Y.; Robert, C.; Wu, B.; Smith, P.D.; et al. MHC Class I-Related Neonatal Fc Receptor for IgG Is Functionally Expressed in Monocytes, Intestinal Macrophages, and Dendritic Cells. J. Immunol. 2001, 166, 3266–3276. [Google Scholar] [CrossRef] [Green Version]
- Beenhouwer, D.; Wallis, R.; Broder, M.E.; Furst, D. Mechanisms of action of tumor necrosis factor antagonist and granulomatous infections. J. Rheumatol. 2004, 31, 1888–1892. [Google Scholar]
- Morales, A.J.; Maldonado-Montoro, M.; de la Plata, J.E.M.; Ramírez, C.P.; Daddaoua, A.; Alarcón-Payer, C.; Msc, M.E.R.; Collado, C.G. FCGR2A/FCGR3A Gene Polymorphisms and Clinical Variables as Predictors of Response to Tocilizumab and Rituximab in Patients with Rheumatoid Arthritis. J. Clin. Pharmacol. 2019, 59, 517–531. [Google Scholar] [CrossRef]
- Quartuccio, L.; Fabris, M.; Pontarini, E.; Salvin, S.; Zabotti, A.; Benucci, M.; Manfredi, M.; Biasi, D.; Ravagnani, V.; Atzeni, F.; et al. The 158VV Fcgamma receptor 3A genotype is associated with response to rituximab in rheumatoid arthritis: Results of an Italian multicentre study. Ann. Rheum. Dis. 2013, 73, 716–721. [Google Scholar] [CrossRef]
- Pál, I.; Szamosi, S.; Hodosi, K.; Szekanecz, Z.; Váróczy, L. Effect of Fcγ-receptor 3a (FCGR3A) gene polymorphisms on rituximab therapy in Hungarian patients with rheumatoid arthritis. RMD Open 2017, 3, e000485. [Google Scholar] [CrossRef] [Green Version]
- Liu, D.; Tian, Y.; Sun, D.; Sun, H.; Jin, Y.; Dong, M. The FCGR3A polymorphism predicts the response to rituximab-based therapy in patients with non-Hodgkin lymphoma: A meta-analysis. Ann. Hematol. 2016, 95, 1483–1490. [Google Scholar] [CrossRef] [PubMed]
- Nimmerjahn, F.; Ravetch, J.V. Fcγ receptors as regulators of immune responses. Nat. Rev. Immunol. 2008, 8, 34–47. [Google Scholar] [CrossRef] [PubMed]
- Nishio, S.; Yamamoto, T.; Kaneko, K.; Tanaka-Matsumoto, N.; Muraoka, S.; Kaburaki, M.; Kusunoki, Y.; Takagi, K.; Kawai, S. Pharmacokinetic study and Fcγ receptor gene analysis in two patients with rheumatoid arthritis controlled by low-dose infliximab. Mod. Rheumatol. 2009, 19, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Antonatos, C.; Stavrou, E.F.; Evangelou, E.; Vasilopoulos, Y. Exploring pharmacogenetic variants for predicting response to anti-TNF therapy in autoimmune diseases: A meta-analysis. Pharmacogenomics 2021, 22, 435–445. [Google Scholar] [CrossRef]
- Van Der Pol, W.-L.; Van De Winkel, J.G.J. IgG receptor polymorphisms: Risk factors for disease. Immunogenetics 1998, 48, 222–232. [Google Scholar] [CrossRef] [PubMed]
- Chai, L.; Song, Y.-Q.; Leung, W.K. Genetic polymorphism studies in periodontitis and Fcγ receptors. J. Periodontal Res. 2011, 47, 273–285. [Google Scholar] [CrossRef] [Green Version]
- Cañete, J.D.; Suarez, B.; Hernandez, M.V.; Sanmarti, R.; Rego, I.; Celis, R.; Moll, C.; Pinto, J.A.; Blanco, F.J.; Lozano, F. Influence of variants of Fcγ receptors IIA and IIIA on the American College of Rheumatology and European League Against Rheumatism responses to anti-tumour necrosis factor α therapy in rheumatoid arthritis. Ann. Rheum. Dis. 2009, 68, 1547–1552. [Google Scholar] [CrossRef] [PubMed]
- Binstadt, B.A.; Geha, R.S.; Bonilla, F.A. IgG Fc receptor polymorphisms in human disease: Implications for intravenous immunoglobulin therapy. J. Allergy Clin. Immunol. 2003, 111, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Hatjiharissi, E.; Xu, L.; Santos, D.D.; Hunter, Z.R.; Ciccarelli, B.T.; Verselis, S.; Modica, M.; Cao, Y.; Manning, R.J.; Leleu, X.; et al. Increased natural killer cell expression of CD16, augmented binding and ADCC activity to rituximab among individuals expressing the FcγRIIIa-158 V/V and V/F polymorphism. Blood 2007, 110, 2561–2564. [Google Scholar] [CrossRef]
- Dávila-Fajardo, C.L.; Van Der Straaten, T.; Baak-Pablo, R.; Caballero, C.M.; Barrera, J.C.; Huizinga, T.W.; Guchelaar, H.-J.; Swen, J.J. FcGR genetic polymorphisms and the response to adalimumab in patients with rheumatoid arthritis. Pharmacogenomics 2015, 16, 373–381. [Google Scholar] [CrossRef]
- Lee, Y.H.; Bae, S.-C. Associations between PTPRC rs10919563 A/G and FCGR2A R131H polymorphisms and responsiveness to TNF blockers in rheumatoid arthritis: A meta-analysis. Rheumatol. Int. 2016, 36, 837–844. [Google Scholar] [CrossRef]
- Tutuncu, Z.; Kavanaugh, A.; Zvaifler, N.; Corr, M.; Deutsch, R.; Boyle, D. Fcgamma receptor type IIIA polymorphisms influence treatment outcomes in patients with inflammatory arthritis treated with tumor necrosis factor alpha-blocking agents. Arthritis Rheum. 2005, 52, 2693–2696. [Google Scholar] [CrossRef]
- Morales-Lara, M.J.; Conesa-Zamora, P.; García-Simón, M.S.; Pedrero, F.; Santaclara, V.; Perez-Guillermo, M.; Soriano-Navarro, E. Association between the FCGR3A V158F polymorphism and the clinical response to infliximab in rheumatoid arthritis and spondyloarthritis patients. Scand. J. Rheumatol. 2010, 39, 518–520. [Google Scholar] [CrossRef]
- Kastbom, A.; Bratt, J.; Ernestam, S.; Lampa, J.; Padyukov, L.; Söderkvist, P.; Skogh, T. Fcgamma receptor type IIIA genotype and response to tumor necrosis factor alpha-blocking agents in patients with rheumatoid arthritis. Arthritis Rheum. 2007, 56, 448–452. [Google Scholar] [CrossRef] [PubMed]
- Okada, Y.; Wu, D.; Trynka, G.; Raj, T.; Terao, C.; Ikari, K.; Kochi, Y.; Ohmura, K.; Suzuki, A.; Yoshida, S.; et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 2014, 506, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Frisell, T.; Askling, J. Comment on: Comparative effectiveness of abatacept, rituximab, tocilizumab and TNFi biologics in RA: Results from the nationwide Swedish register: Reply. Rheumatology 2019, 58, 1507–1509. [Google Scholar] [CrossRef] [Green Version]
- Pete, N.M.; Montoro, M.D.M.M.; Ramírez, C.P.; Martín, A.S.; De La Plata, J.E.M.; Martínez, F.M.; Caliz, R.C.; Daddaoua, A.; Tortosa, M.D.C.R.; Morales, A.J. Impact of Single-Nucleotide Polymorphisms of CTLA-4, CD80 and CD86 on the Effectiveness of Abatacept in Patients with Rheumatoid Arthritis. J. Pers. Med. 2020, 10, 220. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.A.; Saag, K.G.; Bridges, S.L., Jr.; Akl, E.A.; Bannuru, R.R.; Sullivan, M.C.; Vaysbrot, E.; McNaughton, C.; Osani, M.; Shmerling, R.H.; et al. 2015 American College of Rheumatology Guideline for the Treatment of Rheumatoid Arthritis. Arthritis Rheumatol. 2016, 68, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Felson, D.T.; Anderson, J.J. Methodological and statistical approaches to criteria development in rheumatic diseases. Bailliere’s Clin. Rheumatol. 1995, 9, 253–266. [Google Scholar] [CrossRef]
- Wells, G.; Becker, J.-C.; Teng, J.; Dougados, M.; Schiff, M.; Smolen, J.; Aletaha, D.; Van Riel, P.L.C.M. Validation of the 28-joint Disease Activity Score (DAS28) and European League Against Rheumatism response criteria based on C-reactive protein against disease progression in patients with rheumatoid arthritis, and comparison with the DAS28 based on erythrocyte sedimentation rate. Ann. Rheumathol. Dis. 2008, 68, 954–960. [Google Scholar] [CrossRef]
- Prevoo, M.L.L.; Hof, M.A.V.; Kuper, H.H.; Van Leeuwen, M.A.; Van De Putte, L.B.A.; Van Riel, P.L.C.M. Modified disease activity scores that include twenty-eight-joint counts development and validation in a prospective longitudinal study of patients with rheumatoid arthritis. Arthritis Rheumathol. 1995, 38, 44–48. [Google Scholar] [CrossRef] [Green Version]
- Smolen, J.S.; Landewé, R.B.M.; Bijlsma, J.W.J.; Burmester, G.R.; Dougados, M.; Kerschbaumer, A.; McInnes, I.B.; Sepriano, A.; van Vollenhoven, R.F.; de Wit, M.; et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2019 update. Ann. Rheumathol. Dis. 2020, 79, 685–699. [Google Scholar] [CrossRef] [Green Version]
- Van Gestel, A.M.; Anderson, J.J.; Van Riel, P.L.; Boers, M.; Haagsma, C.J.; Rich, B.; Wells, G.; Lange, M.L.; Felson, D. ACR and EULAR improvement criteria have comparable validity in rheumatoid arthritis trials. American College of Rheumatology European League of Associations for Rheumatology. J. Rheumatol. 1999, 26, 705–711. [Google Scholar] [PubMed]
- Felson, D.T.; Smolen, J.S.; Wells, G.; Zhang, B.; Van Tuyl, L.H.D.; Funovits, J.; Aletaha, D.; Allaart, C.F.; Bathon, J.; Bombardieri, S.; et al. American College of Rheumatology/European League Against Rheumatism provisional definition of remission in rheumatoid arthritis for clinical trials. Arthritis Rheumathol. 2011, 63, 573–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Team RC. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2013. [Google Scholar]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [Green Version]
- Zhai, C.; Li, S.; Feng, W.; Shi, W.; Wang, J.; Wang, Q.; Chai, L.; Zhang, Q.; Yan, X.; Li, M. Association of interleukin-17a rs2275913 gene polymorphism and asthma risk: A meta-analysis. Arch. Med. Sci. 2018, 14, 1204–1211. [Google Scholar] [CrossRef]
- Sole, X.; Guinó, E.; Valls, J.; Iniesta, R.; Moreno, V. SNPStats: A web tool for the analysis of association studies. Bioinformatics 2006, 22, 1928–1929. [Google Scholar] [CrossRef] [Green Version]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alyousef, Y.M.; Borgio, J.F.; AbdulAzeez, S.; Al-Masoud, N.; Al-Ali, A.A.; Al-Shwaimi, E.; Al-Ali, A.K. Association of MBL2 Gene Polymorphism with Dental Caries in Saudi Children. Caries Res. 2016, 51, 12–16. [Google Scholar] [CrossRef]
- AbdulAzeez, S.; Al-Nafie, A.N.; Al-Shehri, A.; Borgio, J.F.; Baranova, E.V.; Al-Madan, M.S.; Al-Ali, R.A.; Al-Muhanna, F.; Al-Ali, A.; Al-Mansori, M.; et al. Intronic Polymorphisms in the CDKN2B-AS1 Gene Are Strongly Associated with the Risk of Myocardial Infarction and Coronary Artery Disease in the Saudi Population. Int. J. Mol. Sci. 2016, 17, 395. [Google Scholar] [CrossRef]
- Al Asoom, L.I.; Alsuwat, H.S.; Rafique, N.; Al Makhaita, M.; AlAmoudi, W.; AbdulAzeez, S.; Borgio, J.F. Functional DNA variations associated with Saudi female with low VO2max: A pilot microarray study. Am. J. Transl. Res. 2019, 11, 3659–3670. [Google Scholar]
- Nam, J.; Winthrop, K.; Van Vollenhoven, R.; Pavelka, K.; Valesini, G.; Hensor, E.; Worthy, G.; Landewe, R.; Smolen, J.; Emery, P.; et al. Current evidence for the management of rheumatoid arthritis with biological disease-modifying antirheumatic drugs: A systematic literature review informing the EULAR recommendations for the management of RA. Ann. Rheumathol. Dis. 2010, 69, 976–986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.H.; Bae, S.C. Comparative efficacy and safety of tocilizumab, rituximab, abatacept and tofacitinib in patients with active rheumatoid arthritis that inadequately responds to tumor necrosis factor inhibitors: A Bayesian network meta-analysis of randomized controlled trials. Int. J. Rheum. Dis. 2016, 19, 1103–1111. [Google Scholar] [PubMed]
- Avila-Pedretti, G.; Tornero, J.; Fernández-Nebro, A.; Blanco, F.; Gonzalez-Alvaro, I.; Cañete, J.D.; Maymó, J.; Alperiz, M.; Fernandez-Gutierrez, B.; Olivè, A.; et al. Variation at FCGR2A and Functionally Related Genes Is Associated with the Response to Anti-TNF Therapy in Rheumatoid Arthritis. PLoS ONE 2015, 10, e0122088. [Google Scholar] [CrossRef] [Green Version]
- Cagnotto, G.; Willim, M.; Nilsson, J.; Compagno, M.; Jacobsson, L.T.H.; Saevarsdottir, S.; Turesson, C. Abatacept in rheumatoid arthritis: Survival on drug, clinical outcomes, and their predictors—data from a large national quality register. Arthritis Res. 2020, 22, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrold, L.R.; Litman, H.J.; Connolly, S.E.; Kelly, S.; Hua, W.; Alemao, E.; Rosenblatt, L.; Rebello, S.; Kremer, J.M. A window of opportunity for abatacept in RA: Is disease duration an independent predictor of low disease activity/remission in clinical practice? Clin. Rheumatol. 2017, 36, 1215–1220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, J.J.; Wells, G.; Verhoeven, A.C.; Felson, D.T. Factors predicting response to treatment in rheumatoid arthritis: The importance of disease duration. Arthritis Rheumathol. 2000, 43, 22–29. [Google Scholar] [CrossRef]
- Möttönen, T.; Hannonen, P.; Korpela, M.; Nissilä, M.; Kautiainen, H.; Ilonen, J.; Laasonen, L.; Kaipiainen-Seppänen, O.; Franzen, P.; Helve, T.; et al. Delay to institution of therapy and induction of remission using single-drug or combination-disease-modifying antirheumatic drug therapy in early rheumatoid arthritis. Arthritis Rheumathol. 2002, 46, 894–898. [Google Scholar] [CrossRef]
- Paul, D.; Fazeli, M.S.; Mintzer, L.; Duarte, L.; Gupta, K.; Ferri, L. Comparative efficacy and safety of current therapies for early rheumatoid arthritis: A systematic literature review and network meta-analysis. Clin. Exp. Rheumatol. 2020, 38, 1008–1015. [Google Scholar]
- Lee, J.S.; Ahmad, H.; Shim, S.-C.; Bae, S.-C.; Song, Y.W.; Lee, E.Y. High Proportion of Subjective Component to the Disease Activity Score is Associated with Favorable Response to Abatacept in Rheumatoid Arthritis. Patient Cent. Outcomes Res. 2018, 12, 319–326. [Google Scholar] [CrossRef] [Green Version]
- Jurgens, M.S.; Overman, C.L.; Jacobs, J.W.G.; Geenen, R.; Cuppen, B.V.J.; Marijnissen, A.C.A.; Bijlsma, J.W.J.; Welsing, P.M.J.; Lafeber, F.P.J.G.; Van Laar, J.M.; et al. Contribution of the Subjective Components of the Disease Activity Score to the Response to Biologic Treatment in Rheumatoid Arthritis. Arthritis Rheumathol. 2015, 67, 923–928. [Google Scholar] [CrossRef]
- Alten, R.; Mariette, X.; Lorenz, H.-M.; Galeazzi, M.; Cantagrel, A.; Nüßlein, H.G.; Chartier, M.; Elbez, Y.; Rauch, C.; Le Bars, M. Real-world predictors of 12–month intravenous abatacept retention in patients with rheumatoid arthritis in the ACTION observational study. RMD Open 2017, 3, e000538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maldonado-Montoro, M.; Cañadas-Garre, M.; González-Utrilla, A.; Plaza-Plaza, J.C.; Calleja-Hernández, M. Ÿngel Genetic and clinical biomarkers of tocilizumab response in patients with rheumatoid arthritis. Pharmacol. Res. 2016, 111, 264–271. [Google Scholar] [CrossRef] [PubMed]
- Montes, A.; Perez-Pampin, E.; Narvaez, J.; Cañete, J.D.; Navarro-Sarabia, F.; Moreira, V.; Fernández-Nebro, A.; Ordóñez, M.D.C.; de la Serna, A.R.; Magallares, B.; et al. Association of FCGR2A with the response to infliximab treatment of patients with rheumatoid arthritis. Pharm. Genom. 2014, 24, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Bae, S.-C.; Song, G.G. Functional FCGR3A 158 V/F and IL-6 −174 C/G polymorphisms predict response to biologic therapy in patients with rheumatoid arthritis: A meta-analysis. Rheumatol. Int. 2014, 34, 1409–1415. [Google Scholar] [CrossRef] [PubMed]
- Julià, M.; Guilabert, A.; Lozano, F.; Suarez-Casasús, B.; Moreno, N.; Carrascosa, J.M.; Ferrándiz, C.; Pedrosa, E.; Alsina-Gibert, M.; Mascaró, J.M. The Role of Fcγ Receptor Polymorphisms in the Response to Anti–Tumor Necrosis Factor Therapy in Psoriasis. JAMA Dermatol. 2013, 149, 1033–1039. [Google Scholar] [CrossRef] [PubMed]
- Davis, P.M.; Abraham, R.; Xu, L.; Nadler, S.G.; Suchard, S.J. Abatacept binds to the Fc receptor CD64 but does not mediate complement-dependent cytotoxicity or antibody-dependent cellular cytotoxicity. J. Rheumatol. 2007, 34, 2204–2210. [Google Scholar]
| Variables | Initial Level | ||
|---|---|---|---|
| N | (%) | Mean ± SD/p50(p25–p75) | |
| Sex | 120 | ||
| Women | 89 | 74 | – |
| Smoking | |||
| Smoker | 18 | 15 | – |
| Ex-smoker | 14 | 12 | – |
| Non-smoker | 88 | 73 | – |
| Age at Dx | 120 | – | 45.15 ± 13.72 |
| Years with RA | 120 | – | 24 (9–21) |
| ABA start age | 120 | – | 56.63 ± 13.03 |
| ABA duration | 120 | – | 24.00 (14.75–44.25) |
| Administration | |||
| SC | 68 | 57 | – |
| Concomitant csDMARDs | |||
| MTX | 42 | 35 | – |
| LFN | 14 | 12 | – |
| none | 64 | 53 | – |
| Concomitant GCs | |||
| Yes | 102 | 85 | – |
| Monotherapy | |||
| No | 113 | 94 | – |
| Number previous BTs | 120 | – | 2 (1–3) |
| Duration previous BTs | 120 | – | 36 (24–72) |
| Previous BTs | |||
| Naïve | 15 | 12 | – |
| 1 TNF | 31 | 26 | – |
| 2 TNFs | 34 | 28 | – |
| 3 or more TNFs | 40 | 33 | – |
| Reason for suspension | |||
| Primary failure | 25 | 21 | – |
| Secondary failure | 12 | 10 | – |
| AR | 6 | 5 | – |
| No suspension | 77 | 64 | – |
| RF | |||
| Positive | 96 | 80 | – |
| ACPA | |||
| Positive | 85 | 71 | – |
| DAS28 | 120 | – | 4.70 ± 1.43 |
| Baseline NPJ | 120 | – | 6 (3–10) |
| Baseline NSJ | 120 | – | 3 (0–6) |
| PVAS | 120 | – | 70 (50–80) |
| Baseline CRP | 120 | – | 2 (1–4) |
| Baseline ESR | 120 | – | 22 (10–38) |
| HAQ | 120 | – | 1.75 (1.00–2.00) |
| No-Bionaïve Patients | ||||
|---|---|---|---|---|
| Response Variable | 6 Months | 12 Months | ||
| N | % | N | % | |
| EULAR response | 120 | 105 | ||
| Satisfactory | 38 | 31.67 | 48 | 45.71 |
| Unsatisfactory | 82 | 68.33 | 57 | 54.29 |
| Remission (DAS28 < 2.6) | 18 | 15 | 29 | 27.62 |
| LDA (2.6≤ DAS28≤ 3.2) | 24 | 20 | 24 | 22.86 |
| ABA-bionaïve patients | ||||
| Response variable | 6 months | 12 months | ||
| EULAR response | 15 | 14 | ||
| Satisfactory | 8 | 53.33 | 11 | 78.57 |
| Unsatisfactory | 7 | 46.67 | 3 | 21.43 |
| Remission (DAS28 < 2.6) | 3 | 20 | 9 | 64.29 |
| LDA (2.6≤ DAS28≤ 3.2) | 5 | 33.33 | 3 | 21.43 |
| Response Variable | Independent Variable | B | OR | p-Value (Variable) | 95% CI | R2 | Goodness of Fit |
|---|---|---|---|---|---|---|---|
| 6 MONTHS | |||||||
| EULAR response | |||||||
| Duration previous BTs | −0.017 | 0.98 | 0.006 | 0.97−0.99 | Cox Snell R2 = 0.173 | χ2 = 9.750 | |
| FCGR2A (AA vs. G) | 0.887 | 2.43 | 0.048 | 1.01−5.92 | |||
| Monotherapy (yes vs. no) | 3.199 | 24.53 | 0.006 | 3.46−523.80 | Nagelkerke R2 = 0.243 | p = 0.283 | |
| LDA | |||||||
| Initial PVAS | −0.033 | 0.97 | 0.003 | 0.95−0.99 | Cox Snell R2 = 0.110 | χ2 = 9.606 | |
| FCGR2A (AA vs. G) | 1.149 | 3.16 | 0.022 | 1.19−8.66 | Nagelkerke R2 = 0.174 | p = 0.294 | |
| Remission | |||||||
| ABA duration | 0.023 | 1.02 | 0.026 | 1.01−1.04 | Cox Snell R2 = 0.232 | χ2 = 3.338 | |
| Duration previous BTs | −0.023 | 0.98 | 0.029 | 0.95−0.99 | |||
| Initial ESR | −0.079 | 0.92 | 0.005 | 0.87−0.97 | Nagelkerke R2 = 0.406 | p = 0.911 | |
| Monotherapy (yes vs. no) | 2.956 | 19.22 | 0.019 | 2.05−343.00 | |||
| 12 MONTHS | |||||||
| EULAR response | |||||||
| Initial PVAS | −0.056 | 0.95 | <0.001 | 0.92−0.97 | Cox Snell R2 = 0.248 | χ2 = 13.130 | |
| Duration previous BTs | −0.012 | 0.99 | 0.029 | 0.98−0.99 | Nagelkerke R2 = 0.332 | p = 0.108 | |
| LDA | |||||||
| ABA start age | 0.059 | 1.06 | 0.007 | 1.02−1.11 | Cox Snell R2 = 0.196 | χ2 = 15.030 | |
| Concomitant GCs | −2.149 | 0.12 | 0.004 | 0.02−0.47 | |||
| FCGR2A (AA vs. AG) | 2.551 | 12.82 | 0.002 | 2.95−83.04 | Nagelkerke R2 = 0.297 | p = 0.059 | |
| FCGR2A (AA vs. GG) | 1.890 | 6.62 | 0.036 | 1.25−46.89 | |||
| Remission | |||||||
| Duration previous BTs | −0.019 | 0.98 | 0.006 | 0.97–0.99 | Cox Snell R2 = 0.190 | χ2 = 7.215 | |
| Initial PVAS | −0.042 | 0.96 | <0.001 | 0.94−0.98 | Nagelkerke R2 = 0.274 | p = 0.514 | |
| FCGR2A | FCGR3A | Frequencies | Odds Ratio (95% CI) | p-Value |
|---|---|---|---|---|
| rs1801274 | rs396991 | |||
| A | C | 0.2746 | 1.00 | – |
| G | A | 0.2662 | 1.90 (0.66–5.49) | 0.240 |
| A | A | 0.2629 | 0.90 (0.28–2.91) | 0.860 |
| G | C | 0.1963 | 5.24 (0.98–28.08) | 0.056 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Márquez Pete, N.; Maldonado Montoro, M.d.M.; Pérez Ramírez, C.; Martínez Martínez, F.; Martínez de la Plata, J.E.; Daddaoua, A.; Jiménez Morales, A. Influence of the FCGR2A rs1801274 and FCGR3A rs396991 Polymorphisms on Response to Abatacept in Patients with Rheumatoid Arthritis. J. Pers. Med. 2021, 11, 573. https://doi.org/10.3390/jpm11060573
Márquez Pete N, Maldonado Montoro MdM, Pérez Ramírez C, Martínez Martínez F, Martínez de la Plata JE, Daddaoua A, Jiménez Morales A. Influence of the FCGR2A rs1801274 and FCGR3A rs396991 Polymorphisms on Response to Abatacept in Patients with Rheumatoid Arthritis. Journal of Personalized Medicine. 2021; 11(6):573. https://doi.org/10.3390/jpm11060573
Chicago/Turabian StyleMárquez Pete, Noelia, María del Mar Maldonado Montoro, Cristina Pérez Ramírez, Fernando Martínez Martínez, Juan Enrique Martínez de la Plata, Abdelali Daddaoua, and Alberto Jiménez Morales. 2021. "Influence of the FCGR2A rs1801274 and FCGR3A rs396991 Polymorphisms on Response to Abatacept in Patients with Rheumatoid Arthritis" Journal of Personalized Medicine 11, no. 6: 573. https://doi.org/10.3390/jpm11060573
APA StyleMárquez Pete, N., Maldonado Montoro, M. d. M., Pérez Ramírez, C., Martínez Martínez, F., Martínez de la Plata, J. E., Daddaoua, A., & Jiménez Morales, A. (2021). Influence of the FCGR2A rs1801274 and FCGR3A rs396991 Polymorphisms on Response to Abatacept in Patients with Rheumatoid Arthritis. Journal of Personalized Medicine, 11(6), 573. https://doi.org/10.3390/jpm11060573