Detection of Aberrant Glycosylation of Serum Haptoglobin for Gastric Cancer Diagnosis Using a Middle-Up-Down Glycoproteome Platform
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Serum Samples from Gastric Cancer Patients and Healthy Control Subjects
2.3. Haptoglobin Purification from Serum Samples
2.4. Enzymatic Digestion for Glycopeptide Production
2.5. LC–MS Analysis of Haptoglobin Glycopeptide
2.6. Data Processing, Glycopeptide Identification, and Statistical Analysis
3. Results and Discussion
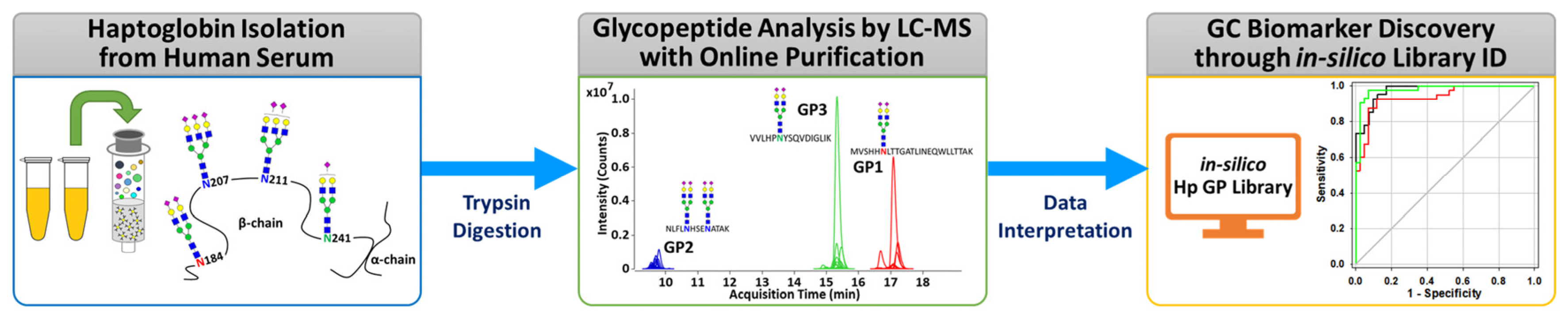
3.1. Analytical Strategy Using a Middle-Up-Down Glycoproteome Approach
3.2. Identification of Glycopeptides Using In Silico Haptoglobin Glycopeptide Library
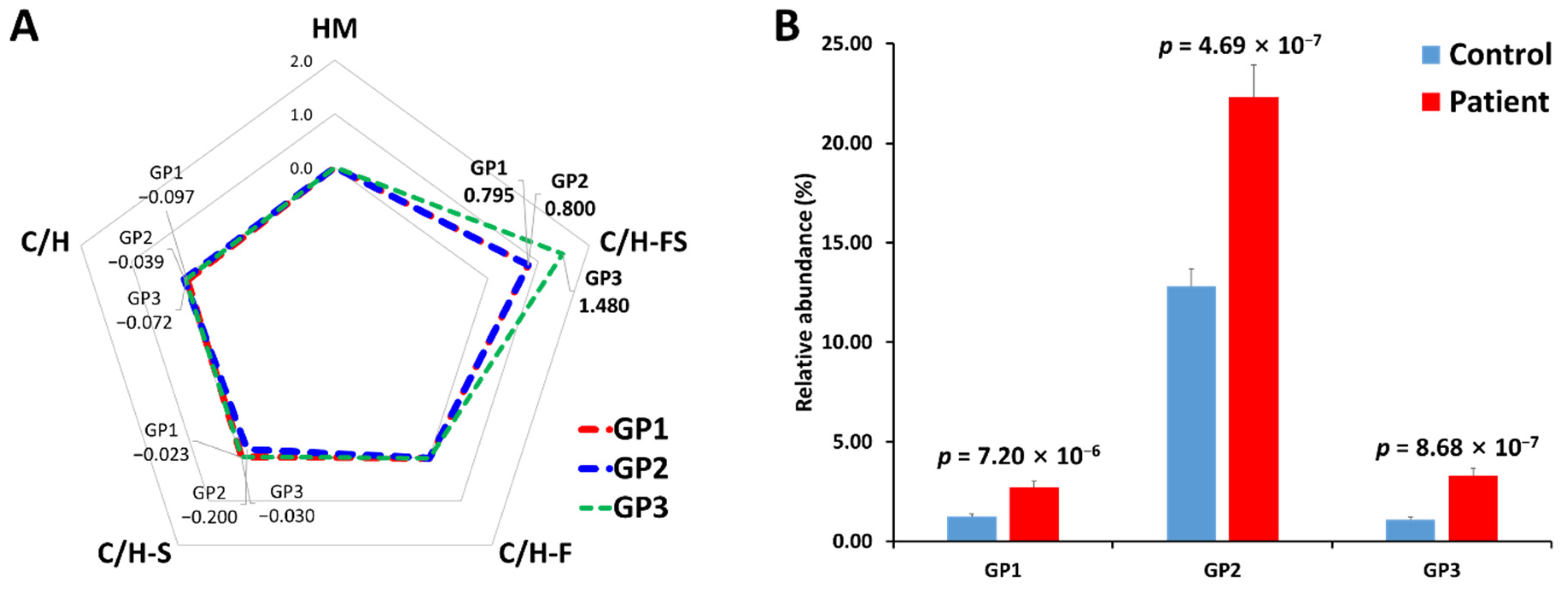
3.3. Gastric Cancer Biomarker Discovery via Middle-Up-Down Glycoproteome Platform
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Jung, K.W.; Won, Y.J.; Kong, H.J.; Lee, E.S. Prediction of Cancer Incidence and Mortality in Korea, 2019. Cancer Res. Treat. 2019, 51, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Torre, L.A.; Siegel, R.L.; Ward, E.M.; Jemal, A. Global Cancer Incidence and Mortality Rates and Trends—An Update. Cancer Epidemiol. Biomark. Prev. 2016, 25, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, T.; Yashiro, M. Biomarkers of gastric cancer: Current topics and future perspective. World J. Gastroenterol. 2018, 24, 2818–2832. [Google Scholar] [CrossRef] [PubMed]
- Duraker, N.; Naci Celik, A.; Gencler, N. The prognostic significance of gastric juice CA 19-9 and CEA levels in gastric carcinoma patients. Eur. J. Surg. Oncol. 2002, 28, 844–849. [Google Scholar] [CrossRef]
- Jeong, S.; Oh, M.J.; Kim, U.; Lee, J.; Kim, J.H.; An, H.J. Glycosylation of serum haptoglobin as a marker of gastric cancer: An overview for clinicians. Expert Rev. Proteom. 2020, 17, 109–117. [Google Scholar] [CrossRef]
- Dube, D.H.; Bertozzi, C.R. Glycans in cancer and inflammation—Potential for therapeutics and diagnostics. Nat. Rev. Drug Discov. 2005, 4, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Fuster, M.M.; Esko, J.D. The sweet and sour of cancer: Glycans as novel therapeutic targets. Nat. Rev. Cancer 2005, 5, 526–542. [Google Scholar] [CrossRef]
- Drake, P.M.; Cho, W.; Li, B.; Prakobphol, A.; Johansen, E.; Anderson, N.L.; Regnier, F.E.; Gibson, B.W.; Fisher, S.J. Sweetening the pot: Adding glycosylation to the biomarker discovery equation. Clin. Chem. 2010, 56, 223–236. [Google Scholar] [CrossRef]
- Hua, S.; An, H.J. Glycoscience aids in biomarker discovery. BMB Rep. 2012, 45, 323–330. [Google Scholar] [CrossRef]
- Powlesland, A.S.; Hitchen, P.G.; Parry, S.; Graham, S.A.; Barrio, M.M.; Elola, M.T.; Mordoh, J.; Dell, A.; Drickamer, K.; Taylor, M.E. Targeted glycoproteomic identification of cancer cell glycosylation. Glycobiology 2009, 19, 899–909. [Google Scholar] [CrossRef]
- An, H.J.; Kronewitter, S.R.; de Leoz, M.L.; Lebrilla, C.B. Glycomics and disease markers. Curr. Opin. Chem. Biol. 2009, 13, 601–607. [Google Scholar] [CrossRef]
- Pinho, S.S.; Reis, C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 2015, 15, 540–555. [Google Scholar] [CrossRef] [PubMed]
- Anderson, N.L.; Anderson, N.G. The human plasma proteome: History, character, and diagnostic prospects. Mol. Cell Proteom. 2002, 1, 845–867. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, J.G.; Balmana, M.; Macedo, J.A.; Pocas, J.; Fernandes, A.; de-Freitas-Junior, J.C.M.; Pinho, S.S.; Gomes, J.; Magalhaes, A.; Gomes, C.; et al. Glycosylation in cancer: Selected roles in tumour progression, immune modulation and metastasis. Cell Immunol. 2018, 333, 46–57. [Google Scholar] [CrossRef]
- de Leoz, M.L.; Young, L.J.; An, H.J.; Kronewitter, S.R.; Kim, J.; Miyamoto, S.; Borowsky, A.D.; Chew, H.K.; Lebrilla, C.B. High-mannose glycans are elevated during breast cancer progression. Mol. Cell Proteom. 2011, 10, M110.002717. [Google Scholar] [CrossRef]
- Josic, D.; Martinovic, T.; Pavelic, K. Glycosylation and metastases. Electrophoresis 2019, 40, 140–150. [Google Scholar] [CrossRef]
- Oliveira-Ferrer, L.; Legler, K.; Milde-Langosch, K. Role of protein glycosylation in cancer metastasis. Semin. Cancer Biol. 2017, 44, 141–152. [Google Scholar] [CrossRef]
- Magalhaes, A.; Duarte, H.O.; Reis, C.A. Aberrant Glycosylation in Cancer: A Novel Molecular Mechanism Controlling Metastasis. Cancer Cell 2017, 31, 733–735. [Google Scholar] [CrossRef]
- Bull, C.; Boltje, T.J.; Wassink, M.; de Graaf, A.M.; van Delft, F.L.; den Brok, M.H.; Adema, G.J. Targeting aberrant sialylation in cancer cells using a fluorinated sialic acid analog impairs adhesion, migration, and in vivo tumor growth. Mol. Cancer Ther. 2013, 12, 1935–1946. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.H.; Misek, D.E. Glycoproteomics-based identification of cancer biomarkers. Int. J. Proteom. 2011, 2011, 601937. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jiao, J.; Yang, P.; Lu, H. Mass spectrometry-based N-glycoproteomics for cancer biomarker discovery. Clin. Proteom. 2014, 11, 18. [Google Scholar] [CrossRef]
- Clark, D.; Mao, L. Cancer biomarker discovery: Lectin-based strategies targeting glycoproteins. Dis. Markers 2012, 33, 1–10. [Google Scholar] [CrossRef]
- Yuan, W.; Benicky, J.; Wei, R.; Goldman, R.; Sanda, M. Quantitative Analysis of Sex-Hormone-Binding Globulin Glycosylation in Liver Diseases by Liquid Chromatography-Mass Spectrometry Parallel Reaction Monitoring. J. Proteome Res. 2018, 17, 2755–2766. [Google Scholar] [CrossRef]
- Kurosky, A.; Barnett, D.R.; Lee, T.H.; Touchstone, B.; Hay, R.E.; Arnott, M.S.; Bowman, B.H.; Fitch, W.M. Covalent structure of human haptoglobin: A serine protease homolog. Proc. Natl. Acad. Sci. USA 1980, 77, 3388–3392. [Google Scholar] [CrossRef]
- Mandato, V.D.; Magnani, E.; Abrate, M.; Casali, B.; Nicoli, D.; Farnetti, E.; Formisano, D.; Pirillo, D.; Ciarlini, G.; De Iaco, P.; et al. Haptoglobin phenotype and epithelial ovarian cancer. Anticancer Res. 2012, 32, 4353–4358. [Google Scholar] [PubMed]
- Morishita, K.; Ito, N.; Koda, S.; Maeda, M.; Nakayama, K.; Yoshida, K.; Takamatsu, S.; Yamada, M.; Eguchi, H.; Kamada, Y.; et al. Haptoglobin phenotype is a critical factor in the use of fucosylated haptoglobin for pancreatic cancer diagnosis. Clin. Chim. Acta 2018, 487, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Fujita, K.; Shimomura, M.; Uemura, M.; Nakata, W.; Sato, M.; Nagahara, A.; Nakai, Y.; Takamatsu, S.; Miyoshi, E.; Nonomura, N. Serum fucosylated haptoglobin as a novel prognostic biomarker predicting high-Gleason prostate cancer. Prostate 2014, 74, 1052–1058. [Google Scholar] [CrossRef]
- Takahashi, S.; Sugiyama, T.; Shimomura, M.; Kamada, Y.; Fujita, K.; Nonomura, N.; Miyoshi, E.; Nakano, M. Site-specific and linkage analyses of fucosylated N-glycans on haptoglobin in sera of patients with various types of cancer: Possible implication for the differential diagnosis of cancer. Glycoconj. J. 2016, 33, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Jeong, S.; Lee, J.; Yeo, I.S.; Oh, M.J.; Kim, U.; Kim, S.; Kim, S.H.; Park, S.Y.; Kim, J.H.; et al. Glycomic profiling of targeted serum haptoglobin for gastric cancer using nano LC/MS and LC/MS/MS. Mol. Biosyst. 2016, 12, 3611–3621. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Lee, S.H.; Choi, S.; Kim, U.; Yeo, I.S.; Kim, S.H.; Oh, M.J.; Moon, H.; Lee, J.; Jeong, S.; et al. Direct analysis of aberrant glycosylation on haptoglobin in patients with gastric cancer. Oncotarget 2017, 8, 11094–11104. [Google Scholar] [CrossRef]
- Lee, J.; Hua, S.; Lee, S.H.; Oh, M.J.; Yun, J.; Kim, J.Y.; Kim, J.H.; Kim, J.H.; An, H.J. Designation of fingerprint glycopeptides for targeted glycoproteomic analysis of serum haptoglobin: Insights into gastric cancer biomarker discovery. Anal. Bioanal. Chem. 2018, 410, 1617–1629. [Google Scholar] [CrossRef]
- Oh, M.J.; Lee, S.H.; Kim, U.; An, H.J. In-depth investigation of altered glycosylation in human haptoglobin associated cancer by mass spectrometry. Mass Spectrom. Rev. 2021. [Google Scholar] [CrossRef]
- Stavenhagen, K.; Hinneburg, H.; Thaysen-Andersen, M.; Hartmann, L.; Varon Silva, D.; Fuchser, J.; Kaspar, S.; Rapp, E.; Seeberger, P.H.; Kolarich, D. Quantitative mapping of glycoprotein micro-heterogeneity and macro-heterogeneity: An evaluation of mass spectrometry signal strengths using synthetic peptides and glycopeptides. J. Mass Spectrom. 2013, 48, 627–639. [Google Scholar] [CrossRef] [PubMed]
- Di Girolamo, F.; Lante, I.; Muraca, M.; Putignani, L. The Role of Mass Spectrometry in the “Omics” Era. Curr. Org. Chem. 2013, 17, 2891–2905. [Google Scholar] [CrossRef] [PubMed]
- Wolyniak, M.J.; Reyna, N.S.; Plymale, R.; Pope, W.H.; Westholm, D.E. Mass Spectrometry as a Tool to Enhance “-omics” Education. J. Microbiol. Biol. Educ. 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Goldenberg, N.A.; Everett, A.D.; Graham, D.; Bernard, T.J.; Nowak-Gottl, U. Proteomic and other mass spectrometry based “omics” biomarker discovery and validation in pediatric venous thromboembolism and arterial ischemic stroke: Current state, unmet needs, and future directions. Proteom. Clin. Appl. 2014, 8, 828–836. [Google Scholar] [CrossRef] [PubMed]
- Varghese, R.S.; Goldman, L.; An, Y.; Loffredo, C.A.; Abdel-Hamid, M.; Kyselova, Z.; Mechref, Y.; Novotny, M.; Drake, S.K.; Goldman, R.; et al. Integrated peptide and glycan biomarker discovery using MALDI-TOF mass spectrometry. In Proceedings of the 30th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Vancouver, BC, Canada, 20–25 August 2008; Volume 2008, pp. 3791–3794. [Google Scholar] [CrossRef]
- Wuhrer, M. Glycomics using mass spectrometry. Glycoconj. J. 2013, 30, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Aizpurua-Olaizola, O.; Torano, J.S.; Falcon-Perez, J.M.; Williams, C.; Reichardt, N.; Boons, G.J. Mass spectrometry for glycan biomarker discovery. TrAC Trend Anal. Chem. 2018, 100, 7–14. [Google Scholar] [CrossRef]
- Lebrilla, C.B.; An, H.J. The prospects of glycan biomarkers for the diagnosis of diseases. Mol. Biosyst. 2009, 5, 17–20. [Google Scholar] [CrossRef] [PubMed]
- Zaia, J. Mass spectrometry and glycomics. OMICS 2010, 14, 401–418. [Google Scholar] [CrossRef] [PubMed]
- Ozcan, S.; Barkauskas, D.A.; Renee Ruhaak, L.; Torres, J.; Cooke, C.L.; An, H.J.; Hua, S.; Williams, C.C.; Dimapasoc, L.M.; Han Kim, J.; et al. Serum glycan signatures of gastric cancer. Cancer Prev. Res. 2014, 7, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Hua, S.; Williams, C.C.; Dimapasoc, L.M.; Ro, G.S.; Ozcan, S.; Miyamoto, S.; Lebrilla, C.B.; An, H.J.; Leiserowitz, G.S. Isomer-specific chromatographic profiling yields highly sensitive and specific potential N-glycan biomarkers for epithelial ovarian cancer. J. Chromatogr. A 2013, 1279, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Hua, S.; Saunders, M.; Dimapasoc, L.M.; Jeong, S.H.; Kim, B.J.; Kim, S.; So, M.; Lee, K.S.; Kim, J.H.; Lam, K.S.; et al. Differentiation of cancer cell origin and molecular subtype by plasma membrane N-glycan profiling. J. Proteome Res. 2014, 13, 961–968. [Google Scholar] [CrossRef]
- Tang, Z.; Varghese, R.S.; Bekesova, S.; Loffredo, C.A.; Hamid, M.A.; Kyselova, Z.; Mechref, Y.; Novotny, M.V.; Goldman, R.; Ressom, H.W. Identification of N-glycan serum markers associated with hepatocellular carcinoma from mass spectrometry data. J. Proteome Res. 2010, 9, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Hua, S.; An, H.J.; Ozcan, S.; Ro, G.S.; Soares, S.; DeVere-White, R.; Lebrilla, C.B. Comprehensive native glycan profiling with isomer separation and quantitation for the discovery of cancer biomarkers. Analyst 2011, 136, 3663–3671. [Google Scholar] [CrossRef] [PubMed]
- Bones, J.; Mittermayr, S.; O’Donoghue, N.; Guttman, A.; Rudd, P.M. Ultra performance liquid chromatographic profiling of serum N-glycans for fast and efficient identification of cancer associated alterations in glycosylation. Anal. Chem. 2010, 82, 10208–10215. [Google Scholar] [CrossRef] [PubMed]
- Schneck, N.A.; Ivleva, V.B.; Cai, C.X.; Cooper, J.W.; Lei, Q.P. Characterization of the furin cleavage motif for HIV-1 trimeric envelope glycoprotein by intact LC-MS analysis. Analyst 2020, 145, 1636–1640. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Chen, Z.; Zhang, J.; An, M.; Wu, J.; Yu, Q.; Skilton, S.J.; Bern, M.; Ilker Sen, K.; Li, L.; et al. Differential Quantitative Determination of Site-Specific Intact N-Glycopeptides in Serum Haptoglobin between Hepatocellular Carcinoma and Cirrhosis Using LC-EThcD-MS/MS. J. Proteome Res. 2019, 18, 359–371. [Google Scholar] [CrossRef]
- Baerenfaenger, M.; Meyer, B. Intact Human Alpha-Acid Glycoprotein Analyzed by ESI-qTOF-MS: Simultaneous Determination of the Glycan Composition of Multiple Glycosylation Sites. J. Proteome Res. 2018, 17, 3693–3703. [Google Scholar] [CrossRef]
- Turiak, L.; Sugar, S.; Acs, A.; Toth, G.; Gomory, A.; Telekes, A.; Vekey, K.; Drahos, L. Site-specific N-glycosylation of HeLa cell glycoproteins. Sci. Rep. 2019, 9, 14822. [Google Scholar] [CrossRef] [PubMed]
- Hever, H.; Darula, Z.; Medzihradszky, K.F. Characterization of Site-Specific N-Glycosylation. Methods Mol. Biol. 2019, 1934, 93–125. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Diedrich, J.K.; Ma, Y.; Wang, N.; Pauthner, M.; Park, S.R.; Delahunty, C.M.; McLellan, J.S.; Burton, D.R.; Yates, J.R.; et al. Global site-specific analysis of glycoprotein N-glycan processing. Nat. Protoc. 2018, 13, 1196–1212. [Google Scholar] [CrossRef]
- Birzele, F.; Csaba, G.; Zimmer, R. Alternative splicing and protein structure evolution. Nucleic Acids Res. 2008, 36, 550–558. [Google Scholar] [CrossRef] [PubMed]
- Rehder, D.S.; Dillon, T.M.; Pipes, G.D.; Bondarenko, P.V. Reversed-phase liquid chromatography/mass spectrometry analysis of reduced monoclonal antibodies in pharmaceutics. J. Chromatogr. A 2006, 1102, 164–175. [Google Scholar] [CrossRef] [PubMed]
- Gomar, J.J.; Bobes-Bascaran, M.T.; Conejero-Goldberg, C.; Davies, P.; Goldberg, T.E. The Alzheimer’s Disease Neuroimaging Initiative. Utility of combinations of biomarkers, cognitive markers, and risk factors to predict conversion from mild cognitive impairment to Alzheimer disease in patients in the Alzheimer’s disease neuroimaging initiative. Arch. Gen. Psychiatry 2011, 68, 961–969. [Google Scholar] [CrossRef] [PubMed]
- Laxman, B.; Morris, D.S.; Yu, J.; Siddiqui, J.; Cao, J.; Mehra, R.; Lonigro, R.J.; Tsodikov, A.; Wei, J.T.; Tomlins, S.A.; et al. A first-generation multiplex biomarker analysis of urine for the early detection of prostate cancer. Cancer Res. 2008, 68, 645–649. [Google Scholar] [CrossRef]
- Mamtani, M.R.; Thakre, T.P.; Kalkonde, M.Y.; Amin, M.A.; Kalkonde, Y.V.; Amin, A.P.; Kulkarni, H. A simple method to combine multiple molecular biomarkers for dichotomous diagnostic classification. BMC Bioinform. 2006, 7, 442. [Google Scholar] [CrossRef] [PubMed]
- Bones, J.; Byrne, J.C.; O’Donoghue, N.; McManus, C.; Scaife, C.; Boissin, H.; Nastase, A.; Rudd, P.M. Glycomic and glycoproteomic analysis of serum from patients with stomach cancer reveals potential markers arising from host defense response mechanisms. J. Proteome Res. 2011, 10, 1246–1265. [Google Scholar] [CrossRef] [PubMed]
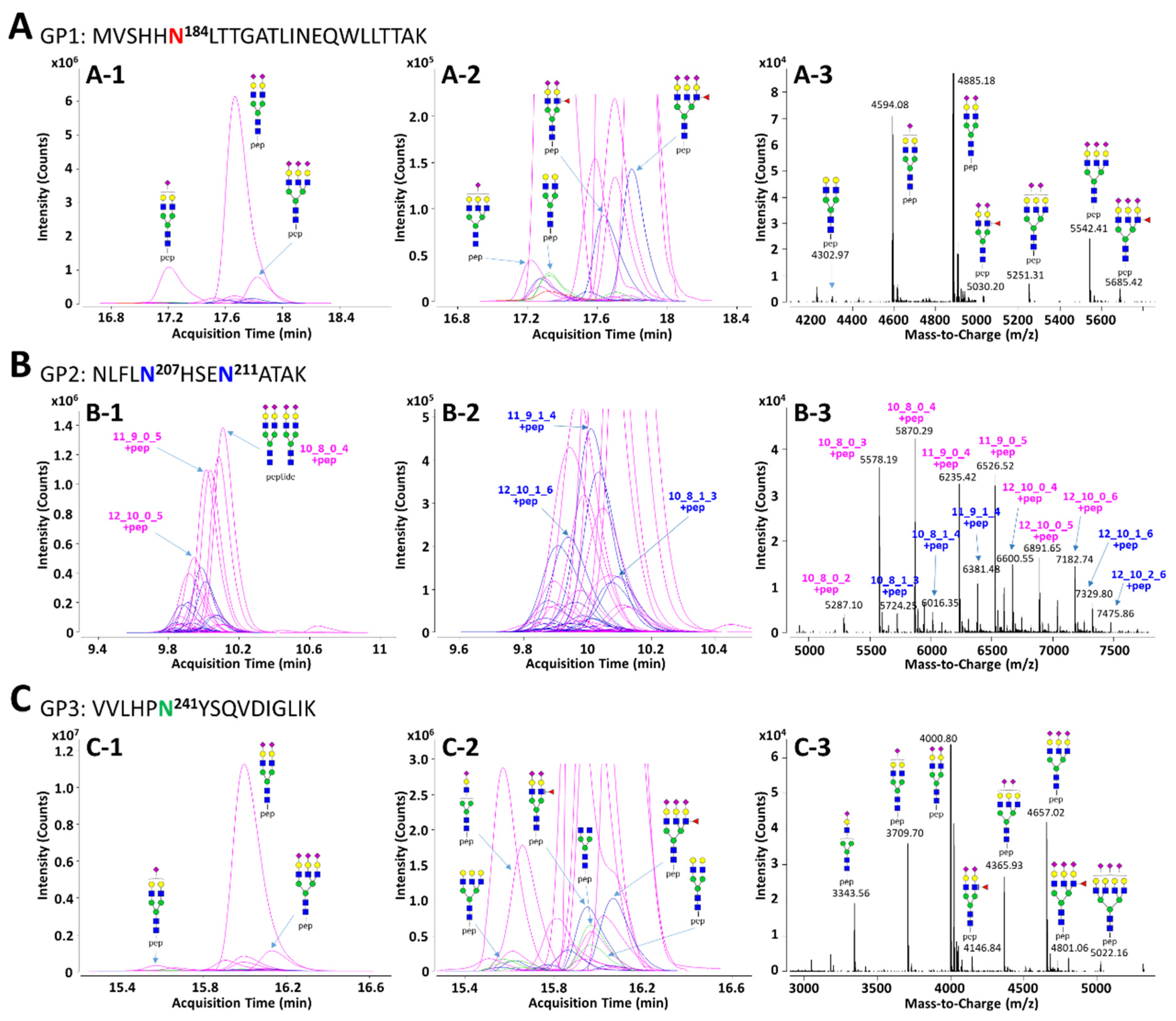

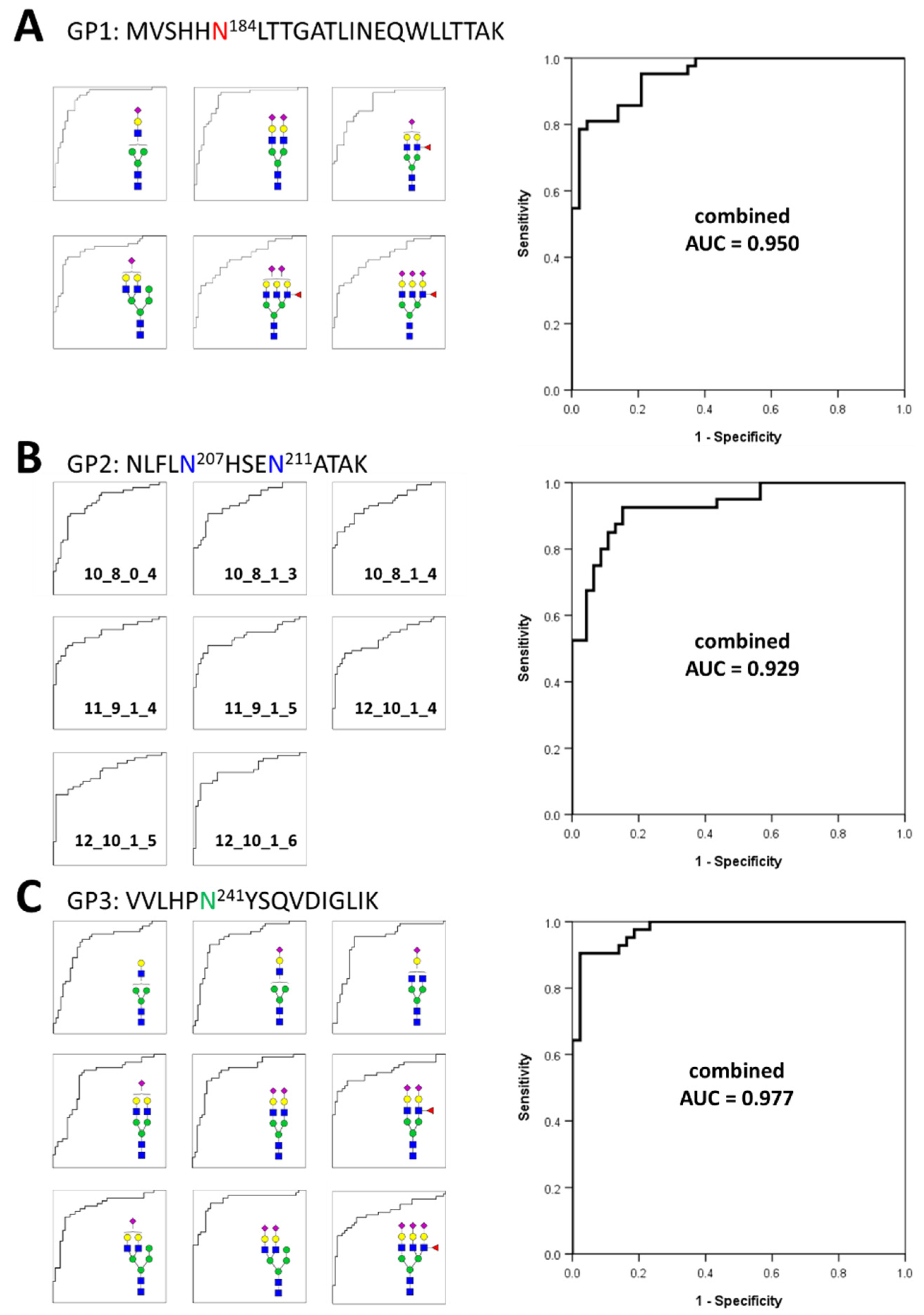
| GP Group a | Mass | N-Glycan Composition b | p-Value | AUC | |||
|---|---|---|---|---|---|---|---|
| Hex | HexNAc | Fuc | NeuAc | ||||
| GP1 | 4226.930 | 4 | 3 | 0 | 1 | 3.11 × 10−11 | 0.895 |
| 4738.120 | 5 | 4 | 1 | 1 | 1.17 × 10−8 | 0.873 | |
| 4754.115 | 6 | 4 | 0 | 1 | 8.63 × 10−11 | 0.864 | |
| 4883.157 | 5 | 4 | 0 | 2 | 3.13 × 10−11 | 0.887 | |
| 5394.348 | 6 | 5 | 1 | 2 | 1.19 × 10−7 | 0.831 | |
| 5685.443 | 6 | 5 | 1 | 3 | 1.12 × 10−6 | 0.803 | |
| GP2 | 5722.234 | 10 | 8 | 1 | 3 | 3.28 × 10−9 | 0.828 |
| 5867.271 | 10 | 8 | 0 | 4 | 8.18 × 10−8 | 0.818 | |
| 6013.329 | 10 | 8 | 1 | 4 | 2.32 × 10−8 | 0.814 | |
| 6378.461 | 11 | 9 | 1 | 4 | 9.32 × 10−11 | 0.848 | |
| 6669.557 | 11 | 9 | 1 | 5 | 6.78 × 10−9 | 0.820 | |
| 6743.593 | 12 | 10 | 1 | 4 | 1.74 × 10−7 | 0.805 | |
| 7034.689 | 12 | 10 | 1 | 5 | 1.86 × 10−9 | 0.823 | |
| 7325.784 | 12 | 10 | 1 | 6 | 7.50 × 10−10 | 0.846 | |
| GP3 | 3051.453 | 4 | 3 | 0 | 0 | 3.23 × 10−7 | 0.803 |
| 3342.549 | 4 | 3 | 0 | 1 | 1.19 × 10−8 | 0.840 | |
| 3545.628 | 4 | 4 | 0 | 1 | 1.33 × 10−7 | 0.827 | |
| 3707.681 | 5 | 4 | 0 | 1 | 3.25 × 10−6 | 0.783 | |
| 3869.734 | 6 | 4 | 0 | 1 | 9.63 × 10−9 | 0.849 | |
| 3998.776 | 5 | 4 | 0 | 2 | 5.54 × 10−9 | 0.844 | |
| 4144.834 | 5 | 4 | 1 | 2 | 3.51 × 10−9 | 0.844 | |
| 4160.829 | 6 | 4 | 0 | 2 | 7.82 × 10−12 | 0.901 | |
| 4801.062 | 6 | 5 | 1 | 3 | 1.75 × 10−6 | 0.805 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeong, S.; Kim, U.; Oh, M.J.; Nam, J.; Park, S.H.; Choi, Y.J.; Lee, D.H.; Kim, J.; An, H.J. Detection of Aberrant Glycosylation of Serum Haptoglobin for Gastric Cancer Diagnosis Using a Middle-Up-Down Glycoproteome Platform. J. Pers. Med. 2021, 11, 575. https://doi.org/10.3390/jpm11060575
Jeong S, Kim U, Oh MJ, Nam J, Park SH, Choi YJ, Lee DH, Kim J, An HJ. Detection of Aberrant Glycosylation of Serum Haptoglobin for Gastric Cancer Diagnosis Using a Middle-Up-Down Glycoproteome Platform. Journal of Personalized Medicine. 2021; 11(6):575. https://doi.org/10.3390/jpm11060575
Chicago/Turabian StyleJeong, Seunghyup, Unyong Kim, Myung Jin Oh, Jihyeon Nam, Se Hoon Park, Yoon Jin Choi, Dong Ho Lee, Jaehan Kim, and Hyun Joo An. 2021. "Detection of Aberrant Glycosylation of Serum Haptoglobin for Gastric Cancer Diagnosis Using a Middle-Up-Down Glycoproteome Platform" Journal of Personalized Medicine 11, no. 6: 575. https://doi.org/10.3390/jpm11060575
APA StyleJeong, S., Kim, U., Oh, M. J., Nam, J., Park, S. H., Choi, Y. J., Lee, D. H., Kim, J., & An, H. J. (2021). Detection of Aberrant Glycosylation of Serum Haptoglobin for Gastric Cancer Diagnosis Using a Middle-Up-Down Glycoproteome Platform. Journal of Personalized Medicine, 11(6), 575. https://doi.org/10.3390/jpm11060575








