Strategies to Integrate Genomic Medicine into Clinical Care: Evidence from the IGNITE Network
Abstract
:1. Introduction
2. Materials and Methods
2.1. Settings
2.2. Frameworks
2.3. Procedures
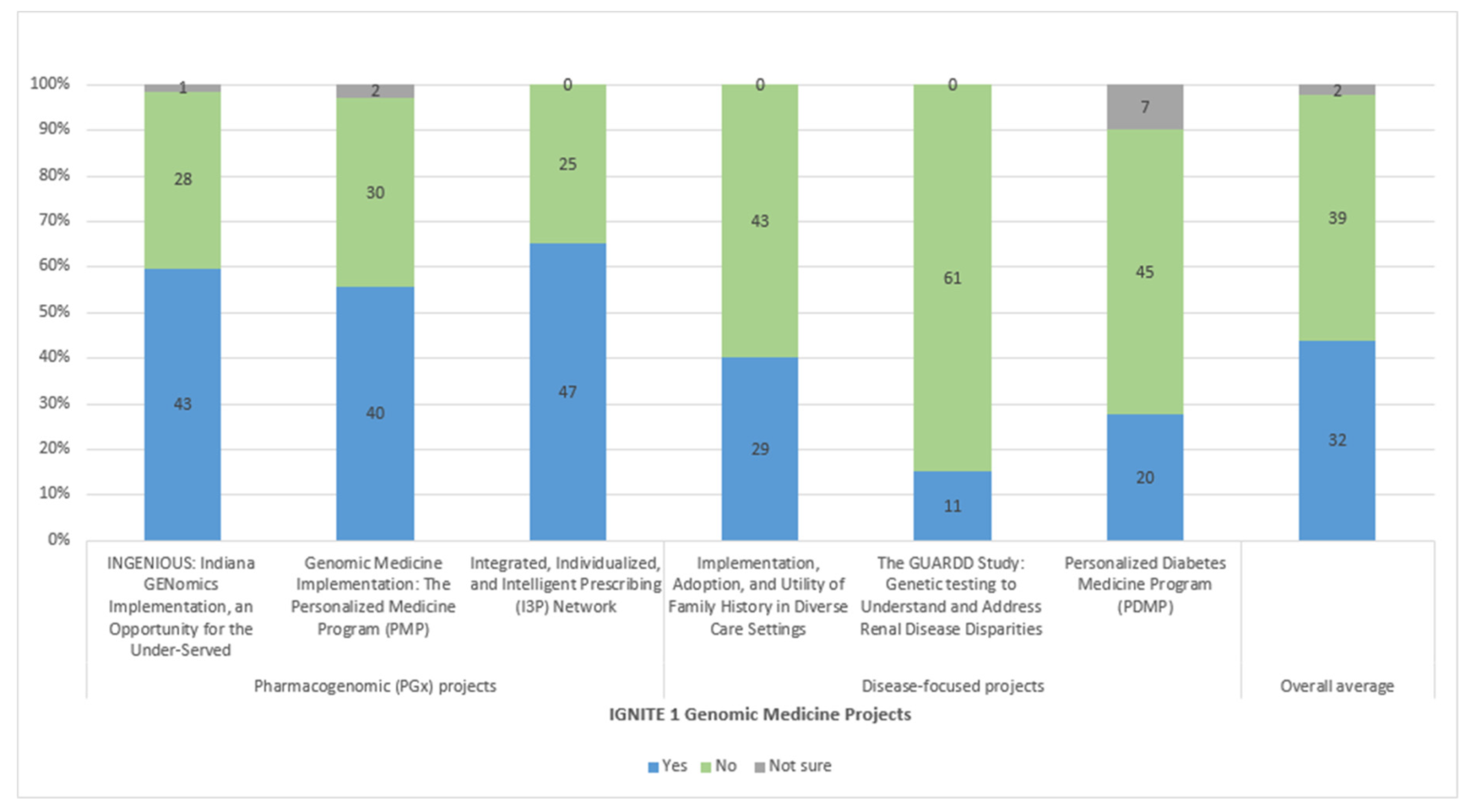
3. Results
3.1. Variety of Implementation Strategies Used across the Network
3.2. Common Implementation Strategies Found among Diverse Implementation Projects
3.2.1. Implementation Strategy 1: Obtaining and Using Stakeholder Feedback (e.g., from Patients, Families, or Providers) to Evaluate and Iteratively Develop the Genomic Program
3.2.2. Implementation Strategy 2: Identifying Early Adopters to Develop Stakeholder Interrelationships to Deliver the Genomic Program
3.2.3. Implementation Strategies 3 and 4: Conducting Educational Meetings and Having an Expert Meet with Clinicians to Train or Educate Providers to Deliver the Genomic Program
3.3. Implementation Outcomes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ginsburg, G.S.; Phillips, K.A. Precision Medicine: From Science to Value. Health Aff. Proj. Hope 2018, 37, 694–701. [Google Scholar] [CrossRef]
- Tcheng, J.E. Optimizing Strategies for Clinical Decision Support: Summary of a Meeting Series; National Academy of Medicine: Washington, DC, USA, 2017. [Google Scholar]
- Overby, C.L.; Kohane, I.; Kannry, J.L.; Williams, M.S.; Starren, J.; Bottinger, E.; Gottesman, O.; Denny, J.C.; Weng, C.; Tarczy-Hornoch, P.; et al. Opportunities for genomic clinical decision support interventions. Genet. Med. 2013, 15, 817–823. [Google Scholar] [CrossRef] [Green Version]
- Welch, B.M.; Kawamoto, K. Clinical decision support for genetically guided personalized medicine: A systematic review. J. Am. Med Inform. Assoc. 2012, 20, 388–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawamoto, K.; Lobach, D.F.; Willard, H.F.; Ginsburg, G.S. A national clinical decision support infrastructure to enable the widespread and consistent practice of genomic and personalized medicine. BMC Med. Inform. Decis. Mak. 2009, 9, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, M.C.; Clyne, M.; Kennedy, A.E.; Chambers, D.A.; Khoury, M.J. The current state of funded NIH grants in implementation science in genomic medicine: A portfolio analysis. Genet. Med. 2019, 21, 1218–1223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, C.G.; Peterson, S.; Khoury, M.J.; Brody, L.C.; McBride, C.M. A scoping review of social and behavioral science research to translate genomic discoveries into population health impact. Transl. Behav. Med. 2020, 11, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Sperber, N.R.; Carpenter, J.S.; Cavallari, L.H.; Damschroder, L.J.; Cooper-DeHoff, R.M.; Denny, J.C.; Ginsburg, G.S.; Guan, Y.; Horowitz, C.R.; Levy, K.D.; et al. Challenges and strategies for implementing genomic services in diverse settings: Experiences from the Implementing GeNomics In pracTicE (IGNITE) network. BMC Med. Genom. 2017, 10, 35. [Google Scholar] [CrossRef] [PubMed]
- Bauer, M.S.; Kirchner, J. Implementation science: What is it and why should I care? Psychiatry Res. 2020, 283, 112376. [Google Scholar] [CrossRef]
- Powell, B.; Waltz, T.; Chinman, M.; Damschroder, L.; Smith, J.; Matthieu, M.; Proctor, E.; Kirchner, J. A refined compilation of implementation strategies: Results from the Expert Recommendations for Implementing Change (ERIC) project. Implement. Sci. 2015, 10, 21. [Google Scholar] [CrossRef] [Green Version]
- Curran, G.M. Implementation science made too simple: A teaching tool. Implement. Sci. Commun. 2020, 1, 27. [Google Scholar] [CrossRef]
- Weitzel, K.W.; Alexander, M.; Bernhardt, B.A.; Calman, N.; Carey, D.J.; Cavallari, L.H.; Field, J.R.; Hauser, D.; Junkins, H.A.; Levin, P.A.; et al. The IGNITE network: A model for genomic medicine implementation and research. BMC Med. Genom. 2016, 9, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Owusu Obeng, A.; Fei, K.; Levy, K.D.; Elsey, A.R.; Pollin, T.I.; Ramirez, A.H.; Weitzel, K.W.; Horowitz, C.R. Physician-Reported Benefits and Barriers to Clinical Implementation of Genomic Medicine: A Multi-Site IGNITE-Network Survey. J. Pers. Med. 2018, 8, 24. [Google Scholar] [CrossRef] [Green Version]
- Levy, K.D.; Blake, K.; Fletcher-Hoppe, C.; Franciosi, J.; Goto, D.; Hicks, J.K.; Holmes, A.M.; Kanuri, S.H.; Madden, E.B.; Musty, M.D. Opportunities to implement a sustainable genomic medicine program: Lessons learned from the IGNITE Network. Genet. Med. 2019, 21, 743–747. [Google Scholar] [CrossRef] [Green Version]
- Damschroder, L.J. Clarity out of chaos: Use of theory in implementation research. Psychiatry Res. 2020, 283. [Google Scholar] [CrossRef] [PubMed]
- Glasgow, R.E.; Harden, S.M.; Gaglio, B.; Rabin, B.; Smith, M.L.; Porter, G.C.; Ory, M.G.; Estabrooks, P.A. RE-AIM Planning and Evaluation Framework: Adapting to New Science and Practice With a 20-Year Review. Front. Public Health 2019, 7. [Google Scholar] [CrossRef] [Green Version]
- Holtrop, J.S.; Rabin, B.A.; Glasgow, R.E. Qualitative approaches to use of the RE-AIM framework: Rationale and methods. BMC Health Serv. Res. 2018, 18, 177. [Google Scholar] [CrossRef]
- Wu, R.R.; Myers, R.A.; Sperber, N.; Voils, C.I.; Neuner, J.; McCarty, C.A.; Haller, I.V.; Harry, M.; Fulda, K.G.; Cross, D.; et al. Implementation, adoption, and utility of family health history risk assessment in diverse care settings: Evaluating implementation processes and impact with an implementation framework. Genet. Med. 2019, 21, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Proctor, E.; Powell, B.; McMillen, J. Implementation strategies: Recommendations for specifying and reporting. Implement Sci. 2013, 8, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sperber, N.R.; Andrews, S.M.; Voils, C.I.; Green, G.L.; Provenzale, D.; Knight, S. Barriers and Facilitators to Adoption of Genomic Services for Colorectal Care within the Veterans Health Administration. J. Pers. Med. 2016, 6, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rogal, S.S.; Yakovchenko, V.; Waltz, T.J.; Powell, B.J.; Gonzalez, R.; Park, A.; Chartier, M.; Ross, D.; Morgan, T.R.; Kirchner, J.E.; et al. Longitudinal assessment of the association between implementation strategy use and the uptake of hepatitis C treatment: Year 2. Implement. Sci. 2019, 14, 36. [Google Scholar] [CrossRef]
- Waltz, T.; Powell, B.; Matthieu, M.; Damschroder, L.; Chinman, M.; Smith, J.; Proctor, E.; Kirchner, J. Use of concept mapping to characterize relationships among implementation strategies and assess their feasibility and importance: Results from the Expert Recommendations for Implementing Change (ERIC) study. Implement. Sci. 2015, 10, 109. [Google Scholar] [CrossRef] [Green Version]
- Damschroder, L.; Aron, D.; Keith, R.; Kirsh, S.; Alexander, J.; Lowery, J. Fostering implementation of health services research findings into practice: A consolidated framework for advancing implementation science. Implement. Sci. 2009, 4, 50. [Google Scholar] [CrossRef] [Green Version]
- Horowitz, C.R.; Sabin, T.; Ramos, M.; Richardson, L.D.; Hauser, D.; Robinson, M.; Fei, K. Successful recruitment and retention of diverse participants in a genomics clinical trial: A good invitation to a great party. Genet. Med. 2019, 21, 2364–2370. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, B.; Caddle-Steele, C.; Chisholm, G.; Esmond, W.A.; Ferryman, K.; Gertner, M.; Goytia, C.; Hauser, D.; Richardson, L.D.; Robinson, M.; et al. A Culture of Understanding: Reflections and Suggestions from a Genomics Research Community Board. Prog. Community Health Partnersh. 2017, 11, 161–165. [Google Scholar] [CrossRef] [Green Version]
- Lewis, C.C.; Boyd, M.R.; Walsh-Bailey, C.; Lyon, A.R.; Beidas, R.; Mittman, B.; Aarons, G.A.; Weiner, B.J.; Chambers, D.A. A systematic review of empirical studies examining mechanisms of implementation in health. Implement. Sci. 2020, 15, 21. [Google Scholar] [CrossRef]
- Perry, C.K.; Damschroder, L.J.; Hemler, J.R.; Woodson, T.T.; Ono, S.S.; Cohen, D.J. Specifying and comparing implementation strategies across seven large implementation interventions: A practical application of theory. Implement. Sci. 2019, 14, 32. [Google Scholar] [CrossRef] [Green Version]
- Hockings, J.K.; Pasternak, A.L.; Erwin, A.L.; Mason, N.T.; Eng, C.; Hicks, J.K. Pharmacogenomics: An evolving clinical tool for precision medicine. Clevel. Clin. J. Med. 2020, 87, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Cavallari, L.H.; Weitzel, K.W.; Elsey, A.R.; Liu, X.; Mosley, S.A.; Smith, D.M.; Staley, B.J.; Winterstein, A.G.; Mathews, C.A.; Franchi, F.; et al. Institutional profile: University of Florida Health Personalized Medicine Program. Pharmacogenomics 2017, 18, 421–426. [Google Scholar] [CrossRef] [PubMed]
- Cavallari, L.H.; Van Driest, S.L.; Prows, C.A.; Bishop, J.R.; Limdi, N.A.; Pratt, V.M.; Ramsey, L.B.; Smith, D.M.; Tuteja, S.; Duong, B.Q.; et al. Multi-site investigation of strategies for the clinical implementation of CYP2D6 genotyping to guide drug prescribing. Genet. Med. 2019, 21, 2255–2263. [Google Scholar] [CrossRef]
- Arwood, M.J.; Chumnumwat, S.; Cavallari, L.H.; Nutescu, E.A.; Duarte, J.D. Implementing Pharmacogenomics at Your Institution: Establishment and Overcoming Implementation Challenges. Clin. Transl. Sci. 2016, 9, 233–245. [Google Scholar] [CrossRef] [PubMed]

| Project | Actor | Temporality | Action | Justification | Target |
|---|---|---|---|---|---|
| Who were the people actively involved and what were their roles? | What can you tell us about when the strategy was used? | What were the steps in using this strategy? | Please briefly describe the rationale for using it. | What were you trying to change? Were there multiple targets you were trying to change? | |
| INGENIOUS: Indiana Genomics Implementation, an Opportunity for the Underserved | Pharmacogenomics experts on the IGNITE team, lab experts, and providers involved with the projects provided feedback on validation of testing, returning results to providers, and clinical actions | Implementation | During team meetings, obtained feedback from lab experts | To unite key stakeholders | Implement an approach to use pharmacogenomics to guide 27 drug therapies |
| Genomic Medicine Implementation: The Personalized Medicine Program (PMP) | Principal investigators, project coordinator, and pharmacists sought feedback from providers/directors of Clinical Translational Science Instiute (CTSI) and pathology lab gave feedback | Pre-implementation and implementation | Informally asked for feedback during regular meetings with directors | To engage the appropriate stakeholder groups to ensure program success | Improve feasibility of ordering genetic test and patient clinical outcomes |
| Integrated, Individualized, and Intelligent Prescribing (I3P) Network | Lab operations, health IT, PGx experts, and clinical champions provided feedback on logistics on implementation | Pre-implementation and implementation | Discussed during regularly scheduled meetings | Pragmatic reason/prior experience | Implement an approach to use PGx in diverse clinical settings |
| Implementation, Adoption, and Utility of Family History in Diverse Care Settings | Genomics Expert Executive Board guided activities/study team tested family history program/Spanish speakers tested Spanish version | Pre-implementation and implementation | Met with clinics to assess implementation readiness and evaluated progress | To ensure the program worked at their site, and to understand how to address clinic barriers | Understand how to incorporate and adapt family history tool in healthcare systems |
| The GUARDD Study: Genetic testing to Understand and Address Renal Disease Disparities | Principal investigator reviewed study tools (recruitment scripts, informed consent) with stakeholder board | Implementation | Met with Stakeholder Board | To improve program success through using influence from similar target populations | Assess recruitment feasibility |
| Personalized Diabetes Medicine Program (PDMP) | Principal investigators obtained feedback from clinicans and patients | Pre-implementation and implementation | Included clinical champions in study; interacted with providers at staff meetings, in clinic, by email; informally asked patients in study and from advocacy groups | To obtain feedback to develop study protocol | Provider knowledge about candidates for genetic testing |
| Project | Actor | Temporality | Action | Justification | Target |
|---|---|---|---|---|---|
| Who were the people actively involved and what were their roles? | What can you tell us about when the strategy was used? | What were steps in using this strategy? | Please briefly describe the rationale for using it. | What were you trying to change? Were there multiple targets you were trying to change? | |
| INGENIOUS: Indiana Genomics Implementation, an Opportunity for the Underserved | Fellows analyzed data/clinicians supervised the fellows | Implementation | Analyzed genetic results, generated recommendations, sent reports to patients’ providers via the Electronic Health Record | To rely on clinicians familiar with recommendations and to have the ability to send EHR notes to providers | Policy change (e.g., generate evidence to get tests reimbursed and clincians to accept it) for 27 different drugs |
| Genomic Medicine Implementation: The Personalized Medicine Program (PMP) | Clinical champion with relevant experience (in chronic pain) helped implement project at multiple clinics by getting buy-in from medical directors, patients, and providers | Pre-implementation/implementation | Educated providers via lunch and learns, met with medical directors to discuss the project, enrolled patients, wrote study protocol | To use the site champion’s experience in chronic pain to get buy-in and program support from medical directors | Provider knowledge about and skill with using pharmacogenomics and using CYP2D6 information in their prescribing |
| Integrated, Individualized, and Intelligent Prescribing (I3P) Network | Clinical champions on the study team obtained buy-in from clinics to recruit patients | Pre-implementation/implementation | Clinical champions helped to educate providers and encourage adoption | Pragmatic and prior experience | Existing clinical processes |
| Implementation, Adoption, and Utility of Family History in Diverse Care Settings | Champions from each site enrolled patients and providers and solved issues in clinic | Pre-implementation/implementation | PI identified site champions who were then trained on the project | To allow site champions to highlight importance of program to other providers | Existing clinical processes |
| The GUARDD Study: Genetic testing to Understand and Address Renal Disease Disparities | Clinical champions on research team obtained buy-in from clinics to recruit patients | Pre-implementation | Clinical site champions presented to providers and answered questions (sometimes these providers became champions) | To make providers aware of the program and how to use program information in their practice | Provider knowledge about genetic testing and acceptability of study |
| Personalized Diabetes Medicine Program (PDMP) | Principal investigator and fellow championed the project in endocrinology clinics | Pre-implementation | Supported project with initial funding to develop various aspects of the projects, including the genetic test, and to educate providers to incorporate into clinic | To leverage the co-PI’s connections in the field and the fellow’s clinical expertise when incorporating the study into the clinic | Effective implementation by helping to develop logistics |
| Project | Actor | Temporality | Action | Justification | Target |
|---|---|---|---|---|---|
| Who were the people actively involved and what were their roles? | What can you tell us about when the strategy was used? | What were steps in using this strategy? | Please briefly describe the rationale for using it. | What were you trying to change? Were there multiple targets you were trying to change? | |
| INGENIOUS: Indiana Genomics Implementation, an Opportunity for the Underserved | Pharmacogenmics experts provided training to project clinicians who helped evaluate and return pharmacognomics results and recommendations to participants’ providers | Early implementation | Described project process at meetings/ involved clinicians in project design | To train the project clinicians to in turn consult with providers | Train multiple clinicians to make clinical recommendations |
| Genomic Medicine Implementation: The Personalized Medicine Program (PMP) | Principal investigator and fellow presented project to providers /pharmacist presented case studies to providers | Project presentation pre-implementation and case studies throughout. | Principal investigator and fellow tailored presentation based on medical director’s knowledge of their patients/pharmacist arranged meeting with providers to present case studies | To ensure providers were engaged and understood the program and how to integrate into workflow | Provider knowledge |
| Integrated, Individualized, and Intelligent Prescribing (I3P) Network | Clinical champions and subject matter experts presented to providers | Early implementation | Presented at exisiting meetings like morning report | Pragmatic | Provider knowledge of study and implementation of PGx testing to encourage buy-in |
| Implementation, Adoption, and Utility of Family History in Diverse Care Settings | Principal investigator, site champions, and project managers visited each site to discuss project and created training videos and informational packets for providers | Pre- and early implementation | Visited each site to provide them with base study protocol, although sites could revise as needed, and provided ongoing educational sessions as needed | To inform sites about the program and allow sites to adjust protocols as needed | Adapt protocol to fit each site/ provider knowledge about how to implement family history assessment |
| The GUARDD Study: Genetic testing to Understand and Address Renal Disease Disparities | Primary care providers (from the research team and sites) ran educational sessions with providers/specialists with relevant experience (nephrologists, geneticists) developed educational materials/genetic counselor trained site coordinator to return results | Pre-implementation | Asked for time at existing meetings | To ensure providers were trained and received training from peers with similar training backgrounds | Provider understanding of project and expectations |
| Personalized Diabetes Medicine Program (PDMP) | Principal investigator and study geneticists promoted the project among clinicians/external speakers provided seminars to clinicians | Early implementation | Conducted educational sessions as needed and expert seminars about project intermittently | To make sure providers understood the project, get their buy-in | Maximize provider uptake by increasing knowledge about monogenic types of diabetes, improving case identification, and in turn increasing clinic referrals |
| Genomic Medicine Project | Implementation Outcomes | Implementaiton Strategies | ||
|---|---|---|---|---|
| Reach 1 | Adoption 1 | Effectiveness 1 | ||
| Who actually was exposed to the service?/ Who is or was intended to benefit from your genomic service? | Where is or was the program applied and who applied it? | What is or was the most important benefit you are or were trying to achieve? Were there negative outcomes? | Summary and interpretation | |
| INGENIOUS: Indiana Genomics Implementation, an Opportunity for the Underserved | 1309/4380 patients newly prescribed one of 27 different drugs that have clinically actionable genetic variants associated with them; approximately 20% of patients would carry an actionable genetic variant and benefit from a change in their therapy | Indiana University Health (state-wide) and Eskenazi Health (county hospital) healthcare systems, delivered by mostly MDs of multiple disciplines | Improved efficacy and reduced side-effects of the drug therapies; no negative outcomes | Pharmacogenomics experts trained study team clinicians to make recommendations through the EHR to providers who had prescribed one of the drugs with actionable variants |
| Genomic Medicine Implementation: The Personalized Medicine Program (PMP) | >5000 patients from diverse backgrounds and settings/NA 2 | Implemented pharmacogenetic testing into clinical practice in 3 hospitals, 23 different clinics, including in academic medical centers, and primary and specialty care settings for 12 different patient populations | Use of genetic testing for drug prescribing (e.g., reduced cardiovascular adverse events); no negative outcomes | Employed relevant multidisciplinary expertise (e.g., clinical champions, pharmacogenomics, pathology, translational medicine) to not only develop project but also engage and educate primary care providers |
| Integrated, Individualized, and Intelligent Prescribing (I3P) Network | 25,777 across four diverse healthcare systems in Tennessee, North and South Dakota, and Wisconsin/NA 2 | VUMC, Advocate Aurora Health, Meharry Medical College, Sanford Health | Uptake in PGx testing and change in treatment; PGx recommendations were followed 50% to 80% of the time; one planned site could not implement PGx testing due to policy issues | Feedback on logistics from diverse stakeholders helped to plan around unique policy and implementation issues at sites, with many difficult to anticipate; providers were receptive to education and PGx recommendations |
| Implementation, Adoption, and Utility of Family History in Diverse Care Settings | 2514/172,160 primary care patients across 5 health systems | 28 primary care practices across 4 major healthcare delivery systems in the United States, generally delivered by primary care providers and other healthcare providers, such as nurses, as desired by sites | Feasiblity of implementing genetic risk testing across diverse settings; two negative outcomes: one site dropped out because of feasiblity issues and one clinic dropped out because of issues with time commitment | Conducted pre-implementation site assessments to support local adaptations across the 5 diverse healthcare systems while maintaining project fidelity |
| The GUARDD Study: Genetic testing to Understand and Address Renal Disease Disparities | 2052/7959 eligible adults identified through the EHR 3 | 15 primary care sites, some part of a large academic institution and others from a network of federal qualified health centers in different NYC neighborhoods, delivered by research team | Systolic blood pressure (SBP) decrease: greatest in APOL1-positive compared to APOL1-negative and control groups at 3 months; improved patient health outcomes through genetic testing and information provided to patients and and their providers | Used participatory research approach to inform study tools and educate providers about genetic risk/testing at all participating clinical sites |
| Personalized Diabetes Medicine Program | 2522 patients with diabetes or prediabetes screened in diabetes clinic waiting rooms or patient portal, referred by providers, or referred by patients themselves/NA 2 | 4 endocrinology clinics across 4 healthcare delivery systems, delivered by research team | Improve the identification and diagnosis of patients with monogenic diabetes to enable individualized treatment | Engaged experts in the specific therapeutic area, monogenic diabetes, as part of the study team, to develop protocol and educate providers on how to identify patients with the screening tool |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sperber, N.R.; Dong, O.M.; Roberts, M.C.; Dexter, P.; Elsey, A.R.; Ginsburg, G.S.; Horowitz, C.R.; Johnson, J.A.; Levy, K.D.; Ong, H.; et al. Strategies to Integrate Genomic Medicine into Clinical Care: Evidence from the IGNITE Network. J. Pers. Med. 2021, 11, 647. https://doi.org/10.3390/jpm11070647
Sperber NR, Dong OM, Roberts MC, Dexter P, Elsey AR, Ginsburg GS, Horowitz CR, Johnson JA, Levy KD, Ong H, et al. Strategies to Integrate Genomic Medicine into Clinical Care: Evidence from the IGNITE Network. Journal of Personalized Medicine. 2021; 11(7):647. https://doi.org/10.3390/jpm11070647
Chicago/Turabian StyleSperber, Nina R., Olivia M. Dong, Megan C. Roberts, Paul Dexter, Amanda R. Elsey, Geoffrey S. Ginsburg, Carol R. Horowitz, Julie A. Johnson, Kenneth D. Levy, Henry Ong, and et al. 2021. "Strategies to Integrate Genomic Medicine into Clinical Care: Evidence from the IGNITE Network" Journal of Personalized Medicine 11, no. 7: 647. https://doi.org/10.3390/jpm11070647
APA StyleSperber, N. R., Dong, O. M., Roberts, M. C., Dexter, P., Elsey, A. R., Ginsburg, G. S., Horowitz, C. R., Johnson, J. A., Levy, K. D., Ong, H., Peterson, J. F., Pollin, T. I., Rakhra-Burris, T., Ramos, M. A., Skaar, T., & Orlando, L. A. (2021). Strategies to Integrate Genomic Medicine into Clinical Care: Evidence from the IGNITE Network. Journal of Personalized Medicine, 11(7), 647. https://doi.org/10.3390/jpm11070647







