Let-7a-5p, miR-100-5p, miR-101-3p, and miR-199a-3p Hyperexpression as Potential Predictive Biomarkers in Early Breast Cancer Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients’ Characteristics and Tumor Specimen Collection
2.2. Purification of miRNA from Paraffin-Embedding Tissue Sections
2.3. MiRNA Discovery
2.3.1. Analysis Procedure
2.3.2. MiRNA Target Prediction
2.4. Statistical Analysis
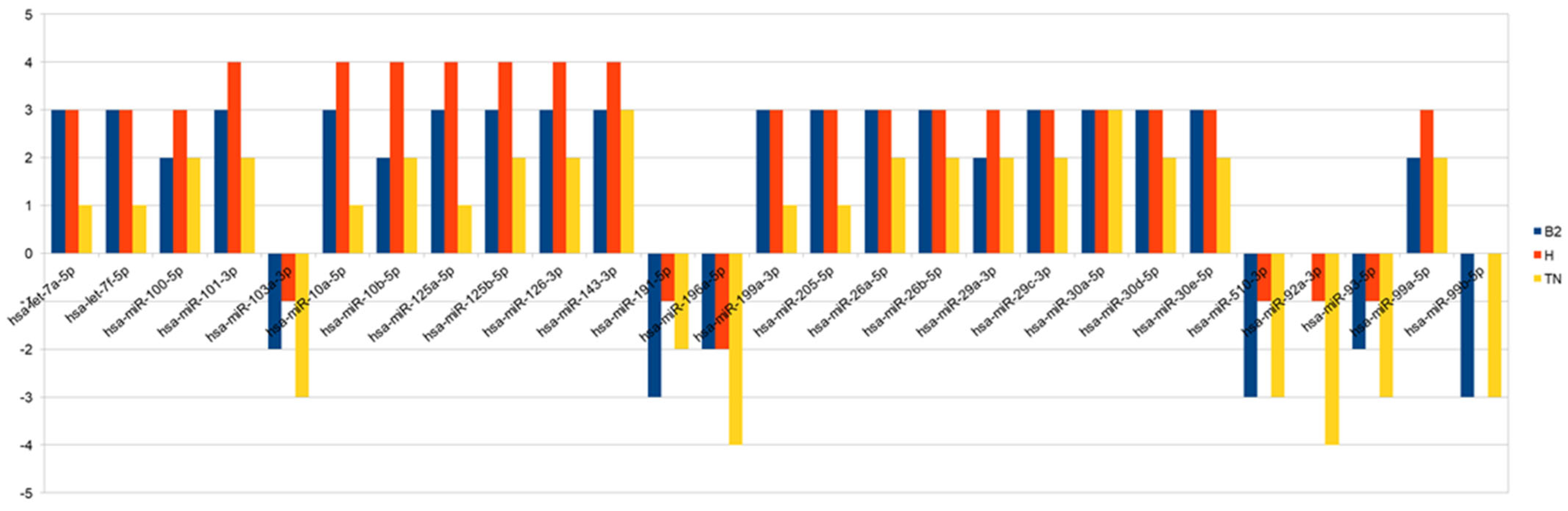
3. Results
3.1. Patients Charachteristics
3.2. Clinicopathological Variables and Outcome
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Mauri, D.; Pavlidis, N.; Ioannidis, J.P. Neoadjuvant versus adjuvant systemic treatment in breast cancer: A meta-analysis. J. Natl. Cancer Inst. 2005, 97, 188–194. [Google Scholar] [CrossRef] [Green Version]
- van der Hage, J.A.; van de Velde, C.J.; Julien, J.P.; Tubiana-Hulin, M.; Vandervelden, C.; Duchateau, L. Preoperative chemotherapy in primary operable breast cancer: Results from the European Organization for Research and Treatment of Cancer trial 10902. J. Clin. Oncol. 2001, 19, 4224. [Google Scholar] [CrossRef]
- Rastogi, P.; Anderson, S.J.; Bear, H.D.; Geyer, C.E.; Kahlenberg, M.S.; Robidoux, A.; Margolese, R.G.; Hoehn, J.L.; Vogel, V.G.; Dakhil, S.R.; et al. Preoperative chemotherapy: Updates of National Surgical Adjuvant Breast and Bowel Project Protocols B-18 and B-27. J. Clin. Oncol. 2008, 26, 778–785. [Google Scholar] [CrossRef] [Green Version]
- Kuerer, H.M.; Newman, L.A.; Smith, T.L.; Ames, F.C.; Hunt, K.K.; Dhingra, K.; Theriault, R.L.; Singh, G.; Binkley, S.M.; Sneige, N.; et al. Clinical course of breast cancer patients with complete pathologic primary tumor and axillary lymph node response to doxorubicin-based neoadjuvant chemotherapy. J. Clin. Oncol. 1999, 17, 460–469. [Google Scholar] [CrossRef]
- von Minckwitz, G.; Untch, M.; Blohmer, J.U.; Costa, S.D.; Eidtmann, H.; Fasching, P.A.; Gerber, B.; Eiermann, W.; Hilfrich, J.; Huober, J.; et al. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J. Clin. Oncol. 2012, 30, 1796–1804. [Google Scholar] [CrossRef] [Green Version]
- von Minckwitz, G.; Untch, M.; Nüesch, E.; Loibl, S.; Kaufmann, M.; Kümmel, S.; Fasching, P.A.; Eiermann, W.; Blohmer, J.U.; Costa, S.D.; et al. Impact of treatment characteristics on response of different breast cancer phenotypes: Pooled analysis of the German neo-adjuvant chemotherapy trials. Breast Cancer Res. Treat. 2011, 125, 145–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gampenrieder, S.P.; Rinnerthaler, G.; Greil, R. Neoadjuvant chemotherapy and targeted therapy in breast cancer: Past, present, and future. J. Oncol. 2013, 732047. [Google Scholar] [CrossRef] [PubMed]
- Bear, H.D.; Anderson, S.; Brown, A.; Smith, R.; Mamounas, E.P.; Fisher, B.; Margolese, R.; Theoret, H.; Soran, A.; Wickerham, D.L.; et al. The effect on tumor response of adding sequential preoperative docetaxel to preoperative doxorubicin and cyclophosphamide: Preliminary results from National Surgical Adjuvant Breast and Bowel Project Protocol B-27. J. Clin. Oncol. 2003, 21, 4165–4174. [Google Scholar] [CrossRef]
- Sataloff, D.M.; Mason, B.A.; Prestipino, A.J.; Seinige, U.L.; Lieber, C.P.; Baloch, Z. Pathologic response to induction chemotherapy in locally advanced carcinoma of the breast: A determinant of outcome. J. Am. Coll. Surg. 1995, 180, 297–306. [Google Scholar]
- Berezikov, E. Evolution of microRNA diversity and regulation in animals. Nat. Rev. Genet. 2011, 12, 846–860. [Google Scholar] [CrossRef]
- Weber, B.; Stresemann, C.; Brueckner, B.; Lyko, F. Methylation of human microRNA genes in normal and neoplastic cells. Cell Cycle 2007, 6, 1001–1005. [Google Scholar] [CrossRef]
- Calin, G.A.; Sevignani, C.; Dumitru, C.D.; Hyslop, T.; Noch, E.; Yendamuri, S.; Shimizu, M.; Rattan, S.; Bullrich, F.; Negrini, M.; et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA 2004, 101, 2999–3004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blenkiron, C.; Goldstein, L.D.; Thorne, N.P.; Spiteri, I.; Chin, S.-F.; Dunning, M.J.; Barbosa-Morais, N.L.; Teschendorff, A.E.; Green, A.R.; Ellis, I.O.; et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007, 8, R214. [Google Scholar] [CrossRef] [Green Version]
- Amin, M.B.; Greene, F.L.; Edge, S.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P.; et al. Cancer Staging Manual, 8th ed.; Springer: New York, NY, USA, 2017. [Google Scholar]
- But-Hadzić, J.; Bilban-Jakopin, C.; Hadzić, V. The role of radiation therapy in locally advanced breast cancer. Breast J. 2010, 16, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Green, M.C.; Buzdar, A.U.; Smith, T.; Ibrahim, N.K.; Valero, V.; Rosales, M.F.; Cristofanilli, M.; Booser, D.J.; Pusztai, L.; Rivera, E.; et al. Weekly paclitaxel improves pathologic complete remission in operable breast cancer when compared with paclitaxel once every 3 weeks. J. Clin. Oncol. 2005, 23, 5983–5992. [Google Scholar] [CrossRef] [PubMed]
- Baselga, J.; Bradbury, I.; Eidtmann, H.; Di Cosimo, S.; Aura, C.; De Azambuja, E.; Gomez, H.; Dinh, P.; Fauria, K.; Van Dooren, V.; et al. Abstract S3-3: First results of the NeoALTTO trial (BIG 01-06/EGF106903): A phase III, randomized, open label, neoadjuvant study of lapatinib, trastuzumab, and their combination plus paclitaxel in women with HER2-positive primary breast cancer. Cancer Res. 2010, 70. [Google Scholar] [CrossRef]
- miRBase: The microRNA Database. Available online: http://www.mirbase.org/ (accessed on 1 August 2021).
- Li, L.; Xu, J.; Yang, D.; Tan, X.; Wang, H. Computational approaches for microRNA studies. Mamm. Genome 2010, 21, 1–12. [Google Scholar] [CrossRef]
- miRDB. Available online: http://www.mirdb.org (accessed on 1 August 2021).
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal. Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [Green Version]
- Bertoli, G.; Cava, C.; Castiglioni, I. MicroRNAs: New Biomarkers for Diagnosis, Prognosis, Therapy Prediction and Therapeutic Tools for Breast Cancer. Theranostics 2015, 5, 1122–1143. [Google Scholar] [CrossRef]
- Bussing, I.; Slack, F.J.; Grosshans, H. let-7 microRNAs in development, stem cells and cancer. Trends Mol. Med. 2008, 14, 400–409. [Google Scholar] [CrossRef]
- Worringer, K.A.; Rand, T.A.; Hayashi, Y.; Sami, S.; Takahashi, K.; Tanabe, K.; Narita, M.; Srivastava, D.; Yamanaka, S. The let-7/LIN-41 pathway regulates reprogramming to human induced pluripotent stem cells by controlling expression of prodifferentiation genes. Cell Stem Cell. 2014, 14, 40–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akao, Y.; Nakagawa, Y.; Naoe, T. Let-7 microrna functions as a potential growth suppressor in human colon cancer cells. Biol. Pharm. Bull. 2006, 29, 903–906. [Google Scholar] [CrossRef] [Green Version]
- Johnson, S.M.; Grosshans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K.L.; Brown, D.; Slack, F.J. Ras is regulated by the let-7 microrna family. Cell 2005, 120, 635–647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, F.; Yao, H.; Zhu, P.; Zhang, X.; Pan, Q.; Gong, C.; Huang, Y.; Hu, X.; Su, F.; Lieberman, J.; et al. Let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 2007, 131, 1109–1123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barh, D.; Parida, S.; Parida, B.P. Let-7, mir -125, mir -205, and mir -296 are prospective therapeutic agents in breast cancer molecular medicine. Gene Ther. Mol. Biol. 2008, 12, 189–206. [Google Scholar]
- Seok-Jun, K.; Ji-Young, S.; Kang-Duck, L.; Bae, Y.-K.; Sung, K.W.; Nam, S.J.; Chun, K.H. MicroRNA let-7a suppresses breast cancer cell migration and invasion through downregulation of C-C chemokine receptor type 7. Breast Cancer Res. 2012, 14, R14. [Google Scholar] [CrossRef] [Green Version]
- Serguienk, A.; Grad, I.; Wennerstrøm, A.B.; Meza-Zepeda, L.A.; Thiede, B.; Stratford, E.W.; Myklebost, O.; Munthe, E. Metabolic reprogramming of metastatic breast cancer and melanoma by let-7a microRNA. Oncotarget 2015, 6, 2451–2465. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Yanping, G.; Kai, Z.; Chen, J.; Han, S.; Feng, B.; Wang, R.; Chen, L. Multiple Roles of MicroRNA-100 in Human Cancer and its Therapeutic Potential. Cell Physiol. Biochem. 2015, 37, 2143–2159. [Google Scholar] [CrossRef]
- Petrelli, A.; Carollo, R.; Cargnelutti, M.; Iovino, F.; Callari, M.; Cimino, D.; Todaro, M.; Mangiapane, L.R.; Giammona, A.; Cordova, A.; et al. By promoting cell differentiation, miR-100 sensitizes basal-like breast cancer stem cells to hormonal therapy. Oncotarget 2015, 6, 2315–2330. [Google Scholar] [CrossRef] [Green Version]
- Lei, Y.; Li, B.; Tong, S.; Qi, L.; Hu, X.; Cui, Y.; Li, Z.; He, W.; Zu, X.; Wang, Z.; et al. miR-101 suppresses vascular endothelial growth factor C that inhibits migration and invasion and enhances cisplatin chemosensitivity of bladder cancer cells. PLoS ONE 2015, 10, e0117809. [Google Scholar] [CrossRef]
- Ye, Z.; Yin, S.; Su, Z.; Bai, M.; Zhang, H.; Hei, Z.; Cai, S. Downregulation of miR-101 contributes to epithelial-mesenchymal transition in cisplatin resistance of NSCLC cells by targeting ROCK2. Oncotarget 2016, 7, 37524–37535. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Zhang, T.; Liu, H.; Lv, T.; Yuan, D.; Yao, Y.; Lv, Y.; Song, Y. MiR-101 and Mcl-1 in non-small cell lung cancer: Expression profile and clinical significance. Med. Oncol. 2012, 29, 1681–1686. [Google Scholar] [CrossRef] [PubMed]
- Li, J.T.; Jia, L.T.; Liu, N.N.; Zhu, X.-S.; Liu, Q.-Q.; Wang, X.-L.; Yu, F.; Liu, Y.-L.; Yang, A.-G.; Gao, C.-F. MiRNA-101 inhibits breast cancer growth and metastasis by targeting CX chemokine receptor 7. Oncotarget 2015, 6, 30818–30830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, F.; Liao, Y.J.; Cai, M.Y.; Liu, T.-H.; Chen, S.-P.; Wu, P.-H.; Wu, L.; Bian, X.-W.; Guan, X.-Y.; Zeng, Y.-X.; et al. Systemic delivery of microRNA-101 potently inhibits hepatocellular carcinoma in vivo by repressing multiple targets. PLoS Genet. 2015, 11, e1004873. [Google Scholar] [CrossRef] [Green Version]
- Slattery, M.L.; Herrick, J.S.; Pellatt, D.F.; Mullany, L.E.; Stevens, J.R.; Wolff, E.; Hoffman, M.D.; Wolff, R.K.; Samowitz, W. Site-specific associations between miRNA expression and survival in colorectal cancer cases. Oncotarget 2016, 7, 60193–60205. [Google Scholar] [CrossRef] [Green Version]
- Varambally, S.; Cao, Q.; Mani, R.S.; Shankar, S.; Wang, X.; Ateeq, B.; Laxman, B.; Cao, X.; Jing, X.; Ramnarayanan, K.; et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 2008, 322, 1695–1699. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.G.; Guo, J.F.; Liu, D.L.; Liu, Q.; Wang, J.-J. MicroRNA-101 exerts tumorsuppressive functions in non-small cell lung cancer through directly targeting enhancer of Zeste homolog 2. J. Thorac. Oncol. 2011, 6, 671–678. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Wu, C.; Zhao, X.; Liu, C. The prognostic value of decreased miR-101 in various cancers: A meta-analysis of 12 studies. Onco Targets Ther. 2017, 10, 3709–3718. [Google Scholar] [CrossRef] [Green Version]
- Xiaoping, L.; Tang, H.; Chen, J.; Song, C.; Yang, L.; Liu, P.; Wang, N.; Xie, X.; Lin, X.; Xie, X. MicroRNA-101 inhibits cell progression and increases paclitaxel sensitivity by suppressing MCL-1 expression in human triple-negative breast cancer. Oncotarget 2015, 6, 20070–20083. [Google Scholar] [CrossRef]
- Rui, W.; Hong-Bin, W.; Chan, J.H.; Cui, Y.; Han, X.-C.; Hu, Y.; Li, F.-F.; Xia, H.-F. Ma XMiR-101 Is Involved in Human Breast Carcinogenesis by Targeting Stathmin. PLoS ONE 2012, 7, e46173. [Google Scholar] [CrossRef]
- Hou, J.; Lin, L.; Zhou, W.; Wang, Z.; Ding, G.; Dong, Q.; Qin, L.; Wu, X.; Zheng, Y.; Yang, Y.; et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell 2011, 19, 232–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, Q.; Wang, X.; Gong, W.; Li, N.; Chen, C.; He, X.; Chen, F.; Yang, L.; Wang, P.; Wang, D.W. ER stress negatively modulates the expression of the miR-199a/214 cluster to regulates tumor survival and progression in human hepatocellular cancer. PLoS ONE 2012, 7, e31518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Ting, Z.; Li, Y.; Chen, G.; Lu, Y.; Hao, X. microRNA-199a is able to reverse cisplatin resistance in human ovarian cancer cells through the inhibition of mammalian target of rapamycin. Oncol. Lett. 2013, 6, 789–794. [Google Scholar] [CrossRef] [Green Version]
- Tsukigi, M.; Bilim, V.; Yuuki, K.; Ugolkov, A.; Naito, S.; Nagaoka, A.; Kato, T.; Motoyama, T.; Tomita, Y. Re-expression of miR-199a suppresses renal cancer cell proliferation and survival by targeting GSK-3beta. Cancer Lett. 2012, 315, 189–197. [Google Scholar] [CrossRef]
- Duan, Z.; Choy, E.; Harmon, D.; Liu, X.; Susa, M.; Mankin, H.; Hornicek, F. MicroRNA199a-3p is downregulated in human osteosarcoma and regulates cell proliferation and migration. Mol. Cancer Ther. 2011, 10, 1337–1345. [Google Scholar] [CrossRef] [Green Version]
- Tian, Y.; Zhang, Y.Z.; Chen, W. MicroRNA-199a-3p and microRNA-34a regulate apoptosis in human osteosarcoma cells. Biosci. Rep. 2014, 34, e00132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minna, E.; Romeo, P.; De Cecco, L.; Dugo, M.; Cassinelli, G.; Pilotti, S.; Degl’Innocenti, D.; Lanzi, C.; Casalini, P.; Pierotti, M.A.; et al. miR-199a-3p displays tumor suppressor functions in papillary thyroid carcinoma. Oncotarget 2014, 5, 2513–2528. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; Huang, H.J.; He, C.N.; Wang, K.-Y. MicroRNA-199a-3p regulates endometrial cancer cell proliferation by targeting mammalian target of rapamycin (mTOR). Int. J. Gynecol. Cancer 2013, 23, 1191–1197. [Google Scholar] [CrossRef] [PubMed]
- Li, S.Q.; Wang, Z.H.; Mi, X.G.; Liu, L.; Tan, Y. MiR-199a/b-3p suppresses migration and invasion of breast cancer cells by downregulating PAK4/MEK/ERK signaling pathway. IUBMB Life 2015, 67, 768–777. [Google Scholar] [CrossRef]
- Xuelong, F.; Shangcheng, Z.; Miao, Z.; Deng, X.; Yi, Y.; Huang, T. MiR-199a-3p enhances breast cancer cell sensitivity to cisplatin by downregulating TFAM (TFAM). Biomed. Pharmacother. 2017, 88, 507–514. [Google Scholar] [CrossRef]
- Ruiyan, H.; Junbai, L.; Feng, P.; Zhang, B.; Yao, Y. The activation of GPER inhibits cells proliferation, invasion and EMT of triple-negative breast cancer via CD151/miR-199a-3p bio-axis. Transl Res. 2020, 12, 32–44. [Google Scholar]
- Krützfeldt, J.; Rajewsky, N.; Braich, R.; Rajeev, K.G.; Tuschl, T.; Manoharan, M.; Stoffel, M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005, 438, 685–689. [Google Scholar] [CrossRef] [PubMed]
- Weiler, J.; Hunziker, J.; Hall, J. Anti-miRNA oligonucleotides (AMOs): Ammunition to target miRNAs implicated in human disease? Gene Ther. 2006, 13, 496–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433, 769–773. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Yao, Y.; Lin, B.; Lin, L.; Yang, M.; Zhang, W.; Chen, W.; Pan, C.; Liu, Q.; Song, E.; et al. MiR-200b and miR-15b regulate chemotherapy-induced epithelial-mesenchymal transition in human tongue cancer cells by targeting BMI1. Oncogene 2012, 31, 432–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, M.; Liaw, C.S.; Ji, S.M.; Tan, H.H.; Wong, C.Y.; Thike, A.A.; Tan, P.H.; Ho, G.H.; Lee, A.S.-G. Identification of circulating microRNA signatures for breast cancer detection. Clin Cancer Res. 2013. [Google Scholar] [CrossRef] [Green Version]

| Characteristics | N | % |
|---|---|---|
| Demographic and clinical | ||
| Age | ||
| Mean (SD) (years) | 49.4 ± 10.4 | |
| ≤40 | 69 | 34.5 |
| >40 | 131 | 65.5 |
| ER status | ||
| Positive | 130 | 65.0 |
| Negative | 55 | 27.5 |
| Unknown | 15 | 7.5 |
| PR status | ||
| Positive | 128 | 64.0 |
| Negative | 58 | 29.0 |
| Unknown | 14 | 7.0 |
| HER2 status | ||
| Positive | 58 | 29.0 |
| Negative | 127 | 63.5 |
| Unknown | 15 | 7.5 |
| Subtype | ||
| Luminal A | 46 | 23.0 |
| Luminal B/HER2-negative | 48 | 24.0 |
| Luminal B/HER2-positive | 37 | 18.5 |
| HER2-positive (non-luminal) | 21 | 10.5 |
| Triple negative | 33 | 16.5 |
| Unknown | 15 | 7.5 |
| Ki 67 | ||
| ≤20% | 59 | 29.5 |
| >20% | 120 | 60.0 |
| Unknown | 21 | 10.5 |
| Grade | ||
| 1 | 0 | 0.0 |
| 2 | 51 | 25.5 |
| 3 | 84 | 42.0 |
| Unknown | 65 | 32.5 |
| 2 Histologic type | ||
| Lobular | 18 | 9.0 |
| Ductal | 150 | 75.0 |
| Other | 28 | 14.0 |
| Unknown | 4 | 2.0 |
| 3Tumor characteristics before treatment | ||
| cT stage | ||
| cTx | 1 | 0.5 |
| cT1 | 11 | 5.5 |
| cT2 | 71 | 35.5 |
| cT3 | 55 | 27.5 |
| cT4 | 48 | 24.0 |
| Unknown | 14 | 7.0 |
| cN stage | ||
| cN0 | 33 | 16.5 |
| cN1 | 106 | 53.0 |
| cN2 | 36 | 18.0 |
| cN3 | 9 | 4.5 |
| Unknown | 16 | 8.0 |
| Clinical AJCC stage | ||
| 0 | 0 | 0.0 |
| I | 1 | 0.5 |
| II | 76 | 38.0 |
| III | 105 | 52.5 |
| IV | 1 | 0.5 |
| Unknown | 17 | 8.5 |
| Treatment | ||
| Neoadjuvant | ||
| TAC | 138 | 69.0 |
| TCH | 54 | 27.0 |
| Other | 8 | 4.0 |
| Adjuvant hormone | ||
| Yes | 130 | 65.0 |
| No | 54 | 27.0 |
| Unknown | 16 | 8.0 |
| Tumor pathology after neoadjuvant treatment | ||
| yT stage | ||
| yT0/is | 42 | 21.0 |
| yT1 | 75 | 37.5 |
| yT2 | 37 | 18.5 |
| yT3 | 13 | 6.5 |
| yT4 | 7 | 3.5 |
| Unknown | 26 | 13.0 |
| yN stage | ||
| yN0 | 83 | 41.5 |
| yN1 | 62 | 31.0 |
| yN2 | 18 | 9.0 |
| yN3 | 12 | 6.0 |
| Unknown | 25 | 12.5 |
| Pathologic yAJCC stage | ||
| 0 | 34 | 17.0 |
| I | 46 | 23.0 |
| II | 55 | 27.5 |
| III | 37 | 18.5 |
| IV | 1 | 0.5 |
| Unknown | 27 | 13.0 |
| Treatment outcomes | ||
| Response to neoadjuvant treatment | ||
| Complete response (R0) | 44 | 22.0 |
| Microscopic residual disease (R1) | 53 | 26.5 |
| Macroscopic residual disease (R2) | 101 | 50.5 |
| Unknown | 2 | 1.0 |
| Events within 3 years | ||
| Distant relapse | 37 | 18.5 |
| Local recurrence | 11 | 5.5 |
| Death | 12 | 6.0 |
| Unknown | 18 | 9.0 |
| Median follow-up, months | 80 | |
| Patients | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age | 42 | 54 | 46 | 40 | 54 | 46 | 45 | 53 | 41 | 68 | 35 | 72 |
| Hystological type | IC | DIC | DIC | DIC | IC | DIC | IC | DIC | IC | DIC | DIC | IC |
| Grade | 3 | 3 | 3 | 3 | 2 | 3 | 2 | |||||
| cKi67 | 65 | 40 | 80 | 80 | 45 | 16 | 60 | 30 | 80 | 80 | 45 | 15 |
| Receptor subtype | B2 | B2 | B2 | B2 | B2 | B2 | B2 | B2 | TN | TN | TN | TN |
| cTMN | cT1N1 | cT2N1 | cT4N1 | cT3N1 | cT3N1 | cT4N1 | cT2N1 | cT2N1 | cT2N1 | cT2N2 | cT2N1 | cT2cN1 |
| Preoperative staging | IIA | IIB | IIIB | IIIA | IIIA | IIIB | IIB | IIB | IIB | IIIA | IIB | IIB |
| Pathological response | R1 | R2 | R2 | R2 | R2 | R2 | R2 | R1 | R1 | R2 | R2 | R2 |
| yKi67 | 70 | 80 | 70 | 45 | 60 | 3 | ||||||
| ypTNM | ypT1N0 | ypT1N1 | ypT2N1 | ypT1N1 | ypT2N1 | ypT2N0 | ypT1N0 | ypT1N0 | ypT1N0 | ypT1N1 | ypT1N1 | ypT1N1 |
| ySTADIO | IA | IIA | IIB | IIA | IIB | IA | IA | IA | IA | IIA | IIA | IIA |
| NAC | TCH | TCH | TCH | TCH | TCH | TCH | TCH | TCH | TAC | TAC | TAC | TC |
| Type of surgery | Q + L | M + L | M + L | Q + L | M + L | M + L | Q + L | M + L | M + L | M + L | Q + L | M + L |
| ADJUVANT CHT | H | H | H | H | H | H | H | H | 0 | 0 | 0 | 0 |
| ET | X | X | X | X | X | X | X | X | ||||
| RT | X | X | X | X |
| Patients | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age | 60 | 52 | 57 | 38 | 67 | 44 | 47 | 40 | 36 | 55 | 57 | 57 |
| Hystological type | DIC | DIC | IC | DIC | DIC | IC | DIC | DIC | DIC | DIC | DIC | DIC |
| Grade | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | |
| cKi67 | 45 | 45 | 90 | 80 | 35 | 35 | 60 | 35 | 40 | 15 | 45 | |
| Receptor subtype | TN | TN | TN | TN | H | H | H | H | H | H | H | H |
| cTMN | cT2N1 | cT1N2 | cT2N1 | cT2N0 | cT2N1 | cT2N1 | cT4N1 | cT4N1 | cT3N1 | cT3N2 | cT4N0 | cT2N0 |
| Preoperative staging | IIB | IIIA | IIB | IIA | IIB | IIB | IIIB | IIIB | IIIA | IIIA | IIIB | IIA |
| Pathological response | R2 | R1 | R2 | R2 | R1 | R2 | R1 | R2 | R2 | R1 | R1 | R1 |
| yKi67 | 85 | 6 | 80 | 70 | 70 | 4 | 2 | 40 | 45 | |||
| ypTNM | ypT2N0 | ypT1N0 | ypT1N1 | ypT1N0 | ypT1N1 | ypT3N0 | ypT1N0 | ypT1N1 | ypT2N1 | ypT1N0 | ypT1N0 | ypT1N0 |
| Pathological staging | IIA | IA | IIA | IA | IIA | IIIA | IA | IIA | IIB | IA | IA | IA |
| NAC | TAC | TAC | TAC | AC-T | TCH | TCH | TCH | TCH | TCH | TCH | TCH | TCH |
| Type of surgery | M + L | Q + L | M + L | M + L | M + L | M + L | M + L | M + L | M + L | M + L | M + L | M + L |
| ADJUVANT CHT | CMF | AC | H | H | H | H | H | H | H | |||
| ET | ||||||||||||
| RT | X | X | X | X | X |
| miRNAs | Gene Target Predicted |
|---|---|
| Let 7a-5p | SMARCAD1 FAM178A LIN28B GATM LRIG3 GNPTAB BZW1 ZNF322 ADAMTS8 C8orf58 ADRB2 DNA2 IGDCC3 TTLL4 NME6 TMPRSS2 HIC2 MAPK6 DMD SCN4B ZFYVE26 FZD3 LIMD2 SMIM3 TMEM2 PCGF3 COL3A1 ZBTB5 ACVR1C EIF4G2 CLP1 SLC25A27 NPHP3 PRTG B3GNT7 COIL CCNJ IGF2BP3 FOXP2 TRIM71 PARD6B FRAS1 MAP4K3 HAND1 UTRN GNG5 NAP1L1 UHRF2 LRIG2 ACER2 RICTOR PRPF38B NR6A1 BEGAIN NHLRC3 IFI44L E2F5 BACH1 PAPPA STK40 SLC5A9 PDP2 RDX THRSP FIGN ZBP1 IGF1R ERCC6 C5orf51 PBX3 RNF20 TGFBR1 C15orf41 ADAMTS15 TSEN34 C14orf28 FIGNL2 ZNF275 CPEB1 ARHGAP28 EDN1 C15orf39 USP38 E2F6 FNDC3A ARG2 SPRYD4 IGDCC4 HIP1 SLC10A7 KCTD21 NDST2 DDI2 TRIM41 SLC20A1 DPP6 PLXNC1 LIPT2 CPA4 FBXL12 PALD1 EEA1 HMGA1 RAB11FIP4 STX3 CEP135 GDF6 TRIM67 SLC5A6 OSBPL3 PLEKHG6 TMEM110 DDX26B PLAGL2 PGRMC1 CLDN12 HMGA2 TOR1AIP2 CLASP2 DDX19A KMT2D RPUSD3 ZNF583 AKAP6 SMAP2 RGS16 TAF9B ESR2 CHD4 MFSD4 PPP1R15B TARBP2 CRTAM ATP8B4 FRMD4B HIF1AN RPUSD2 PARS2 CEP120 USP32 GCNT4 GALNT1 SMC1A NRAS PRRX1 MXD1 TMOD2 RANBP2 KLHL31 FNIP1 ULK2 YOD1 ZSWIM5 FAM104A RALB GLRX APBB3 SLC17A9 DCLRE1B USP24 GALC SERF2 PXT1 CCL7 RRM2 TMEM167A RFX5 TMPPE C9orf40 PPAPDC1B PLEKHA8 SCD AHCTF1 RSPO2 PBX1 ZNF318 ZBTB8B ZNF512B GPR26 SLC2A12 ZNF362 AP1S1 SIGLEC14 RASGRP1 DLST TGFBR3 NGF MTUS1 ZNF10 MED8 GAB2 ESPL1 AMT CRY2 NYNRIN ABCG4 KIAA0930 IQCB1 KLHL23 KLHDC8B COL24A1 GAS7 XKR8 NAA30 ADRB3 ARRDC4 CBX5 CADM2 DDX19B OPA3 RIOK3 TET3 FGD6 SEMA3F GXYLT1 LBR COL4A6 BIN3 CDC34 CLDN1 SNX16 RAB3GAP2 FZD4 MAPK8 PPAPDC2 NNT SDR42E1 RNF5 LOC101930255 LOC102723960 PDPR SEMA4F SLC25A18 GFM2 CASP3 BZW2 CCR7 AGPAT6 THAP9 GRPEL2 MEIS3 CGNL1 ZNF644 SKIL NXT2 TXNDC8 PARP16 LGR4 ARHGEF38 RDH10 PIGA ZNF710 WDR37 AFF2 COL5A2 POLLDOK3 COL1A2 MARS2 MDM4 GAN LRRC17 RFX6 DNAJA2 AMER3 MIB1 IKBKAP MYO1F MGAT4A SEMA4C NKD1 KATNBL1 AGBL1 ABT1 TBC1D13 GGA3 SOX13 FAM210A SESTD1 NRARP NME4 PITPNM3 ANKRD46 KCTD17 SLC52A3 MBTPS2 MAP3K1 DIP2A ABHD14B CCDC141 CBL LOR ABCB9 ASPH USP12 RMI2 ELOVL4 SLC25A24 MTDH MICAL3 TNFRSF1B ZCCHC3 SOCS1 PRKAA2 CHRD ARHGEF15 ZNF516 DCAF15 PLD3 DLGAP4 FMO4 MAB21L3 E2F2 FASLG PEX11B PLA2G3 TIA1 SOWAHA PLXND1 CYP4F2 DCNA BCC5 DUSP22 DAPK1 ZNF879 ELF4 BRWD3 CLDN16 CDKN1A SCN11A KLHL13 MAP4K4 CERCAM ITGB3 CYP46A1 RNMT SLAMF6 GSG1L MC2R ENTPD7 AMOT RUFY3 B3GNT1 KLK10 SCN8A SNX30 EDEM3 FAS KLF9 ATG10 FRMD5 CD86 MMS22L OGG1 AEN LMX1A CCNF ZNF273 CECR6 SUB1 CYB561D1 PRSS22 TBKBP1 DMRT2 DDN SERPINB9 SNAI3 PLA2G15 DAGLA INTS2 FAXC DPP3 C19orf47 GREB1 ERGIC1 LIMK2 ANKRD49 C2 HOOK1 SLC25A40 PARM1 SLC11A2 DPF2 MDFI ABCC10 SMARCC1 IGF2BP1 SPATA2 FAM84B MFSD8 CDC25A C20orf112 SLC6A1 SMCR8 MIER1 IGF2BP2 UBXN2B DZIP1L IRS2 ERCC4 PAG1 CELF3 NEK3 BTBD9 MBD2 ENTHD2 SLC25A12 TMED5 KIAA1429 HDLBP ARPP19 HOXD1 ZBTB39 RAD18 ODF2L CPM TSPAN18 LAMP2 STAT2 CD59 TPK1 RBMS2 DCX ZNF566 IMPG2 MASP1 PNKD NOVA1 SREBF2 SLC25A32 ZC3H3 SPRYD7 SYNPO2L EEF2K LIPH |
| miR-100-5p | KBTBD8 S3ST2 ZZEF1 MTOR MBNL1 TRIB2 SMARCA5 TTC39A ZADH2 RAVER2 PPP3CA AP1AR FGFR3 HS3ST3B1 NOX4 BAZ2A ZNF845 AGO2 PCSK9 NR6A1 TAOK1 FZD8 MTMR3 EPDR1 ETFDH FZD5 CTDSPL |
| miR-101-3p | MPPE1 MOB4 CACNB2 TNPO1 STC1 ABHD17C FLRT3 MYCN TSHZ3 LCOR C3orf58 SOCS5 ZFP36L2 FZD6 REV3L FZD4 RORA TMEM65 ZNF654 FGA RFX3 TGFBR1 ZNF532 CDYL DR1 CPEB3 RANBP9 FOS SCN2A SLC12A2 NLK CDH11 FAT3 ADAMTS17 KBTBD8 FAM214A ATXN1L EZH2 PRKCE PRPF4B USP47 ZFHX4 RASD2 DIP2B INO80D STAG2 UBE2D1 RAP2C ZNF746 MFSD6 UBE2A SMARCA1 ADAMTSL3 ANKRD44 SEL1L MTMR2 ZNF451 SLC1A1 ARID1A EED SMARCD1 ZMAT3 PAPOLG BCL9 EYA1 RAB5A ETV5 SH2B3 EMP2 ICK CBFA2T2 SGK1 SULT4A1 ZEB1 NEK7 ZBTB34 BEAN1 ENY2 ATXN1 ZNF385B HTRA3 PPFIA1 SUB1 TMEM194B MKL2 HSPE1-MOB4 GLCCI1 TET2 PIEZO1 NPNT CTTNBP2 UBE2D2 ING3 TNKS2 BDP1 ZSWIM6 COL10A1 ERBB2IP AJAP1 SHISA6 KIF2A CHAC2 ANKRD11 SSBP2 ASPN CAV3 KIAA1804 KLF3 FBXW7 ETNK1 ANKRD17 GPR85 EXOC5 PCDH8 SLC39A10 MBNL1 UBN2 UNC79 SIX4 SEPT11 EMP1 DUSP1 ZNF207 PLXNA2 FAM46A CAPN2 NR1D2 BTBD3 MTCL1 ZFAND3 ABHD17B CERS2 CEP350 MAGI1 DAG1 GLTSCR1 DIP2C PIP5K1C DISC1 MORN4 MGAT4A ARNTL2 GAB1 NRK IFFO2 PCGF5 PTGS2 MAK PDE4D ARHGEF3 FBN2 B3GALNT2 SCN8A ARAP2 STAMBP STAU2 KLHDC1 LIN7C ZNF518A PHF20L1 POMP RAB39B ZNF217 SLC38A2 LMNB1 UTS2B LRP2 RAB1A AP3D1 ADAMTS3 GSK3B SLC19A2 PPP1R2 DENND1B PPARGC1B RIN2 FBXO30 SLC7A11 MYRIP TCEB1 SYNCRIP DDIT4 ABCC5 FAM83B IMPA1 AP3S1 TGFBR3 DNMT3A FAM114A1 CDK8 CERS6 BICD2 DCBLD2 TAL1 NUPL2 TRPC4 MARK1 NDFIP1 PANK3 DLG5 HELZ CCNJ INPP5F TRIM24 KIAA1244 KCNH7 N4BP2 LRRN1 IKZF2 CPEB2 ADRB1 KAT7 CEP63 TDG RAP1B NOVA1 PPFIA2 SYT4 AEBP2 RSF1 ZDHHC21 PIKFYVE PNISR PABPC5 MED13 SLC39A6 DOT1L SLC2A13 ATP8A1 LRCH1 CAMKK1 SASH1 CLDN11 EVI5 TULP4 PURG DCAF5 KCNA1 ST7 RBM25 DMXL2 PPM1L LHFP ABLIM3 IL1R1 ACAD9 CAMTA1 CIR1 GJA1 ENPP2 ZBTB21 GID4 FKTN MED14OS ZNF557 CYB561D2 RAC1 TFB2M TNRC18 CTNND2 EDEM3 KCTD6 ASAP1 FAM179B PRKAA1 C8orf76 HNRNPA0 PPTC7 RAB4A RAPH1 GCNT3 KLF6 METAP1 TMEM161B TIA1 ZIK1 CDH5 GFRA1 TBRG1 MMGT1 DSC1 ERO1LB SLC30A7 GLRA2 LRCH2 NDST3 CDK5R1 PMPCB POGZ RNF219 KDM3B FAM78A H2AFV UGGT1 SPATA2 MAP3K13 MAML3 MPHOSPH9 AKT3 FA2H PRKD3 MRGBP CEBPA KIAA1586 ASCC3 RAB27A BEGAIN ZNF510 RFPL4B CCDC68 TLK2 TAGAP FUCA2 ZNF549 RAB15 OTUD4 CCSER1 ZBED4 RASGRP3 GRIN2A ANXA10 WWC3 HNRNPF KAT6B HAS2 DCUN1D1 CTCF CCDC88A FAM73A MTSS1L BBX FAM60A RNF19A RCN2 PKD2 ATRX POLR3K MAP3K9 N4BP1 DNM1L MRPL42 KHDRBS2 STX6 CSNK1G3 NOTCH1 GABRB2 SPOP GLIPR1L1 KIF5B C9orf72 DENND2C SACM1L LRRC4 MAP3K2 SPG11 DCAF7 ARHGEF10 KLF2 ZCCHC2 KPNB1 KIAA1432 CRLS1 BTLA NSD1 MAPK1 TMEM167A PDS5B OGT KDM6B GCNT1 C11orf70 ANKZF1 RNF38 ROBO2 SGMS2 EPT1 SLMO2 HIVEP3 FAR1 CAPS2 TMEM231 TKTL1 TMEM68 ZNF469 SGPL1 RXRB WDR72 DESI2 NACA2 MTMR4 LGI2 CREBRF XPO5 PTCH1 NACA GABBR2 PRR11 CTDSPL DCLRE1B DDX3X MAB21L3 MLEC FAM103A1 GNB1 SPATS2L PRRC2C UBR7 FYTTD1 CD86 RIPK1 CNIH3 NAIP MON2 ATRNL1 KIAA1462 BCL2L11 RANBP1 FMNL3 PHTF2 TMF1 LANCL3 ZNF33A TIMM17A PLEKHG1 PBX3 MTX3 UNKL TEX2 RANBP6 AGAP1 ZNF235 CCDC126 FAM169A PTBP3 CADM2 KCNE1 FAM216B OTUD3 MAP10 FLRT2 PIK3C2B PCK1 PYGO1 TMEM201 C7orf73 C1orf52 SPRED1 B3GNT3 NDUFB5 TKTL2 ATP11B NEGR1 CADM1 TMED5 SMARCA4 SMN2 IKZF4 ZNF24 XKR6 PLA2R1 CDKN1A NAV1 PYGO2 NAA15 FRYL PCDH20 KIAA1377 PACRG NF1 SUPT7L C2orf88 RRM1 SMN1 FAM53B INPP4B IPO5 SRPK2 BBS7 STAR GDE1 FBXW11 JDP2 CRISPLD1 MAD2L1 SLTM DPY19L2 TBC1D12 ADH5 VSX1 LONRF1 COTL1 RBBP7 JAK2 SOAT1 NEK4 UBE2F MNX1 AGFG1 PTPRJ KTI12 PHACTR2 C16orf72 ARHGAP32 POGK IQGAP3 FAM122C USP38 CCNT2 DTD2 TMEM170B STMN1 PITPNB PCDH7 ZIC1 LRAT PDP1 CISD2 FOXN2 ZNF260 EPB41L5 DENR SLC25A4 ZC3H7A GRSF1 TMEM132D RHOT1 C10orf12 JAKMIP2 AP1S3 CASP3 BAZ2A |
| miR-199-3a | ETNK1 CELSR2 ADAMTSL3 KLHL3 ACVR2A LRP2 BCAR3 SERPINE2 NOVA1 MAP3K4 FAM110C KIAA0319L RB1 ZHX1 KDM5A PSD2 LIN28B LLGL2 ITGA3 CHMP5 TUBGCP3 FAM60A NLK CD2AP NID2 UTP20 PAK4 C9orf40 KDM6A CDK7 C2orf49 KATNBL1 CDK17 PPP2R2A APLP2 MCFD2 CDNF PRPF40A CXADR PPP2R5E G3BP2 FUBP1 NEDD4 SLC24A2 RASEF SDC2 PDGFRA SCD SUMO3 ITPK1 ARHGEF3 ESRP1 ATAD1 MAP3K5 APLF ASTN1 EMC1 GGNBP2 CYB5R4 PAWR NXPH1 PIP5K1B ATRX NUFIP2 KTN1 RNGTT MDGA2 GORAB PNRC1 VGLL2 FAM199X DEPDC1B GNPTAB NFIA DNHD1 RAPH1 TPPP WDR7 ARL15 ADAM10 NLRP1 CBLB RAPGEF4 SEMA3A COL12A1 TACC2 KLF13 SPIRE1 FAM115C ANKRD44 MS4A7 LRRC1 PTPN3 AEBP2 COL4A5 CBLL1 CISD2 CCDC85C FN1 ATP6V1A NRBP2 PTPRZ1 SP1 ATL1 DNMT3A NET1 FOS PROSER1 RFX3 WFDC8 MFSD6 TAOK1 ZBTB18 PTPRC C20orf194 ITGA6 RPS6KA6 LPAR4 LCOR MAPRE1 CD151 FXR1 PLCB1 MPP7 YWHAE EPG5 SMARCC2 EPB41L5 SLC25A46 C21orf91 SMIM8 GPBP1L1 KIDINS220 GPM6A VPS33A PON2 TMED5 HNF1B WAPAL DCBLD2 CNIH2 C9orf170 RALGPS2 LAMP3 BEND7 FAM129A ITGB8 ANKRD61 CETN3 KCMF1 FAM76B PDE4B HYPK SLC39A10 NAA25 NTRK2 KDM3A GLT8D2 WDR47 MBNL1 MTOR SOWAHC RGS4 FGL2 ALX4 YWHAG STARD9 ENOX2 MAP3K1 GALNT7 YWHAZ CREBRF TENM1 TAB2 EML4 RP1 FMN1 CHKA PVRL2 VAMP3 ZCCHC17 TEAD1 SYNJ1 SLC16A12 PCDH7 ABHD4 DUSP5 KCND2 SECISBP2L DIMT1 PPP1R9A ATP6V1C2 MEIS2 ARG2 CHAD SORL1 RNF216 ELAVL2 CAPRIN1 FCGR3A LONRF3 ADD3 RRM2B CNOT7 SRR IL1RL1 ECM2 MVB12B ADRB1 CLDN8 FCGR3B CCSAP CA5B VLDLR UBQLN1 EFCAB14 TMEM62 PTPRU ABCA1 CABLES1 SH3GLB1 ERO1L ANK2 TMEM218 KIAA0907 ASAP2 ACOX1 SYPL1 BRWD3 DPAGT1 PIK3CB NF1 ZNF614 SLC39A9 SLC5A7 HRNR CYP1B1 ZC3H14 LOC101929844 PCDHB12 HECTD2 PLEKHH1 UCK2 HNMT CDC42BPB RFX7 CCSER1 KCTD7 CITED2 CFL2 RHOT1 UBXN2B HGF KIAA0141 FBXW11 GPR160 KCNH2 TRMT61B GNA12 GRHL1 SLC44A5 PHF6 KLF12 CYP24A1 CDK5R1 MAP3K2 ATP1B4 CCDC88C ADAM22 C10orf2 TXLNG CEP85L KAZN PRKCB BAG4 FAM46D CALCRL PRC1 KIAA1244 SEC16B FKBP14 CDC14A CTNNA2 NAP1L1 UNC45A DDIT4 PAQR3 |
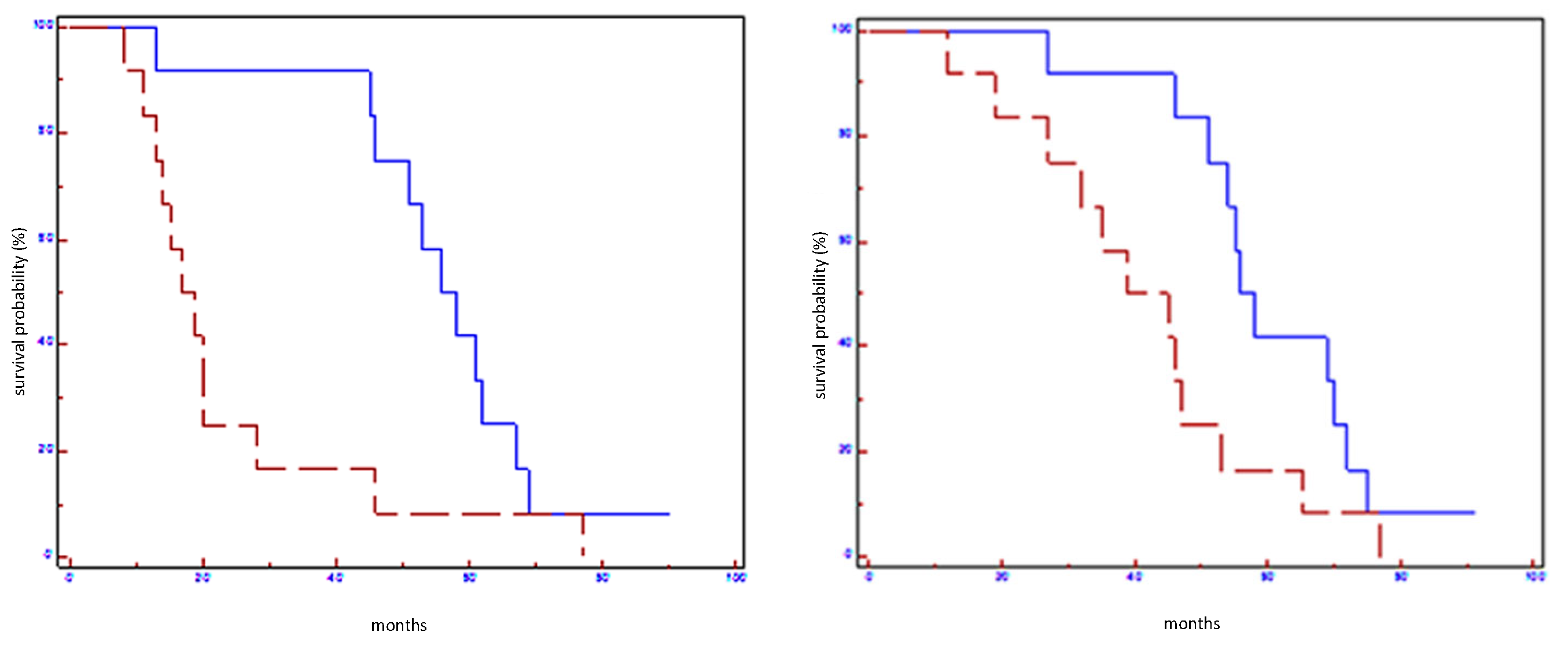
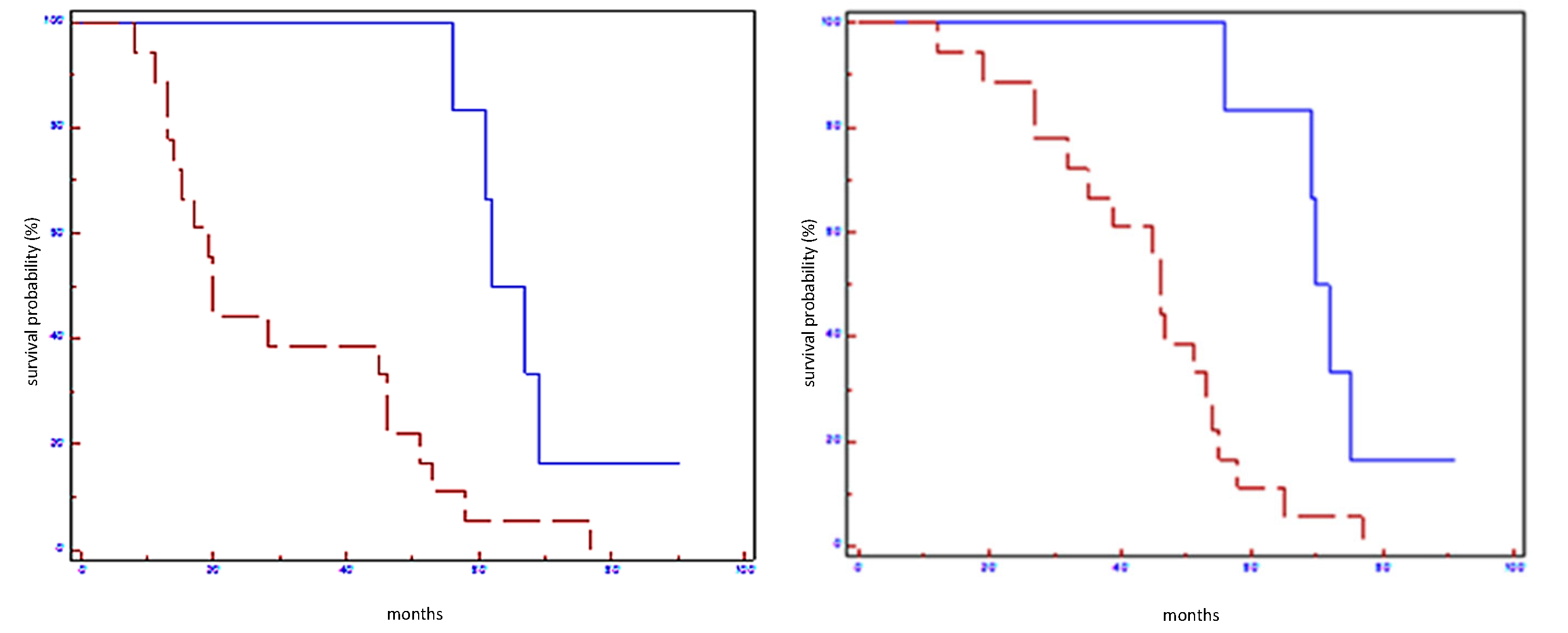
| EFS (Months) | p Value | Hazard Ratio (CI 95%) | OS (Months) | p-Value | Hazard Ratio (CI 95%) | |
|---|---|---|---|---|---|---|
| Let-7a-5p in all populations | 58 vs. 28 | 0.006 | 0.38 (0.08–0.66) | 65 vs. 35 | 0.0001 | 0.27 (0.03–0.33) |
| Let-7a-5p in HER2 non-luminal subtypes | 61 vs. 36 | 0.05 | 0.58 (0.29–6.28) | 71 vs. 44 | 0.05 | 0.31 (0.02–1.0) |
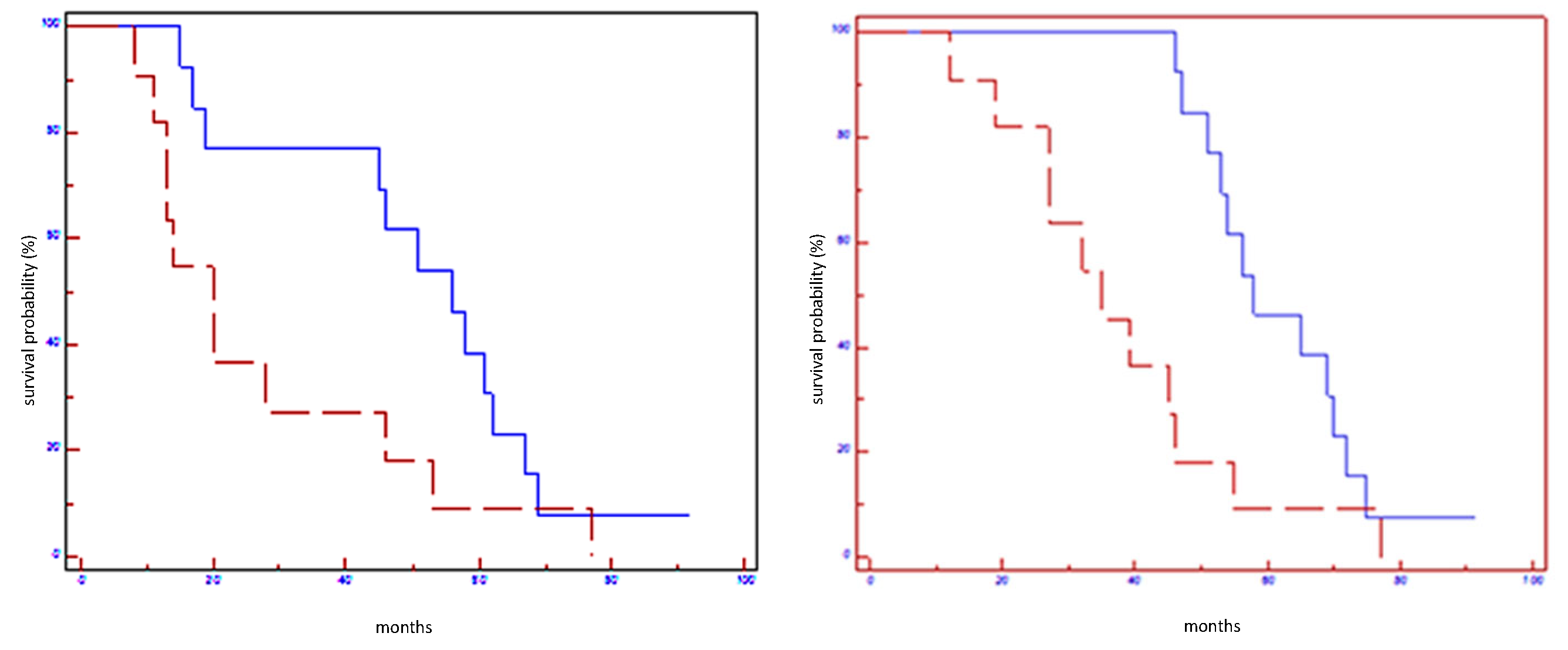
| miR-100-5p in all populations | 56 vs. 17 | 0.01 | 0.39 (0.11–0.75) | 56 vs. 39 | 0.03 | 0.45 (0.15–0.94) |
| miR-100-5p in HER2 non-luminal subtypes | 61 vs. 20 | 0.004 | 0.21 (0.01–0.30) | 70 vs. 35 | 0.004 | 0.19 (0.00–0.30) |
| miR-101-3p in all populations | 56 vs. 20 | 0.05 | 0.48 (0.16–1.03) | 58 vs. 35 | 0.01 | 0.38 (0.10–0.75) |
| miR-101-3p in HER2 non-luminal subtypes | 61 vs. 24 | 0.02 | 0.28 (0.01–0.77) | 71 vs. 40 | 0.02 | 0.27 (0.01–0.77) |
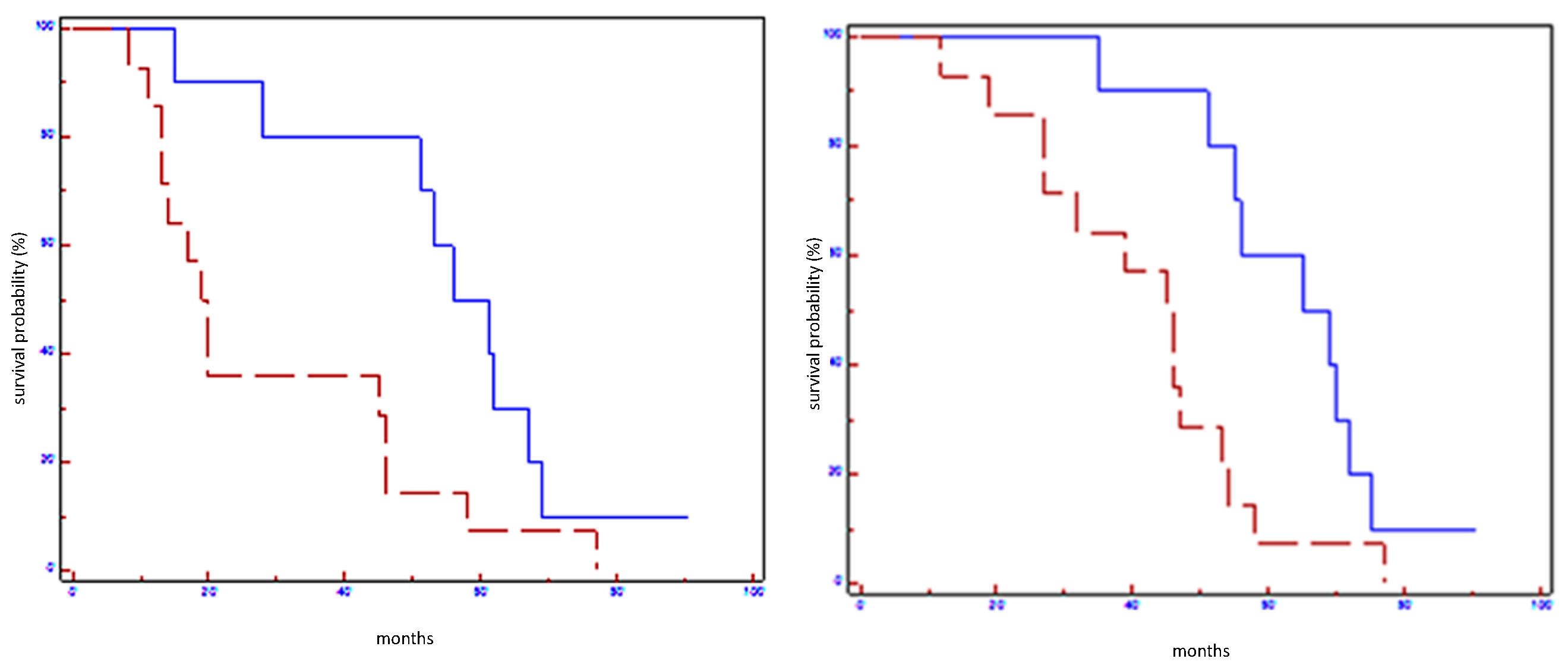
| miR-199a-3p in all populations | 61 vs. 20 | 0.02 | 0.41 (0.14–0.85) | 69 vs. 46 | 0.01 | 0.39 (0.13–0.80) |
| miR199a-3p in HER2 non-luminal subtypes | 61 vs. 20 | 0.02 | 0.27 (0.00–0.70) | 70 vs. 45 | 0.04 | 0.29 (0.01–0.96) |
| Signature in all populations | 64 vs. 20 | 0.004 | 0.31 (0.4–0.66) | 71 vs. 46 | 0.005 | 0.31 (0.11–0.68) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fuso, P.; Di Salvatore, M.; Santonocito, C.; Guarino, D.; Autilio, C.; Mulè, A.; Arciuolo, D.; Rinninella, A.; Mignone, F.; Ramundo, M.; et al. Let-7a-5p, miR-100-5p, miR-101-3p, and miR-199a-3p Hyperexpression as Potential Predictive Biomarkers in Early Breast Cancer Patients. J. Pers. Med. 2021, 11, 816. https://doi.org/10.3390/jpm11080816
Fuso P, Di Salvatore M, Santonocito C, Guarino D, Autilio C, Mulè A, Arciuolo D, Rinninella A, Mignone F, Ramundo M, et al. Let-7a-5p, miR-100-5p, miR-101-3p, and miR-199a-3p Hyperexpression as Potential Predictive Biomarkers in Early Breast Cancer Patients. Journal of Personalized Medicine. 2021; 11(8):816. https://doi.org/10.3390/jpm11080816
Chicago/Turabian StyleFuso, Paola, Mariantonietta Di Salvatore, Concetta Santonocito, Donatella Guarino, Chiara Autilio, Antonino Mulè, Damiano Arciuolo, Antonina Rinninella, Flavio Mignone, Matteo Ramundo, and et al. 2021. "Let-7a-5p, miR-100-5p, miR-101-3p, and miR-199a-3p Hyperexpression as Potential Predictive Biomarkers in Early Breast Cancer Patients" Journal of Personalized Medicine 11, no. 8: 816. https://doi.org/10.3390/jpm11080816
APA StyleFuso, P., Di Salvatore, M., Santonocito, C., Guarino, D., Autilio, C., Mulè, A., Arciuolo, D., Rinninella, A., Mignone, F., Ramundo, M., Di Stefano, B., Orlandi, A., Capoluongo, E., Nicolotti, N., Franceschini, G., Sanchez, A. M., Tortora, G., Scambia, G., Barone, C., & Cassano, A. (2021). Let-7a-5p, miR-100-5p, miR-101-3p, and miR-199a-3p Hyperexpression as Potential Predictive Biomarkers in Early Breast Cancer Patients. Journal of Personalized Medicine, 11(8), 816. https://doi.org/10.3390/jpm11080816










