Analysis of SYK Gene as a Prognostic Biomarker and Suggested Potential Bioactive Phytochemicals as an Alternative Therapeutic Option for Colorectal Cancer: An In-Silico Pharmaco-Informatics Investigation
Abstract
1. Introduction
2. Methods and Materials
2.1. The Analysis of SYK Gene Expression in Colorectal Cancer
2.2. Mutation and Copy Number Alteration Determination in the SYK Gene
2.3. Survival Assay Analysis
2.4. Correlation Analysis and Interaction Network
2.5. Target Identification of the SYK Gene for Colorectal Cancer
2.6. Pharmacokinetics Analysis
2.7. Extracting Lead Molecule for Optimization
2.8. Extraction of Proteins and Preparation for Docking
2.9. Molecular Docking and Post-Docking Data Visualization
2.10. Molecular Dynamics Simulation
3. Results
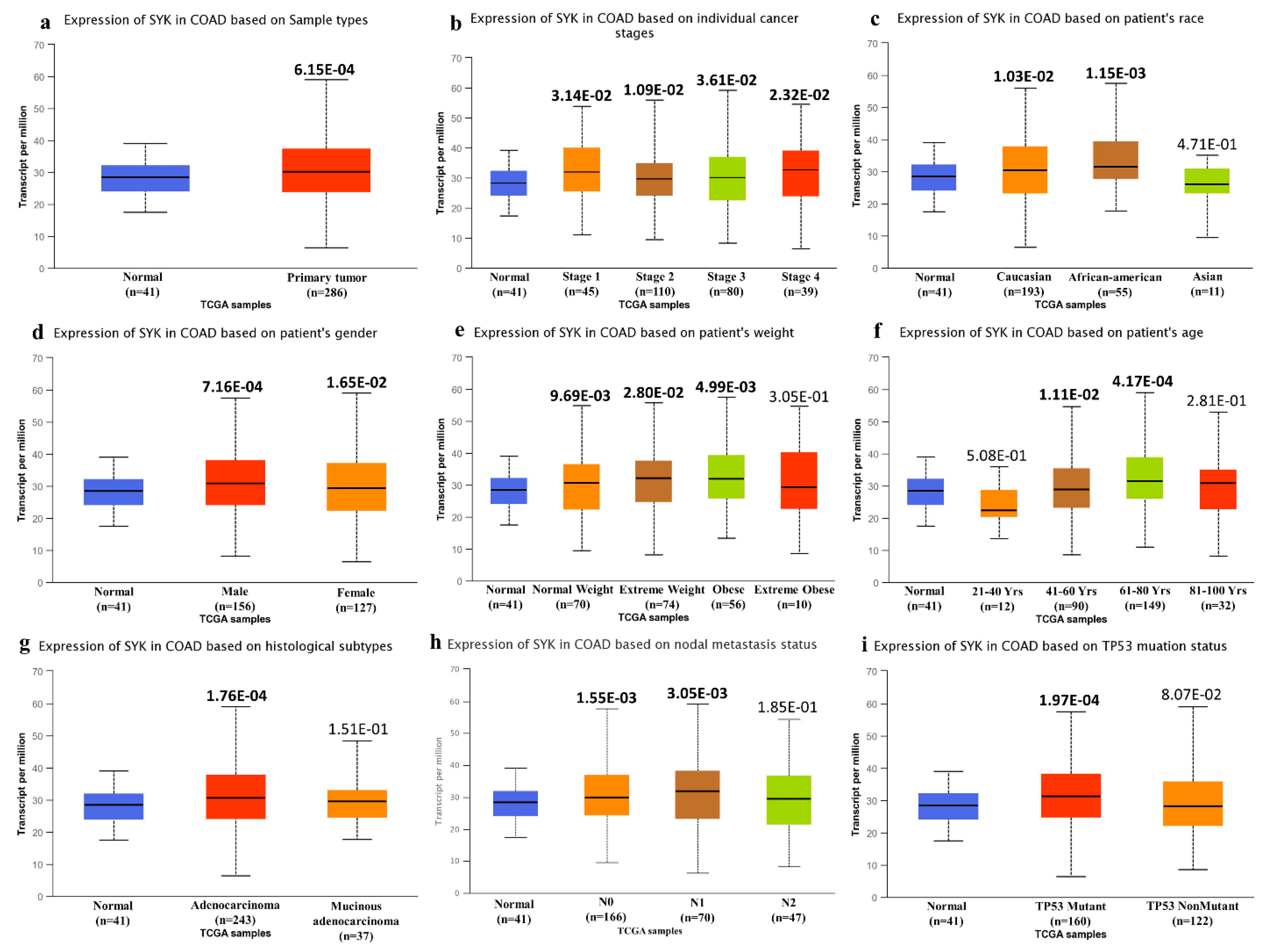
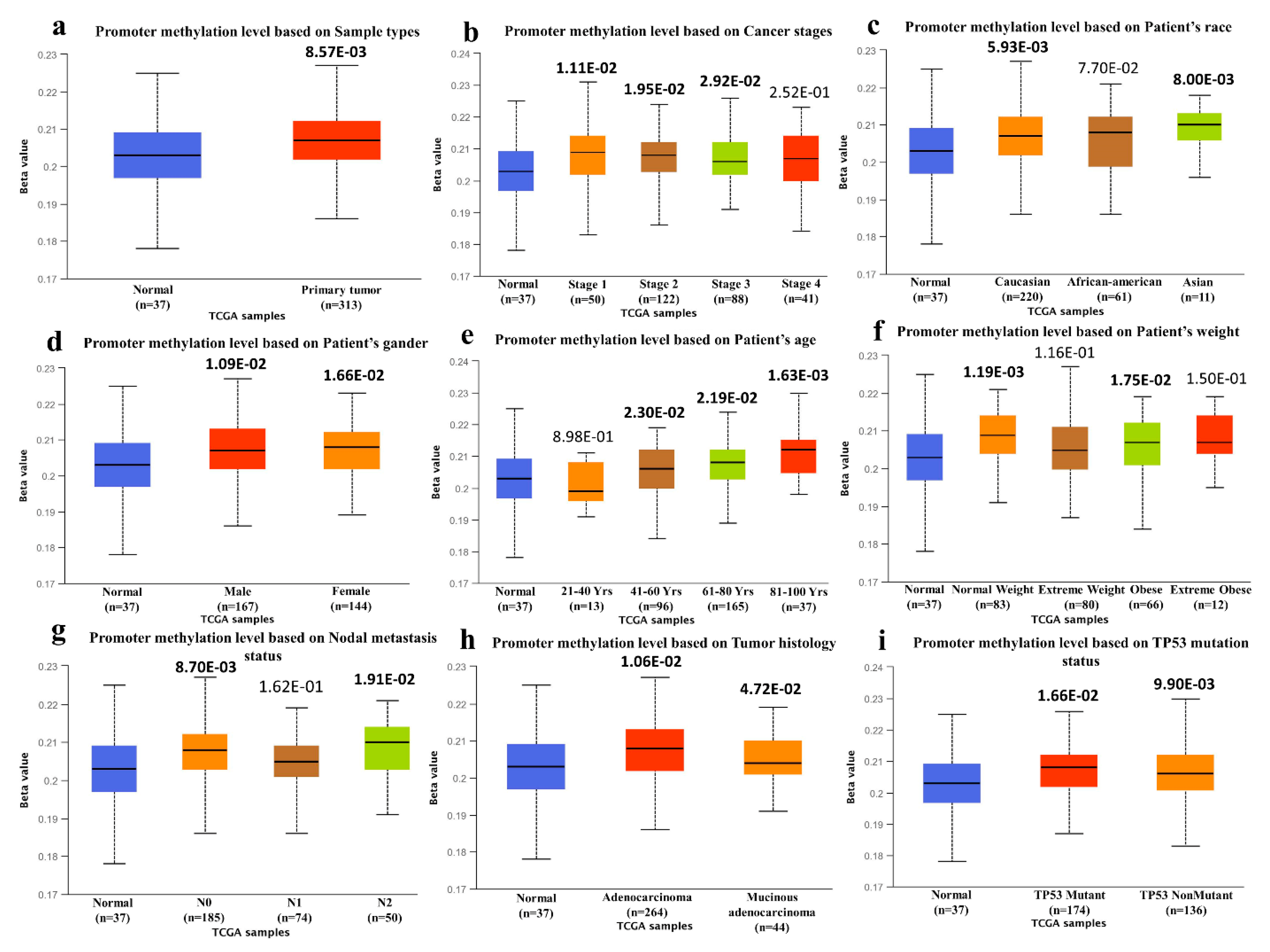
3.1. Expression Analysis of the SYK Gene
3.2. Genetic Alteration Analysis in SYK Protein Sequences Associated with Colorectal Cancer Development
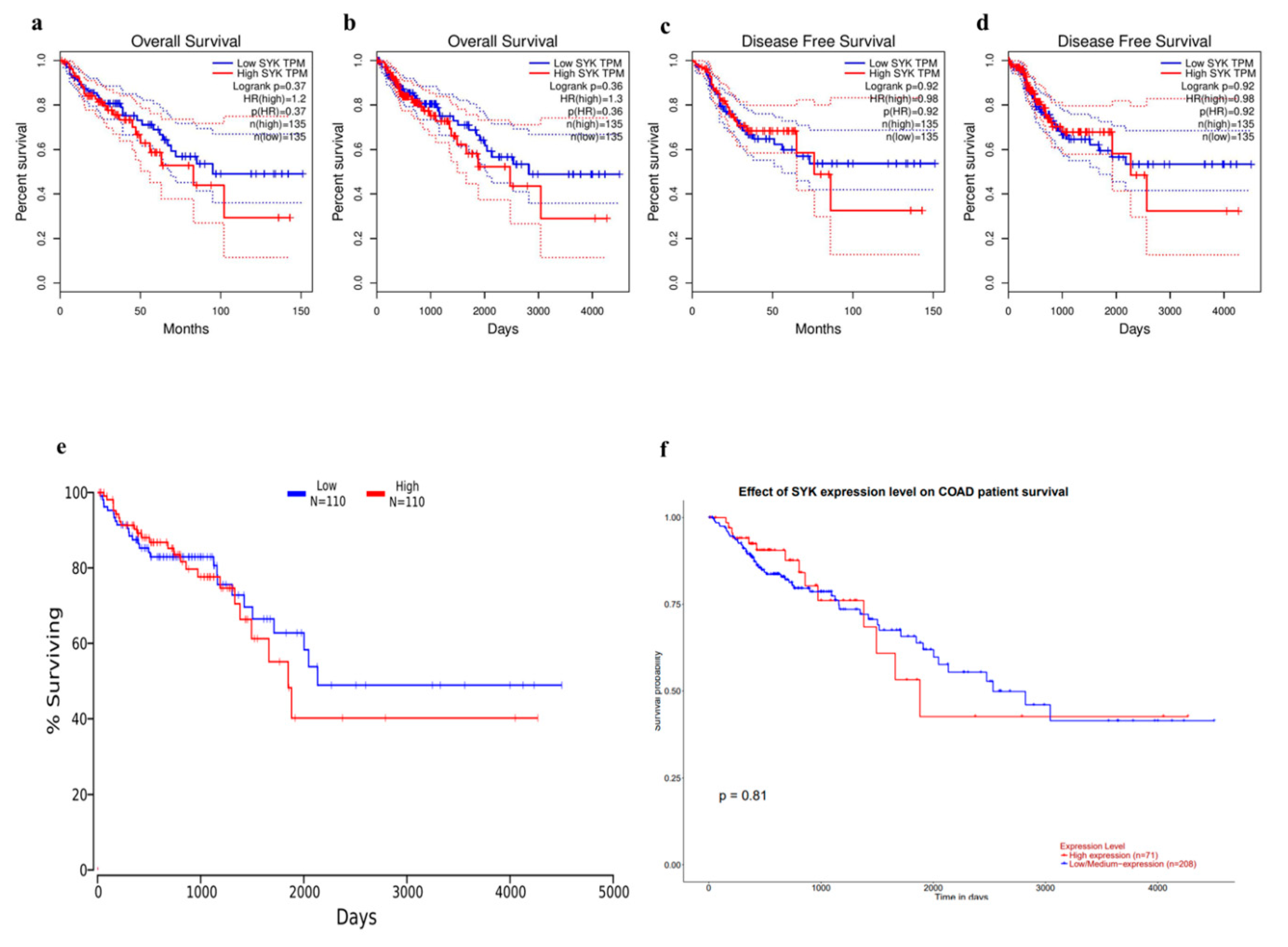
3.3. Prognostic Value of SYK and Survival Analysis
3.4. Analysis of Correlated Genes and Preparation of an Interaction Network
3.5. Pharmacokinetics Analysis
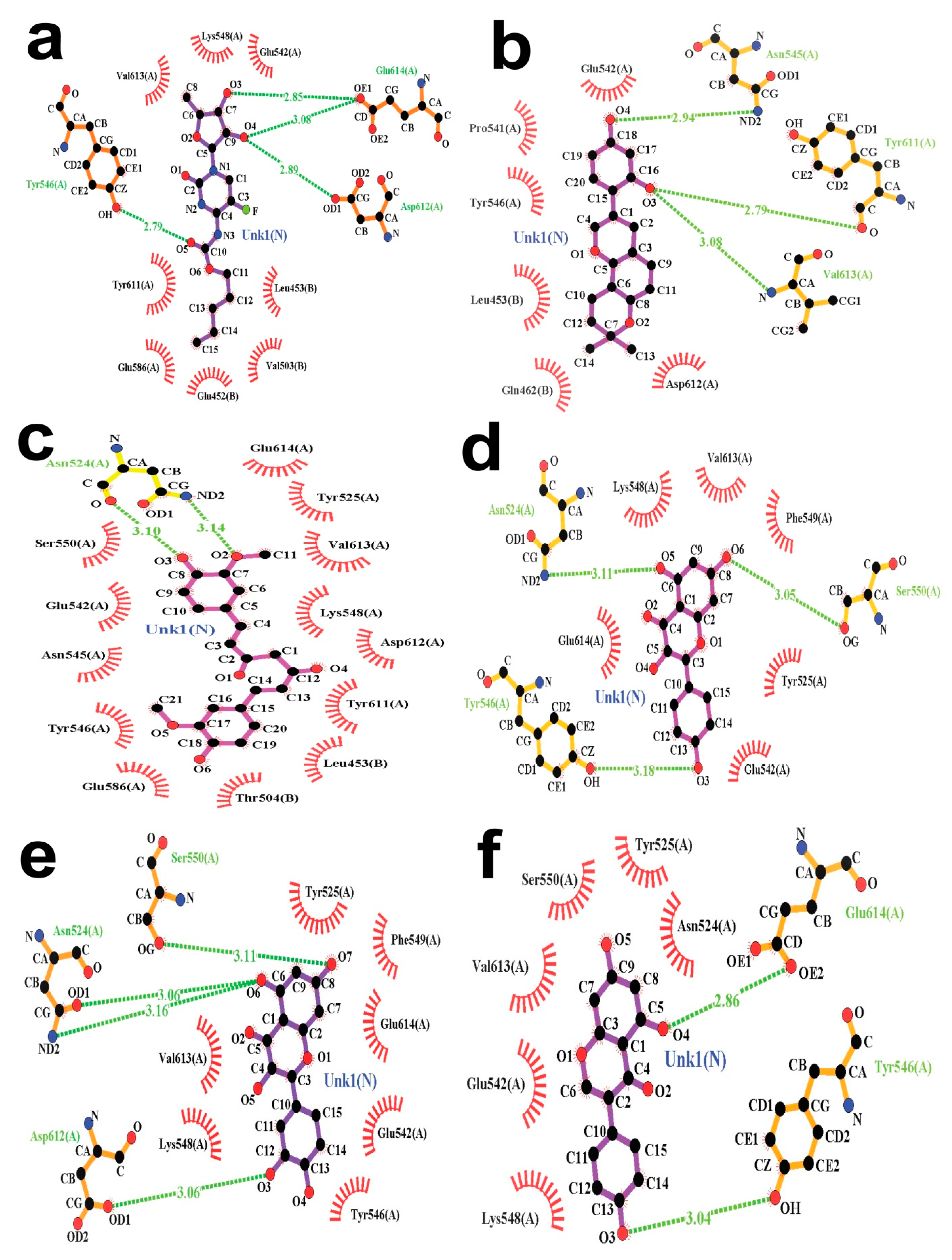
3.6. Molecular Docking and Post-Docking Data Analysis
3.7. Molecular Dynamics Simulation (MDS)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gao, D.; Wang, L.; Zhang, H.; Yan, X.; Yang, J.; Zhou, R.; Chang, X.; Sun, Y.; Tian, S.; Yao, Z.; et al. Spleen tyrosine kinase SYK(L) interacts with YY1 and coordinately suppresses SNAI2 transcription in lung cancer cells. FEBS J. 2018, 285, 4229–4245. [Google Scholar] [CrossRef]
- Blancato, J.; Graves, A.; Rashidi, B.; Moroni, M.; Tchobe, L.; Ozdemirli, M.; Kallakury, B.; Makambi, K.H.; Marian, C.; Mueller, S.C. SYK allelic loss and the role of Syk-regulated genes in breast cancer survival. PLoS ONE 2014, 9, e87610. [Google Scholar] [CrossRef]
- Coopman, P.J.; Mueller, S.C. The Syk tyrosine kinase: A new negative regulator in tumor growth and progression. Cancer Lett. 2006, 241, 159–173. [Google Scholar] [CrossRef]
- Coopman, P.J.; Do, M.T.; Barth, M.; Bowden, E.T.; Hayes, A.J.; Basyuk, E.; Blancato, J.K.; Vezza, P.R.; McLeskey, S.W.; Mangeat, P.H.; et al. The Syk tyrosine kinase suppresses malignant growth of human breast cancer cells. Nature 2000, 406, 742–747. [Google Scholar] [CrossRef]
- Krisenko, M.O.; Geahlen, R.L. Calling in SYK: SYK’s dual role as a tumor promoter and tumor suppressor in cancer. Biochim. Biophys. Acta 2015, 1853, 254–263. [Google Scholar] [CrossRef]
- Repana, K.; Papazisis, K.; Foukas, P.; Valeri, R.; Kortsaris, A.; Deligiorgi, E.; Kyriakidis, D. Expression of Syk in invasive breast cancer: Correlation to proliferation and invasiveness. Anticancer Res. 2006, 26, 4949–4954. [Google Scholar]
- Ogane, S.; Onda, T.; Takano, N.; Yajima, T.; Uchiyama, T.; Shibahara, T. Spleen tyrosine kinase as a novel candidate tumor suppressor gene for human oral squamous cell carcinoma. Int. J. Cancer 2009, 124, 2651–2657. [Google Scholar] [CrossRef] [PubMed]
- Layton, T.; Stalens, C.; Gunderson, F.; Goodison, S.; Silletti, S. Syk tyrosine kinase acts as a pancreatic adenocarcinoma tumor suppressor by regulating cellular growth and invasion. Am. J. Pathol. 2009, 175, 2625–2636. [Google Scholar] [CrossRef]
- Wang, S.; Ding, Y.B.; Chen, G.Y.; Xia, J.G.; Wu, Z.Y. Hypermethylation of Syk gene in promoter region associated with oncogenesis and metastasis of gastric carcinoma. World J. Gastroenterol. 2004, 10, 1815–1818. [Google Scholar] [CrossRef]
- Yang, Z.; Huo, L.; Chen, H.; Ni, B.; Xiang, J.; Kang, L.; Wang, L.; Peng, J.; Yuan, Y.; Wang, J. Hypermethylation and prognostic implication of Syk gene in human colorectal cancer. Med. Oncol. 2013, 30, 586. [Google Scholar] [CrossRef]
- Bu, F.; Zhu, X.; Liu, S.; Lin, K.; Zhu, J.; Huang, J. Comprehensive analysis of Syk gene methylation in colorectal cancer. Immun. Inflamm. Dis. 2021, 9, 923–931. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017, 66, 683–691. [Google Scholar] [CrossRef]
- Miller, K.D.; Nogueira, L.; Mariotto, A.B.; Rowland, J.H.; Yabroff, K.R.; Alfano, C.M.; Jemal, A.; Kramer, J.L.; Siegel, R.L. Cancer treatment and survivorship statistics, 2019. CA A Cancer J. Clin. 2019, 69, 363–385. [Google Scholar] [CrossRef]
- Rawla, P.; Sunkara, T.; Barsouk, A. Epidemiology of colorectal cancer: Incidence, mortality, survival, and risk factors. Prz. Gastroenterol. 2019, 14, 89–103. [Google Scholar] [CrossRef]
- Manson, M.M.; Gescher, A.; Hudson, E.A.; Plummer, S.M.; Squires, M.S.; Prigent, S.A. Blocking and suppressing mechanisms of chemoprevention by dietary constituents. Toxicol. Lett. 2000, 112, 499–505. [Google Scholar] [CrossRef]
- Owuor, E.D.; Kong, A.N. Antioxidants and oxidants regulated signal transduction pathways. Biochem. Pharmacol. 2002, 64, 765–770. [Google Scholar] [CrossRef]
- Ranjan, A.; Ramachandran, S.; Gupta, N.; Kaushik, I.; Wright, S.; Srivastava, S.; Das, H.; Srivastava, S.; Prasad, S.; Srivastava, S.K. Role of phytochemicals in cancer prevention. Int. J. Mol. Sci. 2019, 20, 4981. [Google Scholar] [CrossRef]
- Jang, Y.G.; Ko, E.B.; Choi, K.C. Gallic acid, a phenolic acid, hinders the progression of prostate cancer by inhibition of histone deacetylase 1 and 2 expression. J. Nutr. Biochem. 2020, 84, 108444. [Google Scholar] [CrossRef] [PubMed]
- Hussain, H.; Hamdan, N.; Sim, E.U.-H. Anticancer and antimicrobial peptides from medicinal plants of Borneo island in Sarawak. Adv. Tradit. Med. 2021, 21, 189–197. [Google Scholar] [CrossRef]
- Tsai, Y.M.; Yang, C.J.; Hsu, Y.L.; Wu, L.Y.; Tsai, Y.C.; Hung, J.Y.; Lien, C.T.; Huang, M.S.; Kuo, P.L. Glabridin inhibits migration, invasion, and angiogenesis of human non-small cell lung cancer A549 cells by inhibiting the FAK/rho signaling pathway. Integr. Cancer Ther. 2011, 10, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Hasima, N.; Aggarwal, B.B. Targeting proteasomal pathways by dietary curcumin for cancer prevention and treatment. Curr. Med. Chem. 2014, 21, 1583–1594. [Google Scholar] [CrossRef]
- Imran, M.; Salehi, B.; Sharifi-Rad, J.; Aslam Gondal, T.; Saeed, F.; Imran, A.; Shahbaz, M.; Tsouh Fokou, P.V.; Umair Arshad, M.; Khan, H.; et al. Kaempferol: A key emphasis to its anticancer potential. Molecules 2019, 24, 2277. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.Y.; Chen, Y.C. A review of the dietary flavonoid, kaempferol on human health and cancer chemoprevention. Food Chem. 2013, 138, 2099–2107. [Google Scholar] [CrossRef]
- Spagnuolo, C.; Russo, M.; Bilotto, S.; Tedesco, I.; Laratta, B.; Russo, G.L. Dietary polyphenols in cancer prevention: The example of the flavonoid quercetin in leukemia. Ann. N. Y. Acad. Sci. 2012, 1259, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Kim, H.S.; Song, Y.S. Genistein as a potential anticancer agent against ovarian cancer. J. Tradit. Complementary Med. 2012, 2, 96–104. [Google Scholar] [CrossRef]
- Devanaboyina, N.; Kishore, Y.S.; Pushpalatha, P.; Mamatha, N.; Venkatesh, P. Development and validation of new RP HPLC method for analysis of capecitabine in pharmaceutical dosage form. Int. J. Sci. Invent. Today 2013, 2, 21–30. [Google Scholar]
- Satake, H.; Iwatsuki, M.; Uenosono, Y.; Shiraishi, T.; Tanioka, H.; Saeki, H.; Sugimachi, K.; Kitagawa, D.; Shimokawa, M.; Oki, E.; et al. Phase II trial of capecitabine plus modified cisplatin (mXP) as first-line therapy in Japanese patients with metastatic gastric cancer (KSCC1104). Cancer Chemother. Pharmacol. 2017, 79, 147–153. [Google Scholar] [CrossRef]
- Rhodes, D.R.; Kalyana-Sundaram, S.; Mahavisno, V.; Varambally, R.; Yu, J.; Briggs, B.B.; Barrette, T.R.; Anstet, M.J.; Kincead-Beal, C.; Kulkarni, P.; et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007, 9, 166–180. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Yoon, B.H.; Kim, S.K.; Kim, S.Y. GENT2: An updated gene expression database for normal and tumor tissues. BMC Med. Genom. 2019, 12, 101. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Anaya, J.J.P.C.S. OncoLnc: Linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Comput. Sci. 2016, 2, e67. [Google Scholar] [CrossRef]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef]
- Flück, M.; Zürcher, G.; Andres, A.C.; Ziemiecki, A. Molecular characterization of the murine syk protein tyrosine kinase cDNA, transcripts and protein. Biochem. Biophys. Res. Commun. 1995, 213, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, S.; Yanagi, S.; Inatome, R.; Ding, J.; Hermann, P.; Tsujimura, T.; Matsui, N.; Yamamura, H. Purification of a 72-kDa protein-tyrosine kinase from rat liver and its identification as Syk: Involvement of Syk in signaling events of hepatocytes. J. Biochem. 2000, 127, 321–327. [Google Scholar] [CrossRef]
- Dejmek, J.; Leandersson, K.; Manjer, J.; Bjartell, A.; Emdin, S.O.; Vogel, W.F.; Landberg, G.; Andersson, T. Expression and signaling activity of Wnt-5a/discoidin domain receptor-1 and Syk plays distinct but decisive roles in breast cancer patient survival. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2005, 11, 520–528. [Google Scholar]
- Okamura, S.; Ng, C.C.; Koyama, K.; Takei, Y.; Arakawa, H.; Monden, M.; Nakamura, Y. Identification of seven genes regulated by wild-type p53 in a colon cancer cell line carrying a well-controlled wild-type p53 expression system. Oncol. Res. 1999, 11, 281–285. [Google Scholar] [PubMed]
- Alagumuthu, M.; Muralidharan, V.P.; Andrew, M.; Ahmed, M.H.; Iyer, S.K.; Arumugam, S. Computational approaches to develop isoquinoline based antibiotics through DNA gyrase inhibition mechanisms unveiled through antibacterial evaluation and molecular docking. Mol. Inform. 2018, 37, e1800048. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Lasker, K.; Schneidman-Duhovny, D.; Webb, B.; Huang, C.C.; Pettersen, E.F.; Goddard, T.D.; Meng, E.C.; Sali, A.; Ferrin, T.E. UCSF Chimera, MODELLER, and IMP: An integrated modeling system. J. Struct. Biol. 2012, 179, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Dey, D.; Paul, P.K.; Al Azad, S.; Al Mazid, M.F.; Khan, A.M.; Sharif, M.A.; Rahman, M.H. Molecular optimization, docking, and dynamic simulation profiling of selective aromatic phytochemical ligands in blocking the SARS-CoV-2 S protein attachment to ACE2 receptor: An in silico approach of targeted drug designing. J. Adv. Vet. Anim. Res. 2021, 8, 24–35. [Google Scholar] [CrossRef]
- Subasri, S.; Viswanathan, V.; Manish, K.; Velmurugan, D.J.C.P.B. Phytochemical analysis, molecular docking and molecular dynamics simulations of selected phytoconstituents from four herbs as anti-dotes for snake bites. Clin. Proteom. Bioinform. 2016, 1, 1–13. [Google Scholar] [CrossRef]
- Prasanth, D.; Murahari, M.; Chandramohan, V.; Panda, S.P.; Atmakuri, L.R.; Guntupalli, C. In silico identification of potential inhibitors from Cinnamon against main protease and spike glycoprotein of SARS CoV-2. J. Biomol. Struct. Dyn. 2021, 39, 4618–4632. [Google Scholar] [CrossRef]
- Jain, D.; Hossain, R.; Khan, R.A.; Dey, D.; Toma, T.R.; Islam, M.T.; Janmeda, P.; Hakeem, K.R. Computer-Aided Evaluation of Anti-SARS-CoV-2 (3-chymotrypsin-like Protease and Transmembrane Protease Serine 2 Inhibitors) Activity of Cepharanthine: An In Silico Approach; Biointerface Research Applied Chemistry: Bucuresti, Romania, 2022. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Modeling 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Dickson, C.J.; Madej, B.D.; Skjevik, A.A.; Betz, R.M.; Teigen, K.; Gould, I.R.; Walker, R.C. Lipid14: The amber lipid force field. J. Chem. Theory Comput. 2014, 10, 865–879. [Google Scholar] [CrossRef]
- Land, H.; Humble, M.S. YASARA: A tool to obtain structural guidance in biocatalytic investigations. Methods Mol. Biol. 2018, 1685, 43–67. [Google Scholar] [CrossRef]
- Yadav, I.S.; Nandekar, P.P.; Srivastavaa, S.; Sangamwar, A.; Chaudhury, A.; Agarwal, S.M. Ensemble docking and molecular dynamics identify knoevenagel curcumin derivatives with potent anti-EGFR activity. Gene 2014, 539, 82–90. [Google Scholar] [CrossRef]
- Swargiary, A.; Mahmud, S.; Saleh, M.A. Screening of phytochemicals as potent inhibitor of 3-chymotrypsin and papain-like proteases of SARS-CoV2: An in silico approach to combat COVID-19. J. Biomol. Struct. Dyn. 2020, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.S.; Mahmud, S.; Sultana, R.; Dong, W.J.A.J.o.C. Identification and in silico molecular modelling study of newly isolated Bacillus subtilis SI-18 strain against S9 protein of Rhizoctonia solani. Arab. J. Chem. 2020, 13, 8600–8612. [Google Scholar] [CrossRef]
- Krieger, E.; Vriend, G. New ways to boost molecular dynamics simulations. J. Comput. Chem. 2015, 36, 996–1007. [Google Scholar] [CrossRef]
- Zhao, S.; Sun, G.; Tony, P.W.; Ma, D.; Zhao, C. Expression and methylation status of the Syk gene in cervical carcinoma. Arch. Gynecol. Obstet. 2011, 283, 1113–1119. [Google Scholar] [CrossRef]
- Dong, S.W.; Ma, L.; Xu, N.; Yan, H.Q.; Liu, H.Y.; Li, Y.W.; Zhang, P. Research on the reactivation of Syk expression caused by the inhibition of DNA promoter methylation in the lung cancer. Neoplasma 2011, 58, 89–95. [Google Scholar] [CrossRef][Green Version]
- Beroukhim, R.; Mermel, C.H.; Porter, D.; Wei, G.; Raychaudhuri, S.; Donovan, J.; Barretina, J.; Boehm, J.S.; Dobson, J.; Urashima, M.; et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010, 463, 899–905. [Google Scholar] [CrossRef]
- Zeng, Z.; Yang, Y.; Qing, C.; Hu, Z.; Huang, Y.; Zhou, C.; Li, D.; Jiang, Y. Distinct expression and prognostic value of members of SMAD family in non-small cell lung cancer. Medicine 2020, 99, e19451. [Google Scholar] [CrossRef]
- Gyorffy, B.; Lánczky, A.; Szállási, Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr. Relat. Cancer 2012, 19, 197–208. [Google Scholar] [CrossRef]
- Toyama, T.; Iwase, H.; Yamashita, H.; Hara, Y.; Omoto, Y.; Sugiura, H.; Zhang, Z.; Fujii, Y. Reduced expression of the Syk gene is correlated with poor prognosis in human breast cancer. Cancer Lett. 2003, 189, 97–102. [Google Scholar] [CrossRef]
- Zhou, F.; Tang, D.; Xu, Y.; He, H.; Wu, Y.; Lin, L.; Dong, J.; Tan, W.; Dai, Y. Identification of microRNAs and their Endonucleolytic Cleavaged target mRNAs in colorectal cancer. BMC Cancer 2020, 20, 242. [Google Scholar] [CrossRef]
- Goldenberg-Furmanov, M.; Stein, I.; Pikarsky, E.; Rubin, H.; Kasem, S.; Wygoda, M.; Weinstein, I.; Reuveni, H.; Ben-Sasson, S.A. Lyn is a target gene for prostate cancer: Sequence-based inhibition induces regression of human tumor xenografts. Cancer Res. 2004, 64, 1058–1066. [Google Scholar] [CrossRef]
- Su, N.; Peng, L.; Xia, B.; Zhao, Y.; Xu, A.; Wang, J.; Wang, X.; Jiang, B. Lyn is involved in CD24-induced ERK1/2 activation in colorectal cancer. Mol. Cancer 2012, 11, 43. [Google Scholar] [CrossRef]
- Dillon, L.M.; Miller, T.W. Therapeutic targeting of cancers with loss of PTEN function. Curr. Drug Targets 2014, 15, 65–79. [Google Scholar] [CrossRef]
- Staub, E.; Groene, J.; Heinze, M.; Mennerich, D.; Roepcke, S.; Klaman, I.; Hinzmann, B.; Castanos-Velez, E.; Pilarsky, C.; Mann, B.; et al. An expression module of WIPF1-coexpressed genes identifies patients with favorable prognosis in three tumor types. J. Mol. Med. 2009, 87, 633–644. [Google Scholar] [CrossRef]
- Sundvall, M.; Iljin, K.; Kilpinen, S.; Sara, H.; Kallioniemi, O.P.; Elenius, K. Role of ErbB4 in breast cancer. J. Mammary Gland. Biol. Neoplasia 2008, 13, 259–268. [Google Scholar] [CrossRef]
- Paolino, M.; Choidas, A.; Wallner, S.; Pranjic, B.; Uribesalgo, I.; Loeser, S.; Jamieson, A.M.; Langdon, W.Y.; Ikeda, F.; Fededa, J.P.; et al. The E3 ligase Cbl-b and TAM receptors regulate cancer metastasis via natural killer cells. Nature 2014, 507, 508–512. [Google Scholar] [CrossRef]
- Chan, K.K.; Matchett, K.B.; Coulter, J.A.; Yuen, H.F.; McCrudden, C.M.; Zhang, S.D.; Irwin, G.W.; Davidson, M.A.; Rülicke, T.; Schober, S.; et al. Erythropoietin drives breast cancer progression by activation of its receptor EPOR. Oncotarget 2017, 8, 38251–38263. [Google Scholar] [CrossRef]
- Lu, P.; Qiao, J.; He, W.; Wang, J.; Jia, Y.; Sun, Y.; Tang, S.; Fu, L.; Qin, Y. Genome-wide gene expression profile analyses identify CTTN as a potential prognostic marker in esophageal cancer. PLoS ONE 2014, 9, e88918. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, K.; Zhang, T.; Wang, Z.; Qin, X.; Jing, X.; Wu, H.; Ji, X.; He, Y.; Zhao, R. Cortactin promotes colorectal cancer cell proliferation by activating the EGFR-MAPK pathway. Oncotarget 2017, 8, 1541–1554. [Google Scholar] [CrossRef]
- Kapetanovic, I.M. Computer-aided drug discovery and development (CADDD): In silico-chemico-biological approach. Chemico-Biol. Interact. 2008, 171, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Mannhold, R.; Kubinyi, H.; Folkers, G. Pharmacokinetics and Metabolism in Drug Design; John Wiley & Sons: Hoboken, NJ, USA, 2012; Volume 51. [Google Scholar]
- Notari, R.E. Pharmacokinetics and molecular modification: Implications in drug design and evaluation. J. Pharm. Sci. 1973, 62, 865–881. [Google Scholar] [CrossRef]
- Matin, M.M.; Roshid, M.H.; Bhattacharjee, S.C.; Azad, A.K.J.M.R.A. PASS predication, antiviral, in vitro Antimicrobial, and ADMET studies of rhamnopyranoside esters. Med. Res. Arch. 2020, 8, 1–13. [Google Scholar] [CrossRef]
- Doss, C.G.; Chakraborty, C.; Chen, L.; Zhu, H. Integrating in silico prediction methods, molecular docking, and molecular dynamics simulation to predict the impact of ALK missense mutations in structural perspective. BioMed Res. Int. 2014, 2014, 895831. [Google Scholar] [CrossRef]
- George Priya Doss, C.; Rajith, B.; Chakraboty, C.; Balaji, V.; Magesh, R.; Gowthami, B.; Menon, S.; Swati, M.; Trivedi, M.; Paul, J.; et al. In silico profiling and structural insights of missense mutations in RET protein kinase domain by molecular dynamics and docking approach. Mol. Biosyst. 2014, 10, 421–436. [Google Scholar] [CrossRef]
- Krieger, E.; Koraimann, G.; Vriend, G. Increasing the precision of comparative models with YASARA NOVA--a self-parameterizing force field. Proteins 2002, 47, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Sargsyan, K.; Grauffel, C.; Lim, C. How molecular size impacts RMSD applications in molecular dynamics simulations. J. Chem. Theory Comput. 2017, 13, 1518–1524. [Google Scholar] [CrossRef]
- Joshi, T.; Joshi, T.; Sharma, P.; Chandra, S.; Pande, V. Molecular docking and molecular dynamics simulation approach to screen natural compounds for inhibition of Xanthomonas oryzae pv. Oryzae by targeting peptide deformylase. J. Biomol. Struct. Dyn. 2021, 39, 823–840. [Google Scholar] [CrossRef]
- Alamri, M.A.; Altharawi, A.; Alabbas, A.B.; Alossaimi, M.A.; Alqahtani, S.M.J.A.J.o.C. Structure-based virtual screening and molecular dynamics of phytochemicals derived from Saudi medicinal plants to identify potential COVID-19 therapeutics. Arab. J. Chem. 2020, 13, 7224–7234. [Google Scholar] [CrossRef]
- Kousar, K.; Majeed, A.; Yasmin, F.; Hussain, W.; Rasool, N. Phytochemicals from selective plants have promising potential against SARS-CoV-2: Investigation and corroboration through molecular docking, MD simulations, and quantum computations. BioMed Res. Int. 2020, 2020, 6237160. [Google Scholar] [CrossRef]
- Elfiky, A.A.; Elshemey, W.M. Molecular dynamics simulation revealed binding of nucleotide inhibitors to ZIKV polymerase over 444 nanoseconds. J. Med. Virol. 2018, 90, 13–18. [Google Scholar] [CrossRef]
- Mahmud, S.; Rahman, E.; Nain, Z.; Billah, M.; Karmakar, S.; Mohanto, S.C.; Paul, G.K.; Amin, A.; Acharjee, U.K.; Saleh, M.A. Computational discovery of plant-based inhibitors against human carbonic anhydrase IX and molecular dynamics simulation. J. Biomol. Struct. Dyn. 2021, 39, 2754–2770. [Google Scholar] [CrossRef] [PubMed]
- Carbone, P.; Müller-Plathe, F.J.S.M. Molecular dynamics simulations of polyaminoamide (PAMAM) dendrimer aggregates: Molecular shape, hydrogen bonds and local dynamics. Soft Matter 2009, 5, 2638–2647. [Google Scholar] [CrossRef]




| Cancer Study | Sample Size | Protein Change | Mutation Type | Sample ID |
|---|---|---|---|---|
| Colorectal Adenocarcinoma (Genentech, Nature 2012) | 74 | A52T | Missense | 587376 |
| V633M | Missense | 587342 | ||
| P85L | Missense | 587332 | ||
| Colorectal Adenocarcinoma (DFCI, Cell Reports 2016) | 619 | A353V | Missense | coadread_dfci_2016_68 |
| S511N | Missense | coadread_dfci_2016_1762 | ||
| L143M | Missense | coadread_dfci_2016_197 | ||
| N406Tfs*13 | FS del | coadread_dfci_2016_207430 | ||
| N406Tfs*13 | FS del | coadread_dfci_2016_2944 | ||
| R520H | Missense | coadread_dfci_2016_2372 | ||
| P430L | Missense | coadread_dfci_2016_3048 | ||
| K397E | Missense | coadread_dfci_2016_3670 | ||
| R109Q | Missense | coadread_dfci_2016_62 | ||
| Colorectal Adenocarcinoma (TCGA, Firehose Legacy) | 640 | M166Nfs*14 | FS ins | TCGA-AG-A02N-01 |
| K387N | Missense | TCGA-AF-3913-01 | ||
| R574* | Nonsense | TCGA-AG-A002-01 | ||
| Colorectal Adenocarcinoma (TCGA, Nature 2012) | 276 | M166Nfs*14 | FS ins | TCGA-AG-A02N-01 |
| K387N | Missense | TCGA-AF-3913-01 | ||
| R574* | Nonsense | TCGA-AG-A002-01 | ||
| Colorectal Adenocarcinoma (TCGA, PanCancer Atlas) | 594 | A353T | Missense | TCGA-D5-6922-01 |
| M166Nfs*14 | FS ins | TCGA-AG-A02N-01 | ||
| R574* | Nonsense | TCGA-AG-A002-01 | ||
| R574* | Nonsense | TCGA-F5-6814-01 | ||
| F549L | Missense | TCGA-F5-6814-01 | ||
| K105N | Missense | TCGA-AG-A00Y-01 | ||
| D344G | Missense | TCGA-F5-6814-01 | ||
| P119S | Missense | TCGA-A6-2686-01 | ||
| Y91C | Missense | TCGA-AY-6197-01 | ||
| T345R | Missense | TCGA-G4-6586-01 | ||
| E442Sfs*31 | FS del | TCGA-WS-AB45-01 | ||
| Metastatic Colorectal Cancer (MSKCC, Cancer Cell 2018) | 1134 | R42C | Missense | P-0004602-T01-IM5 |
| R42C | Missense | P-0005230-T01-IM5 | ||
| R45C | Missense | P-0013492-T01-IM5 | ||
| G33Afs*2 | FS del | P-0006365-T01-IM5 | ||
| G33Afs*2 | FS del | P-0013876-T01-IM5 | ||
| A286V | Missense | P-0010929-T01-IM5 | ||
| E442K | Missense | P-0010587-T01-IM5 | ||
| A282V | Missense | P-0005443-T01-IM5 | ||
| S84T | Missense | P-0005823-T01-IM5 | ||
| E144G | Missense | P-0010581-T01-IM5 | ||
| R367* | Nonsense | P-0006960-T01-IM5 | ||
| P411L | Missense | P-0006960-T01-IM5 | ||
| V560A | Missense | P-0006960-T01-IM5 | ||
| H62R | Missense | P-0005455-T01-IM5 | ||
| V433M | Missense | P-0005455-T01-IM5 | ||
| G185* | Nonsense | P-0002671-T01-IM3 | ||
| R175Gfs*4 | FS del | P-0002671-T01-IM3 | ||
| Y47N | Missense | P-0000769-T01-IM3 | ||
| K509R | Missense | P-0001500-T03-IM5 | ||
| M166Nfs*14 | FS ins | P-0007831-T01-IM5 | ||
| E230G | Missense | P-0013227-T01-IM5 | ||
| Rectal Cancer (MSK, Nature Medicine 2019) | 339 | V633M | Missense | RC-MSK-008-pt |
| V633M | Missense | RC-MSK-008-tm | ||
| K509R | Missense | P-0001500-T03-IM5 | ||
| Colon Cancer (CPTAC-2 Prospective, Cell 2019) | 110 | G33Afs*2 | FS del | 05CO041 |
| A83T | Missense | 01CO014 | ||
| A412S | Missense | 05CO015 | ||
| Y47C | Missense | 11CO059 |
| Name of Phytochemicals/ADMET Values | Physiochemical Properties | Pharmacokinetics Properties | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MO | HBA | HBD | Log P | RB | IAb | TCl | LD50 | HPT | AMT | MTHD | CaP | CTOR | |
| Capecitabine (Control) | 359.354 | 8 | 3 | 0.7602 | 6 | 68.027 | 1.054 | 2.459 | Yes | No | 1.051 | 0.255 | 2.401 |
| Glabridin | 324.376 | 4 | 2 | 4.0007 | 1 | 94.164 | 0.121 | 2.523 | No | No | −0.395 | 1.284 | 1.148 |
| Curcumin | 368.385 | 6 | 2 | 3.3699 | 8 | 82.19 | −0.002 | 1.833 | No | No | 0.081 | −0.093 | 2.228 |
| Kaempferol | 286.23 | 6 | 4 | 2.2824 | 1 | 74.29 | 0.477 | 2.449 | No | No | 0.531 | 0.032 | 2.505 |
| Quercetin | 302.238 | 7 | 5 | 1.988 | 1 | 77.207 | 0.407 | 2.471 | No | No | 0.499 | −0.229 | 2.612 |
| Genistein | 270.24 | 5 | 3 | 2.5768 | 1 | 93.387 | 0.151 | 2.268 | No | No | 0.478 | 0.9 | 2.189 |
| Ligands Name | Binding Affinity (Kcal/mol) | Amino Acid Involved Interaction | |
|---|---|---|---|
| Hydrogen Bond Interaction | Hydrophobic Bond Interaction | ||
| Capecitabine (Control) | −6.5 | Asp612 (2.89 Å), Glu614 (2.85 Å), Glu614 (3.08 Å) and Tyr546 (2.79 Å) | Glu452, Glu586, Glu542, Leu453, Lys548, Tyr611, Val613, Val503 |
| Glabridin | −8.2 | Asn545 (2.94 Å), Tyr611 (2.79 Å) and Val613 (3.08 Å) | Asp612, Gln462, Glu542, Leu453, Pro541, Tyr546 |
| Curcumin | −8.0 | Asn524 (3.10 Å) andAsn524 (3.14 Å) | Asn545, Asp612, Glu542, Glu586, Glu614, Leu453, Lys548, Ser550, Thr504, Tyr525, Tyr546, Tyr611, Val613 |
| Kaempferol | −7.3 | Asn524 (3.11 Å), Ser550 (3.05 Å) and Tyr546 (3.18 Å) | Glu614, Glu542, Lys548, Phe549, Tyr525, Val613 |
| Quercetin | −7.2 | Asn524 (3.06 Å), Asn524 (3.16 Å), Asp612 (3.06 Å), and Ser550 (3.11 Å) | Glu542, Glu614, Lys548, Phe549, Tyr525, Tyr546, Val613 |
| Genistein | −7.1 | Glu614 (2.86 Å) and Tyr546 (3.04 Å) | Asn524, Glu542, Lys548, Ser550, Tyr525, Val613. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Biswas, P.; Dey, D.; Rahman, A.; Islam, M.A.; Susmi, T.F.; Kaium, M.A.; Hasan, M.N.; Rahman, M.H.; Mahmud, S.; Saleh, M.A.; et al. Analysis of SYK Gene as a Prognostic Biomarker and Suggested Potential Bioactive Phytochemicals as an Alternative Therapeutic Option for Colorectal Cancer: An In-Silico Pharmaco-Informatics Investigation. J. Pers. Med. 2021, 11, 888. https://doi.org/10.3390/jpm11090888
Biswas P, Dey D, Rahman A, Islam MA, Susmi TF, Kaium MA, Hasan MN, Rahman MH, Mahmud S, Saleh MA, et al. Analysis of SYK Gene as a Prognostic Biomarker and Suggested Potential Bioactive Phytochemicals as an Alternative Therapeutic Option for Colorectal Cancer: An In-Silico Pharmaco-Informatics Investigation. Journal of Personalized Medicine. 2021; 11(9):888. https://doi.org/10.3390/jpm11090888
Chicago/Turabian StyleBiswas, Partha, Dipta Dey, Atikur Rahman, Md. Aminul Islam, Tasmina Ferdous Susmi, Md. Abu Kaium, Md. Nazmul Hasan, MD. Hasanur Rahman, Shafi Mahmud, Md. Abu Saleh, and et al. 2021. "Analysis of SYK Gene as a Prognostic Biomarker and Suggested Potential Bioactive Phytochemicals as an Alternative Therapeutic Option for Colorectal Cancer: An In-Silico Pharmaco-Informatics Investigation" Journal of Personalized Medicine 11, no. 9: 888. https://doi.org/10.3390/jpm11090888
APA StyleBiswas, P., Dey, D., Rahman, A., Islam, M. A., Susmi, T. F., Kaium, M. A., Hasan, M. N., Rahman, M. H., Mahmud, S., Saleh, M. A., Paul, P., Rahman, M. R., Al Saber, M., Song, H., Rahman, M. A., & Kim, B. (2021). Analysis of SYK Gene as a Prognostic Biomarker and Suggested Potential Bioactive Phytochemicals as an Alternative Therapeutic Option for Colorectal Cancer: An In-Silico Pharmaco-Informatics Investigation. Journal of Personalized Medicine, 11(9), 888. https://doi.org/10.3390/jpm11090888













