Interrogating Epigenome toward Personalized Approach in Cutaneous Melanoma
Abstract
1. Introduction
2. Epigenetics: Another Layer of Information in Gene Expression Regulation
3. CM Epigenetics
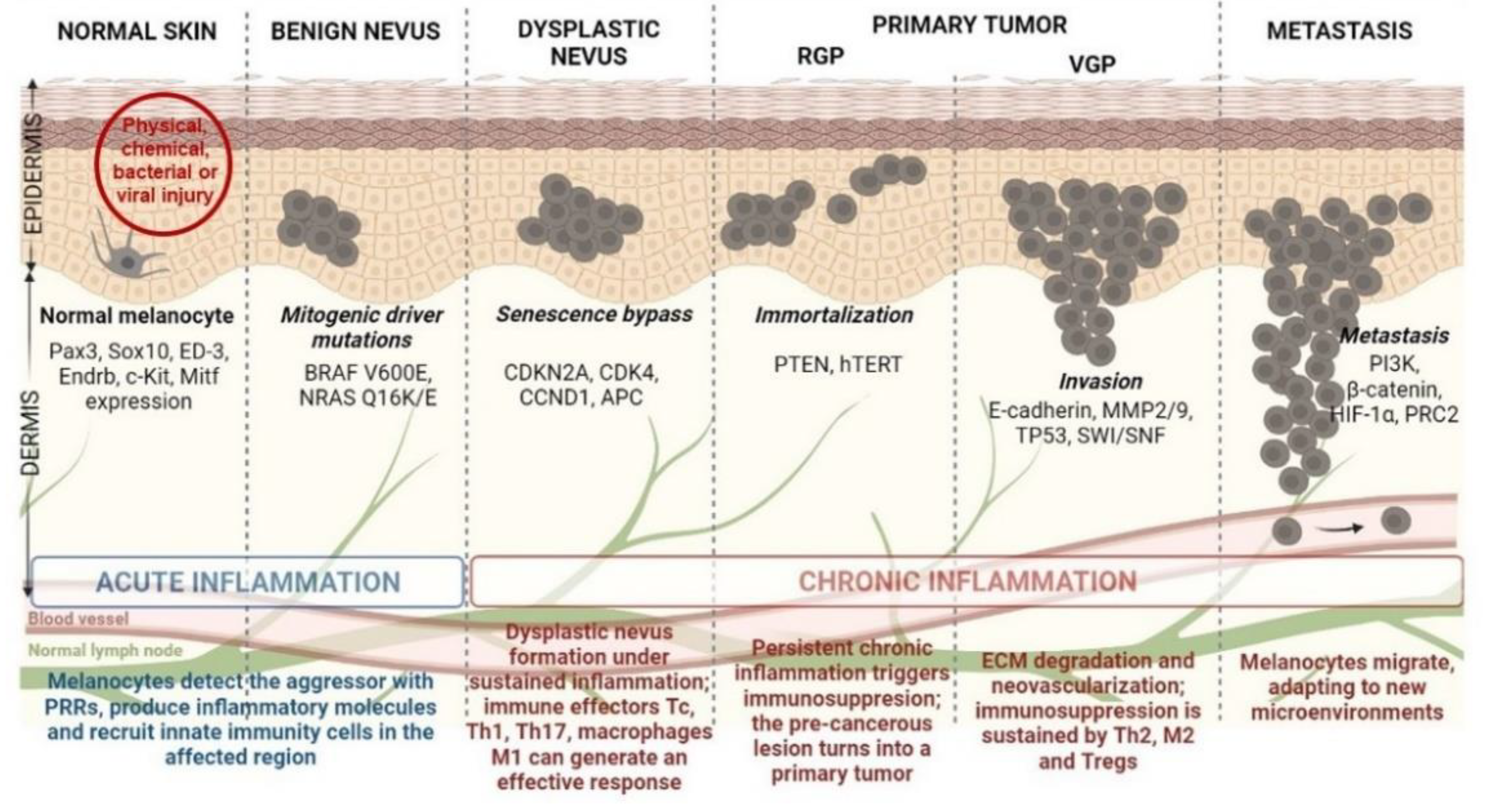
4. Epigenetic Alterations Driving CM initiation and Progression
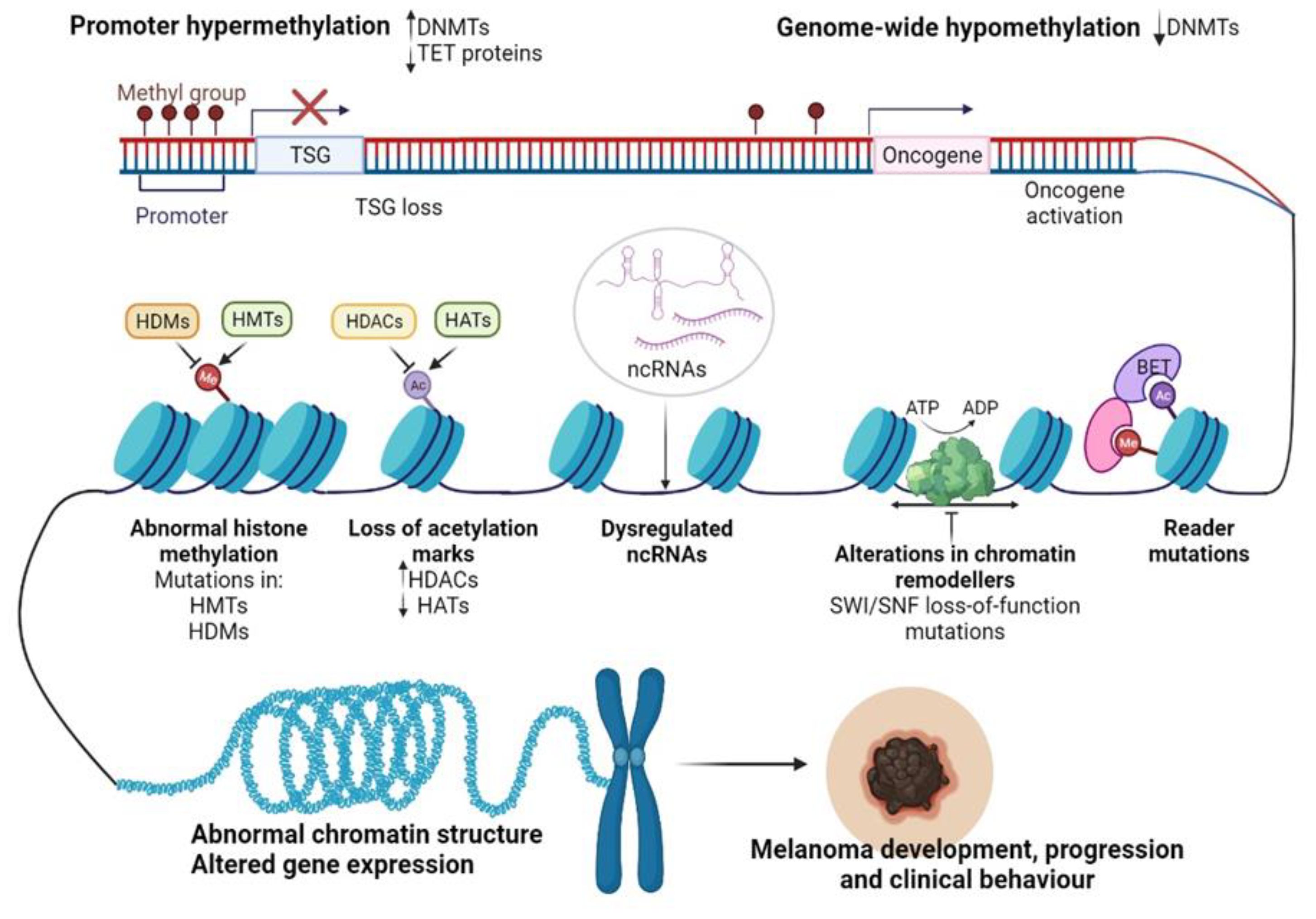
4.1. DNA Methylation in CM Development
4.2. Histone-Modifying Enzymes and PTMs in CM Development
4.2.1. Histone Modifications “Writers”
H3K4 Methyltransferases (KMT2D)
The Roles of H3K4 Methylation Marks
H3K27 Methyltransferases (EZH2)
H3K9 Methyltransferases
4.2.2. Histone Modifications “Readers”
4.2.3. Histone Modifications “Erasers”
Histone Deacetylases (HDACs)
Histone Demethylases (HDMs)
5. Epigenetic Alterations Involved in CM Drug Resistance
5.1. Resistance to MAPK Inhibitors (MAPKi)
5.1.1. DNA Methylation and MAPKi Resistance
5.1.2. Histone-Modifying Enzymes and PTMs Involved in MAPKi Resistance
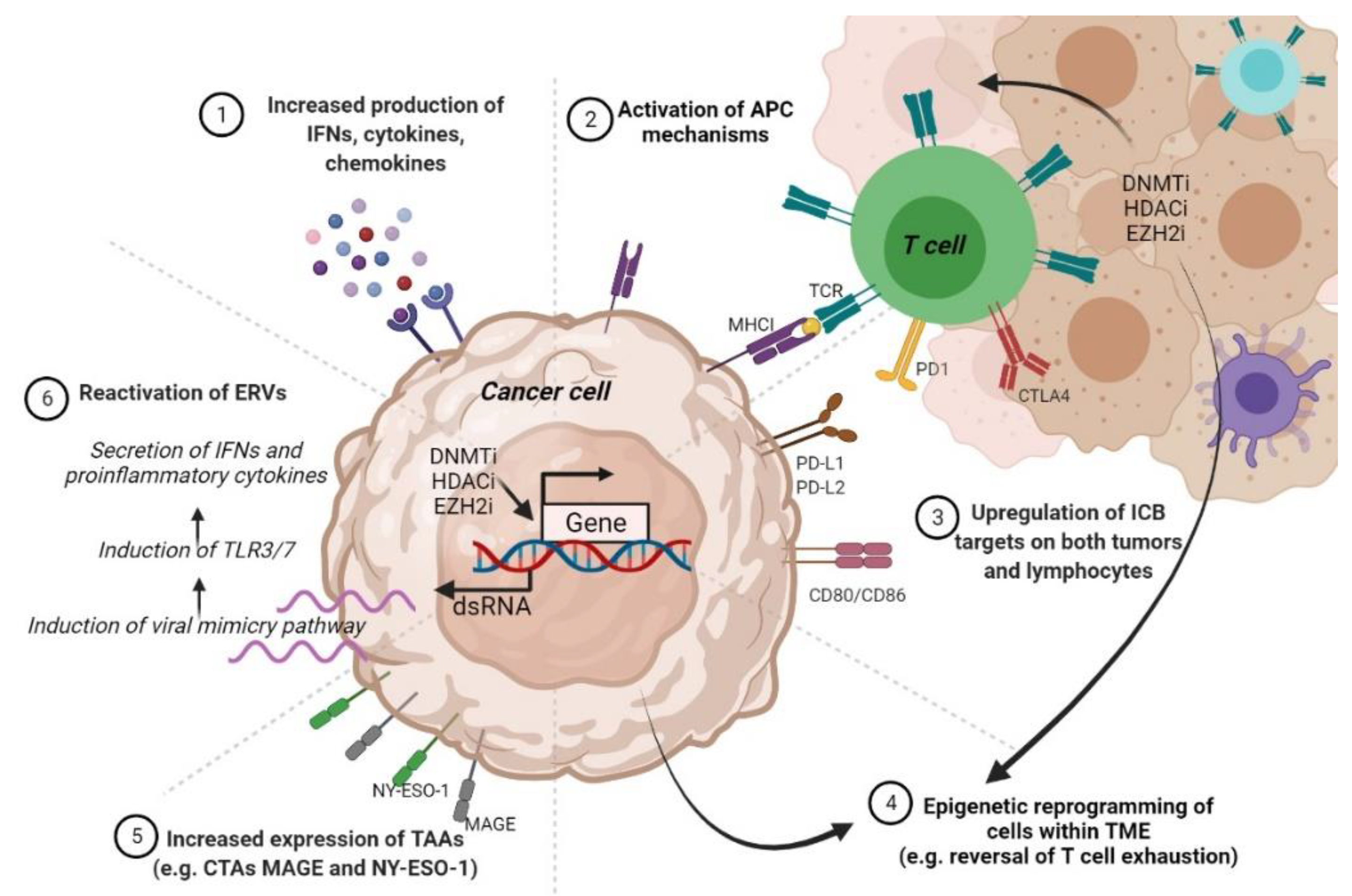
5.2. Resistance to Immunotherapy
5.2.1. DNA Methylation and Resistance to Immunotherapy
5.2.2. Histone-Modifying Enzymes and PTMs Involved in Immunotherapy Resistance
6. Epigenetics-Based Therapies for CM
6.1. Epigenetic Drugs as Monotherapies in CM
6.1.1. DNMTi
6.1.2. HDACi
6.1.3. Next-Generation Epigenetic Agents
6.2. Combinatorial Therapies in CM
6.2.1. Combinations with Targeted Therapy
6.2.2. Combinations with Immunotherapy
7. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Shain, A.H.; Bastian, B.C. From melanocytes to melanomas. Nat. Rev. Cancer 2016, 16, 345–358. [Google Scholar] [CrossRef] [PubMed]
- Leonardi, G.C.; Falzone, L.; Salemi, R.; Zanghì, A.; Spandidos, D.A.; Mccubrey, J.A.; Candido, S.; Libra, M. Cutaneous melanoma: From pathogenesis to therapy (Review). Int. J. Oncol. 2018, 52, 1071–1080. [Google Scholar] [CrossRef] [PubMed]
- Gershenwald, J.E.; Guy, G.P. Stemming the Rising Incidence of Melanoma: Calling Prevention to Action. J. Natl. Cancer Inst. 2016, 108, 381. [Google Scholar] [CrossRef]
- Rebecca, V.W.; Somasundaram, R.; Herlyn, M. Pre-clinical modeling of cutaneous melanoma. Nat. Commun. 2020, 11, 2858. [Google Scholar] [CrossRef] [PubMed]
- Chapman, P.B.; Hauschild, A.; Robert, C.; Haanen, J.B.; Ascierto, P.; Larkin, J.; Dummer, R.; Garbe, C.; Testori, A.; Maio, M.; et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 2011, 364, 2507–2516. [Google Scholar] [CrossRef]
- Long, G.V.; Weber, J.S.; Infante, J.R.; Kim, K.B.; Daud, A.; Gonzalez, R.; Sosman, J.A.; Hamid, O.; Schuchter, L.; Cebon, J.; et al. Overall Survival and Durable Responses in Patients With BRAF V600-Mutant Metastatic Melanoma Receiving Dabrafenib Combined With Trametinib. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2016, 34, 871–878. [Google Scholar] [CrossRef] [PubMed]
- Long, G.V.; Schachter, J.; Ribas, A.; Arance, A.M.; Grob, J.-J.; Mortier, L.; Daud, A.; Carlino, M.S.; McNeil, C.M.; Lotem, M.; et al. 4-year survival and outcomes after cessation of pembrolizumab (pembro) after 2-years in patients (pts) with ipilimumab (ipi)-naive advanced melanoma in KEYNOTE-006. J. Clin. Oncol. 2018, 36, 9503. [Google Scholar] [CrossRef]
- Hamid, O.; Robert, C.; Daud, A.; Hodi, F.S.; Hwu, W.J.; Kefford, R.; Wolchok, J.D.; Hersey, P.; Joseph, R.; Weber, J.S.; et al. Five-year survival outcomes for patients with advanced melanoma treated with pembrolizumab in KEYNOTE-001. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2019, 30, 582–588. [Google Scholar] [CrossRef]
- Gallagher, S.J.; Tiffen, J.C.; Hersey, P. Histone Modifications, Modifiers and Readers in Melanoma Resistance to Targeted and Immune Therapy. Cancers 2015, 7, 1959–1982. [Google Scholar] [CrossRef]
- Song, L.-B.; Zhang, Q.-J.; Hou, X.-Y.; Xiu, Y.-Y.; Chen, L.; Song, N.-H.; Lu, Y. A twelve-gene signature for survival prediction in malignant melanoma patients. Ann. Transl. Med. 2020, 8, 312. [Google Scholar] [CrossRef]
- Reuben, A.; Spencer, C.N.; Prieto, P.A.; Gopalakrishnan, V.; Reddy, S.M.; Miller, J.P.; Mao, X.; De Macedo, M.P.; Chen, J.; Song, X.; et al. Genomic and immune heterogeneity are associated with differential responses to therapy in melanoma. NPJ Genom. Med. 2017, 2, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Uslu, U.; Schliep, S.; Schliep, K.; Erdmann, M.; Koch, H.-U.; Parsch, H.; Rosenheinrich, S.; Anzengruber, D.; Bosserhoff, A.K.; Schuler, G.; et al. Comparison of the Serum Tumor Markers S100 and Melanoma-inhibitory Activity (MIA) in the Monitoring of Patients with Metastatic Melanoma Receiving Vaccination Immunotherapy with Dendritic Cells. Anticancer Res. 2017, 37, 5033–5037. [Google Scholar] [CrossRef]
- Neagu, M.; Constantin, C.; Zurac, S. Immune parameters in the prognosis and therapy monitoring of cutaneous melanoma patients: Experience, role, and limitations. Biomed Res. Int. 2013, 2013, 107940. [Google Scholar] [CrossRef] [PubMed]
- Petrelli, F.; Ardito, R.; Merelli, B.; Lonati, V.; Cabiddu, M.; Seghezzi, S.; Barni, S.; Ghidini, A. Prognostic and predictive role of elevated lactate dehydrogenase in patients with melanoma treated with immunotherapy and BRAF inhibitors: A systematic review and meta-analysis. Melanoma Res. 2019, 29, 1–12. [Google Scholar] [CrossRef]
- Georgescu, S.R.; Tampa, M.; Mitran, C.I.; Mitran, M.I.; Caruntu, C.; Caruntu, A.; Lupu, M.; Matei, C.; Constantin, C.; Neagu, M. Tumour Microenvironment in Skin Carcinogenesis. Adv. Exp. Med. Biol. 2020, 1226, 123–142. [Google Scholar] [CrossRef] [PubMed]
- Clark, W.H.J.; Elder, D.E.; Guerry, D., 4th; Epstein, M.N.; Greene, M.H.; Van Horn, M. A study of tumor progression: The precursor lesions of superficial spreading and nodular melanoma. Hum. Pathol. 1984, 15, 1147–1165. [Google Scholar] [CrossRef]
- Schiferle, E.B.; Cheon, S.Y.; Ham, S.; Son, H.G.; Messerschmidt, J.L.; Lawrence, D.P.; Cohen, J.V.; Flaherty, K.T.; Moon, J.J.; Lian, C.G.; et al. Rejection of benign melanocytic nevi by nevus-resident CD4(+) T cells. Sci. Adv. 2021, 7. [Google Scholar] [CrossRef] [PubMed]
- Neagu, M.; Constantin, C.; Engin, A.B.; Popescu, I. Snapshot-changing melanocyte identity in melanoma developing route, 2020. J. Cell Identity 2020, 1, 33–47. [Google Scholar] [CrossRef]
- Reiter, O.; Kurtansky, N.; Nanda, J.K.; Busam, K.J.; Scope, A.; Musthaq, S.; Marghoob, A.A. The differences in clinical and dermoscopic features between in situ and invasive nevus-associated melanomas and de novo melanomas. J. Eur. Acad. Dermatol. Venereol. 2021, 35, 1111–1118. [Google Scholar] [CrossRef]
- Pampena, R.; Kyrgidis, A.; Lallas, A.; Moscarella, E.; Argenziano, G.; Longo, C. A meta-analysis of nevus-associated melanoma: Prevalence and practical implications. J. Am. Acad. Dermatol. 2017, 77, 938–945.e4. [Google Scholar] [CrossRef]
- Vezzoni, R.; Conforti, C.; Vichi, S.; Giuffrida, R.; Retrosi, C.; Magaton-Rizzi, G.; Di Meo, N.; Pizzichetta, M.A.; Zalaudek, I. Is There More Than One Road to Nevus-Associated Melanoma? Dermatol. Pract. Concept. 2020, 10, e2020028. [Google Scholar] [CrossRef] [PubMed]
- Cymerman, R.M.; Shao, Y.; Wang, K.; Zhang, Y.; Murzaku, E.C.; Penn, L.A.; Osman, I.; Polsky, D. De Novo vs. Nevus-Associated Melanomas: Differences in Associations With Prognostic Indicators and Survival. J. Natl. Cancer Inst. 2016, 108. [Google Scholar] [CrossRef] [PubMed]
- de Souza, C.F.; Morais, A.S.; Jasiulionis, M.G. Biomarkers as Key Contributors in Treating Malignant Melanoma Metastases. Dermatol. Res. Pract. 2012, 2012, 156068. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, A.P.; Ohlsson, R.; Henikoff, S. The epigenetic progenitor origin of human cancer. Nat. Rev. Genet. 2006, 7, 21–33. [Google Scholar] [CrossRef]
- Bruschi, M. The Epigenetic Progenitor Origin of Cancer Reassessed: DNA Methylation Brings Balance to the Stem Force. Epigenomes 2020, 4, 8. [Google Scholar] [CrossRef]
- Voiculescu, V.M.; Lisievici, C.V.; Lupu, M.; Vajaitu, C.; Draghici, C.C.; Popa, A.V.; Solomon, I.; Sebe, T.I.; Constantin, M.M.; Caruntu, C. Mediators of Inflammation in Topical Therapy of Skin Cancers. Mediat. Inflamm. 2019, 2019, 8369690. [Google Scholar] [CrossRef] [PubMed]
- Ciążyńska, M.; Olejniczak-Staruch, I.; Sobolewska-Sztychny, D.; Narbutt, J.; Skibińska, M.; Lesiak, A. Ultraviolet Radiation and Chronic Inflammation-Molecules and Mechanisms Involved in Skin Carcinogenesis: A Narrative Review. Life 2021, 11, 326. [Google Scholar] [CrossRef]
- Neagu, M.; Constantin, C.; Caruntu, C.; Dumitru, C.; Surcel, M.; Zurac, S. Inflammation: A key process in skin tumorigenesis. Oncol. Lett. 2019, 17, 4068–4084. [Google Scholar] [CrossRef] [PubMed]
- Sang, Y.; Deng, Y. Current insights into the epigenetic mechanisms of skin cancer. Dermatol. Ther. 2019, 32, e12964. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Cheng, P.; Jiang, J.; Ren, Y.; Wu, D.; Xue, D. Epigenomic and genomic analysis of transcriptome modulation in skin cutaneous melanoma. Aging 2020, 12, 12703–12725. [Google Scholar] [CrossRef]
- Lee, J.J.; Murphy, G.F.; Lian, C.G. Melanoma epigenetics: Novel mechanisms, markers, and medicines. Lab. Investig. 2014, 94, 822–838. [Google Scholar] [CrossRef]
- Besaratinia, A.; Tommasi, S. Epigenetics of human melanoma: Promises and challenges. J. Mol. Cell Biol. 2014, 6, 356–367. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lauss, M.; Ringnér, M.; Karlsson, A.; Harbst, K.; Busch, C.; Geisler, J.; Lønning, P.E.; Staaf, J.; Jönsson, G. DNA methylation subgroups in melanoma are associated with proliferative and immunological processes. BMC Med. Genom. 2015, 8, 73. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Matsusaka, K.; Fukuyo, M.; Rahmutulla, B.; Matsue, H.; Kaneda, A. Higher methylation subtype of malignant melanoma and its correlation with thicker progression and worse prognosis. Cancer Med. 2020, 9, 7194–7204. [Google Scholar] [CrossRef] [PubMed]
- Wilmott, J.S.; Colebatch, A.J.; Kakavand, H.; Shang, P.; Carlino, M.S.; Thompson, J.F.; Long, G.V.; Scolyer, R.A.; Hersey, P. Expression of the class 1 histone deacetylases HDAC8 and 3 are associated with improved survival of patients with metastatic melanoma. Mod. Pathol. Off. J. USA Can. Acad. Pathol. Inc. 2015, 28, 884–894. [Google Scholar] [CrossRef] [PubMed]
- Park, W.-Y.; Hong, B.-J.; Lee, J.; Choi, C.; Kim, M.-Y. H3K27 Demethylase JMJD3 Employs the NF-κB and BMP Signaling Pathways to Modulate the Tumor Microenvironment and Promote Melanoma Progression and Metastasis. Cancer Res. 2016, 76, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.J.; Sholl, L.M.; Lindeman, N.I.; Granter, S.R.; Laga, A.C.; Shivdasani, P.; Chin, G.; Luke, J.J.; Ott, P.A.; Hodi, F.S.; et al. Targeted next-generation sequencing reveals high frequency of mutations in epigenetic regulators across treatment-naïve patient melanomas. Clin. Epigenetics 2015, 7, 59. [Google Scholar] [CrossRef] [PubMed]
- Bossi, D.; Cicalese, A.; Dellino, G.I.; Luzi, L.; Riva, L.; D’Alesio, C.; Diaferia, G.R.; Carugo, A.; Cavallaro, E.; Piccioni, R.; et al. In Vivo Genetic Screens of Patient-Derived Tumors Revealed Unexpected Frailty of the Transformed Phenotype. Cancer Discov. 2016, 6, 650–663. [Google Scholar] [CrossRef] [PubMed]
- Morel, D.; Jeffery, D.; Aspeslagh, S.; Almouzni, G.; Postel-Vinay, S. Combining epigenetic drugs with other therapies for solid tumours past lessons and future promise. Nat. Rev. Clin. Oncol. 2020, 17, 91–107. [Google Scholar] [CrossRef]
- Sharma, S.; Kelly, T.K.; Jones, P.A. Epigenetics in cancer. Carcinogenesis 2010, 31, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Payer, B.; Lee, J.T.; Namekawa, S.H. X-inactivation and X-reactivation: Epigenetic hallmarks of mammalian reproduction and pluripotent stem cells. Hum. Genet. 2011, 130, 265–280. [Google Scholar] [CrossRef]
- Dobre, E.-G.; Dinescu, S.; Costache, M. Connecting the missing dots: Ncrnas as critical regulators of therapeutic susceptibility in breast cancer. Cancers 2020, 12, 2698. [Google Scholar] [CrossRef] [PubMed]
- Ferrand, J.; Rondinelli, B.; Polo, S.E. Histone Variants: Guardians of Genome Integrity. Cells 2020, 9, 2424. [Google Scholar] [CrossRef]
- Hasan, N.; Ahuja, N. The Emerging Roles of ATP-Dependent Chromatin Remodeling Complexes in Pancreatic Cancer. Cancers 2019, 11, 1859. [Google Scholar] [CrossRef]
- Dueñas-González, A.; Lizano, M.; Candelaria, M.; Cetina, L.; Arce, C.; Cervera, E. Epigenetics of cervical cancer. An overview and therapeutic perspectives. Mol. Cancer 2005, 4, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Mitsis, T.; Efthimiadou, A.; Bacopoulou, F.; Vlachakis, D.; Chrousos, G.; Eliopoulos, E. Transcription factors and evolution: An integral part of gene expression (Review). World Acad. Sci. J. 2020, 2, 3–8. [Google Scholar] [CrossRef]
- Gillette, T.G.; Hill, J.A. Readers, writers, and erasers: Chromatin as the whiteboard of heart disease. Circ. Res. 2015, 116, 1245–1253. [Google Scholar] [CrossRef]
- Hoffmann, F.; Zarbl, R.; Niebel, D.; Sirokay, J.; Fröhlich, A.; Posch, C.; Holderried, T.A.W.; Brossart, P.; Saavedra, G.; Kuster, P.; et al. Prognostic and predictive value of PD-L2 DNA methylation and mRNA expression in melanoma. Clin. Epigenetics 2020, 12, 94. [Google Scholar] [CrossRef] [PubMed]
- Grigore, F.; Yang, H.; Hanson, N.D.; VanBrocklin, M.W.; Sarver, A.L.; Robinson, J.P. BRAF inhibition in melanoma is associated with the dysregulation of histone methylation and histone methyltransferases. Neoplasia 2020, 22, 376–389. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M. CpG island hypermethylation and tumor suppressor genes: A booming present, a brighter future. Oncogene 2002, 21, 5427–5440. [Google Scholar] [CrossRef]
- Fatemi, M.; Pao, M.M.; Jeong, S.; Gal-Yam, E.N.; Egger, G.; Weisenberger, D.J.; Jones, P.A. Footprinting of mammalian promoters: Use of a CpG DNA methyltransferase revealing nucleosome positions at a single molecule level. Nucleic Acids Res. 2005, 33, e176. [Google Scholar] [CrossRef]
- Shen, H.; Laird, P.W. Interplay between the cancer genome and epigenome. Cell 2013, 153, 38–55. [Google Scholar] [CrossRef]
- Jin, B.; Robertson, K.D. DNA methyltransferases, DNA damage repair, and cancer. Adv. Exp. Med. Biol. 2013, 754, 3–29. [Google Scholar] [CrossRef] [PubMed]
- Kulis, M.; Esteller, M. DNA methylation and cancer. Adv. Genet. 2010, 70, 27–56. [Google Scholar] [CrossRef]
- Kohli, R.M.; Zhang, Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature 2013, 502, 472–479. [Google Scholar] [CrossRef]
- Stenz, L.; Schechter, D.S.; Serpa, S.R.; Paoloni-Giacobino, A. Intergenerational Transmission of DNA Methylation Signatures Associated with Early Life Stress. Curr. Genom. 2018, 19, 665–675. [Google Scholar] [CrossRef]
- Emran, A.A.; Chatterjee, A.; Rodger, E.J.; Tiffen, J.C.; Gallagher, S.J.; Eccles, M.R.; Hersey, P. Targeting DNA Methylation and EZH2 Activity to Overcome Melanoma Resistance to Immunotherapy. Trends Immunol. 2019, 40, 328–344. [Google Scholar] [CrossRef]
- Gross, D.S.; Chowdhary, S.; Anandhakumar, J.; Kainth, A.S. Chromatin. Curr. Biol. 2015, 25, R1158–R1163. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hammerlindl, H.; Schaider, H. Epigenetics in Melanoma Development and Drug Resistance. In Human Skin Cancers Pathways, Mechanisms, Targets and Treatments; InTech: Tokyo, Japan, 2018. [Google Scholar]
- Greer, E.L.; Shi, Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012, 13, 343–357. [Google Scholar] [CrossRef]
- Aparicio Pelaz, D.; Yerkesh, Z.; Kirchgäßner, S.; Mahler, H.; Kharchenko, V.; Azhibek, D.; Jaremko, M.; Mootz, H.D.; Jaremko, Ł.; Schwarzer, D.; et al. Examining histone modification crosstalk using immobilized libraries established from ligation-ready nucleosomes. Chem. Sci. 2020, 11, 9218–9225. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Shilatifard, A. Epigenetic modifications of histones in cancer. Genome Biol. 2019, 20, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Pradeepa, M.M. Causal role of histone acetylations in enhancer function. Transcription 2017, 8, 40–47. [Google Scholar] [CrossRef]
- Kumar, D.; Jothi, R. Bivalent chromatin protects reversibly repressed genes from irreversible silencing. bioRxiv 2020. [Google Scholar] [CrossRef]
- Dunican, D.S.; Mjoseng, H.K.; Duthie, L.; Flyamer, I.M.; Bickmore, W.A.; Meehan, R.R. Bivalent promoter hypermethylation in cancer is linked to the H327me3/H3K4me3 ratio in embryonic stem cells. BMC Biol. 2020, 18, 25. [Google Scholar] [CrossRef]
- Ghosh, K.; O’Neil, K.; Capell, B.C. Histone modifiers: Dynamic regulators of the cutaneous transcriptome. J. Dermatol. Sci. 2018, 89, 226–232. [Google Scholar] [CrossRef]
- Calcagno, D.Q.; Gigek, C.O.; Chen, E.S.; Burbano, R.R.; de Arruda Cardoso Smith, M. DNA and histone methylation in gastric carcinogenesis. World J. Gastroenterol. 2013, 19, 1182–1192. [Google Scholar] [CrossRef] [PubMed]
- Yeon, M.; Kim, Y.; Jung, H.S.; Jeoung, D. Histone Deacetylase Inhibitors to Overcome Resistance to Targeted and Immuno Therapy in Metastatic Melanoma. Front. Cell Dev. Biol. 2020, 8, 486. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, H.; Pessoa, G.C.; De Luna Vitorino, F.N.; Nsengimana, J.; Newton-Bishop, J.; Reis, E.M.; Da Cunha, J.P.C.; Jasiulionis, M.G. Gene co-expression and histone modification signatures are associated with melanoma progression, epithelial-to-mesenchymal transition, and metastasis. Clin. Epigenetics 2020, 12, 127. [Google Scholar] [CrossRef] [PubMed]
- Farooqi, A.A.; Fayyaz, S.; Poltronieri, P.; Calin, G.; Mallardo, M. Epigenetic deregulation in cancer: Enzyme players and non-coding RNAs. Semin. Cancer Biol. 2020. [Google Scholar] [CrossRef]
- de Unamuno Bustos, B.; Murria Estal, R.; Pérez Simó, G.; Simarro Farinos, J.; Pujol Marco, C.; Navarro Mira, M.; Alegre de Miquel, V.; Ballester Sánchez, R.; Sabater Marco, V.; Llavador Ros, M.; et al. Aberrant DNA methylation is associated with aggressive clinicopathological features and poor survival in cutaneous melanoma. Br. J. Dermatol. 2018, 179, 394–404. [Google Scholar] [CrossRef]
- Kampilafkos, P.; Melachrinou, M.; Kefalopoulou, Z.; Lakoumentas, J.; Sotiropoulou-Bonikou, G. Epigenetic modifications in cutaneous malignant melanoma: EZH2, H3K4me2, and H3K27me3 immunohistochemical expression is enhanced at the invasion front of the tumor. Am. J. Dermatopathol. 2015, 37, 138–144. [Google Scholar] [CrossRef]
- Maitituoheti, M.; Keung, E.Z.; Tang, M.; Yan, L.; Alam, H.; Han, G.; Singh, A.K.; Raman, A.T.; Terranova, C.; Sarkar, S.; et al. Enhancer Reprogramming Confers Dependence on Glycolysis and IGF Signaling in KMT2D Mutant Melanoma. Cell Rep. 2020, 33, 108293. [Google Scholar] [CrossRef] [PubMed]
- Neagu, M.; Constantin, C.; Cretoiu, S.M.; Zurac, S. miRNAs in the Diagnosis and Prognosis of Skin Cancer. Front. Cell Dev. Biol. 2020, 8, 71. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Schleich, K.; Yue, B.; Ji, S.; Lohneis, P.; Kemper, K.; Silvis, M.R.; Qutob, N.; van Rooijen, E.; Werner-Klein, M.; et al. Targeting the Senescence-Overriding Cooperative Activity of Structurally Unrelated H3K9 Demethylases in Melanoma. Cancer Cell 2018, 33, 322–336.e8. [Google Scholar] [CrossRef] [PubMed]
- Zingg, D.; Debbache, J.; Schaefer, S.M.; Tuncer, E.; Frommel, S.C.; Cheng, P.; Arenas-Ramirez, N.; Haeusel, J.; Zhang, Y.; Bonalli, M.; et al. The epigenetic modifier EZH2 controls melanoma growth and metastasis through silencing of distinct tumour suppressors. Nat. Commun. 2015, 6, 6051. [Google Scholar] [CrossRef] [PubMed]
- Zingg, D.; Debbache, J.; Peña-Hernández, R.; Antunes, A.T.; Schaefer, S.M.; Cheng, P.F.; Zimmerli, D.; Haeusel, J.; Calçada, R.R.; Tuncer, E.; et al. EZH2-Mediated Primary Cilium Deconstruction Drives Metastatic Melanoma Formation. Cancer Cell 2018, 34, 69–84.e14. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, S.-M.; McGeary, M.K.; Krykbaeva, I.; Lai, L.; Jansen, D.J.; Kales, S.C.; Simeonov, A.; Hall, M.D.; Kelly, D.P.; et al. KDM5B Promotes Drug Resistance by Regulating Melanoma-Propagating Cell Subpopulations. Mol. Cancer Ther. 2019, 18, 706–717. [Google Scholar] [CrossRef] [PubMed]
- Licht, J.D.; Bennett, R.L. Leveraging epigenetics to enhance the efficacy of immunotherapy. Clin. Epigenetics 2021, 13, 115. [Google Scholar] [CrossRef]
- Maiuri, A.R.; O’Hagan, H.M. Interplay Between Inflammation and Epigenetic Changes in Cancer. Prog. Mol. Biol. Transl. Sci. 2016, 144, 69–117. [Google Scholar] [CrossRef]
- Maes, K.; Mondino, A.; Lasarte, J.J.; Agirre, X.; Vanderkerken, K.; Prosper, F.; Breckpot, K. Epigenetic Modifiers: Anti-Neoplastic Drugs With Immunomodulating Potential. Front. Immunol. 2021, 12, 652160. [Google Scholar] [CrossRef] [PubMed]
- Kominsky, D.J.; Campbell, E.L.; Colgan, S.P. Metabolic shifts in immunity and inflammation. J. Immunol. 2010, 184, 4062–4068. [Google Scholar] [CrossRef]
- Johnson, C.; Warmoes, M.O.; Shen, X.; Locasale, J.W. Epigenetics and cancer metabolism. Cancer Lett. 2015, 356, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Franco, R.; Schoneveld, O.; Georgakilas, A.G.; Panayiotidis, M.I. Oxidative stress, DNA methylation and carcinogenesis. Cancer Lett. 2008, 266, 6–11. [Google Scholar] [CrossRef]
- Fan, Y.; Mao, R.; Yang, J. NF-κB and STAT3 signaling pathways collaboratively link inflammation to cancer. Protein Cell 2013, 4, 176–185. [Google Scholar] [CrossRef]
- Micevic, G.; Theodosakis, N.; Bosenberg, M. Aberrant DNA methylation in melanoma: Biomarker and therapeutic opportunities. Clin. Epigenetics 2017, 9, 34. [Google Scholar] [CrossRef]
- Howell, P.M.J.; Liu, S.; Ren, S.; Behlen, C.; Fodstad, O.; Riker, A.I. Epigenetics in human melanoma. Cancer Control 2009, 16, 200–218. [Google Scholar] [CrossRef] [PubMed]
- Hoon, D.S.B.; Spugnardi, M.; Kuo, C.; Huang, S.K.; Morton, D.L.; Taback, B. Profiling epigenetic inactivation of tumor suppressor genes in tumors and plasma from cutaneous melanoma patients. Oncogene 2004, 23, 4014–4022. [Google Scholar] [CrossRef] [PubMed]
- Palanca-Ballester, C.; Rodriguez-Casanova, A.; Torres, S.; Calabuig-Fariñas, S.; Exposito, F.; Serrano, D.; Redin, E.; Valencia, K.; Jantus-Lewintre, E.; Diaz-Lagares, A.; et al. Cancer Epigenetic Biomarkers in Liquid Biopsy for High Incidence Malignancies. Cancers 2021, 13, 3016. [Google Scholar] [CrossRef] [PubMed]
- Tanemura, A.; Terando, A.M.; Sim, M.-S.; van Hoesel, A.Q.; de Maat, M.F.G.; Morton, D.L.; Hoon, D.S.B. CpG island methylator phenotype predicts progression of malignant melanoma. Clin. cancer Res. an Off. J. Am. Assoc. Cancer Res. 2009, 15, 1801–1807. [Google Scholar] [CrossRef]
- Garcia-Alvarez, A.; Ortiz, C.; Muñoz-Couselo, E. Current Perspectives and Novel Strategies of NRAS-Mutant Melanoma. Onco. Targets. Ther. 2021, 14, 3709–3719. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, M. DNA hypomethylation in cancer cells. Epigenomics 2009, 1, 239–259. [Google Scholar] [CrossRef]
- Ponomaryova, A.A.; Rykova, E.Y.; Gervas, P.A.; Cherdyntseva, N.V.; Mamedov, I.Z.; Azhikina, T.L. Aberrant Methylation of LINE-1 Transposable Elements: A Search for Cancer Biomarkers. Cells 2020, 9, 2017. [Google Scholar] [CrossRef] [PubMed]
- Ecsedi, S.I.; Hernandez-Vargas, H.; Lima, S.C.; Herceg, Z.; Adany, R.; Balazs, M. Transposable hypomethylation is associated with metastatic capacity of primary melanomas. Int. J. Clin. Exp. Pathol. 2013, 6, 2943–2948. [Google Scholar] [PubMed]
- De Araújo, É.S.S.; Kashiwabara, A.Y.; Achatz, M.I.W.; Moredo, L.F.; De Sá, B.C.S.; Duprat, J.P.; Rosenberg, C.; Carraro, D.M.; Krepischi, A.C. V LINE-1 hypermethylation in peripheral blood of cutaneous melanoma patients is associated with metastasis. Melanoma Res. 2015, 25, 173–177. [Google Scholar] [CrossRef] [PubMed]
- Sigalotti, L.; Fratta, E.; Bidoli, E.; Covre, A.; Parisi, G.; Colizzi, F.; Coral, S.; Massarut, S.; Kirkwood, J.M.; Maio, M. Methylation levels of the “long interspersed nucleotide element-1” repetitive sequences predict survival of melanoma patients. J. Transl. Med. 2011, 9, 78. [Google Scholar] [CrossRef] [PubMed]
- Tonella, L.; Pala, V.; Ponti, R.; Rubatto, M.; Gallo, G.; Mastorino, L.; Avallone, G.; Merli, M.; Agostini, A.; Fava, P.; et al. Prognostic and Predictive Biomarkers in Stage III Melanoma: Current Insights and Clinical Implications. Int. J. Mol. Sci. 2021, 22, 4561. [Google Scholar] [CrossRef] [PubMed]
- Colemon, A.; Harris, T.M.; Ramanathan, S. DNA hypomethylation drives changes in MAGE-A gene expression resulting in alteration of proliferative status of cells. Genes Environ. Off. J. Japanese Environ. Mutagen Soc. 2020, 42, 24. [Google Scholar] [CrossRef]
- Faramarzi, S.; Ghafouri-Fard, S. Melanoma: A prototype of cancer-testis antigen-expressing malignancies. Immunotherapy 2017, 9, 1103–1113. [Google Scholar] [CrossRef]
- Sigalotti, L.; Coral, S.; Nardi, G.; Spessotto, A.; Cortini, E.; Cattarossi, I.; Colizzi, F.; Altomonte, M.; Maio, M. Promoter methylation controls the expression of MAGE2, 3 and 4 genes in human cutaneous melanoma. J. Immunother. 2002, 25, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Sigalotti, L.; Covre, A.; Zabierowski, S.; Himes, B.; Colizzi, F.; Natali, P.G.; Herlyn, M.; Maio, M. Cancer testis antigens in human melanoma stem cells: Expression, distribution, and methylation status. J. Cell. Physiol. 2008, 215, 287–291. [Google Scholar] [CrossRef]
- Danilova, A.; Misyurin, V.; Novik, A.; Girdyuk, D.; Avdonkina, N.; Nekhaeva, T.; Emelyanova, N.; Pipia, N.; Misyurin, A.; Baldueva, I. Cancer/testis antigens expression during cultivation of melanoma and soft tissue sarcoma cells. Clin. Sarcoma Res. 2020, 10, 3. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, S.; Nagai, H.; Jimbo, H.; Jimbo, N.; Tanaka, T.; Inoie, M.; Nishigori, C. Gene Expression and Methylation Analysis in Melanomas and Melanocytes From the Same Patient: Loss of NPM2 Expression Is a Potential Immunohistochemical Marker for Melanoma. Front. Oncol. 2018, 8, 675. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; van den Hurk, K.; Moerkerk, P.T.M.; Goeman, J.J.; Beck, S.; Gruis, N.A.; van den Oord, J.J.; Winnepenninckx, V.J.; van Engeland, M.; van Doorn, R. Promoter CpG island hypermethylation in dysplastic nevus and melanoma: CLDN11 as an epigenetic biomarker for malignancy. J. Invest. Dermatol. 2014, 134, 2957–2966. [Google Scholar] [CrossRef]
- Marzese, D.M.; Scolyer, R.A.; Huynh, J.L.; Huang, S.K.; Hirose, H.; Chong, K.K.; Kiyohara, E.; Wang, J.; Kawas, N.P.; Donovan, N.C.; et al. Epigenome-wide DNA methylation landscape of melanoma progression to brain metastasis reveals aberrations on homeobox D cluster associated with prognosis. Hum. Mol. Genet. 2014, 23, 226–238. [Google Scholar] [CrossRef] [PubMed]
- Lian, C.G.; Xu, Y.; Ceol, C.; Wu, F.; Larson, A.; Dresser, K.; Xu, W.; Tan, L.; Hu, Y.; Zhan, Q.; et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell 2012, 150, 1135–1146. [Google Scholar] [CrossRef] [PubMed]
- Saldanha, G.; Joshi, K.; Lawes, K.; Bamford, M.; Moosa, F.; Teo, K.W.; Pringle, J.H. 5-Hydroxymethylcytosine is an independent predictor of survival in malignant melanoma. Mod. Pathol. Off. J. United States Can. Acad. Pathol. Inc 2017, 30, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Khaliq, M.; Fallahi-Sichani, M. Epigenetic Mechanisms of Escape from BRAF Oncogene Dependency. Cancers 2019, 11, 1480. [Google Scholar] [CrossRef] [PubMed]
- Strub, T.; Ballotti, R.; Bertolotto, C. The “ART” of Epigenetics in Melanoma: From histone “Alterations, to Resistance and Therapies”. Theranostics 2020, 10, 1777–1797. [Google Scholar] [CrossRef] [PubMed]
- Frantz, W.T.; Ceol, C.J. From Tank to Treatment: Modeling Melanoma in Zebrafish. Cells 2020, 9, 1289. [Google Scholar] [CrossRef] [PubMed]
- Patton, E.E.; Widlund, H.R.; Kutok, J.L.; Kopani, K.R.; Amatruda, J.F.; Murphey, R.D.; Berghmans, S.; Mayhall, E.A.; Traver, D.; Fletcher, C.D.M.; et al. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr. Biol. 2005, 15, 249–254. [Google Scholar] [CrossRef]
- Scahill, C.M.; Digby, Z.; Sealy, I.M.; Wojciechowska, S.; White, R.J.; Collins, J.E.; Stemple, D.L.; Bartke, T.; Mathers, M.E.; Patton, E.E.; et al. Loss of the chromatin modifier Kdm2aa causes BrafV600E-independent spontaneous melanoma in zebrafish. PLoS Genet. 2017, 13, e1006959. [Google Scholar] [CrossRef]
- Herz, H.-M.; Mohan, M.; Garruss, A.S.; Liang, K.; Takahashi, Y.-H.; Mickey, K.; Voets, O.; Verrijzer, C.P.; Shilatifard, A. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev. 2012, 26, 2604–2620. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Lee, J.-E.; Lai, B.; Macfarlan, T.S.; Xu, S.; Zhuang, L.; Liu, C.; Peng, W.; Ge, K. Enhancer priming by H3K4 methyltransferase MLL4 controls cell fate transition. Proc. Natl. Acad. Sci. USA 2016, 113, 11871–11876. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.S.; Rai, K.; Garber, M.; Hollinger, A.; Robbins, D.; Anderson, S.; Macbeth, A.; Tzou, A.; Carneiro, M.O.; Raychowdhury, R.; et al. Semiconductor-based DNA sequencing of histone modification states. Nat. Commun. 2013, 4, 2672. [Google Scholar] [CrossRef] [PubMed]
- Gu, B.; Lee, M.G. Histone H3 lysine 4 methyltransferases and demethylases in self-renewal and differentiation of stem cells. Cell Biosci. 2013, 3, 39. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, B.E.; Kamal, M.; Lindblad-Toh, K.; Bekiranov, S.; Bailey, D.K.; Huebert, D.J.; McMahon, S.; Karlsson, E.K.; Kulbokas, E.J., 3rd; Gingeras, T.R.; et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 2005, 120, 169–181. [Google Scholar] [CrossRef]
- Uzdensky, A.; Demyanenko, S.; Bibov, M.; Sharifulina, S.; Kit, O.; Przhedetski, Y.; Pozdnyakova, V. Expression of proteins involved in epigenetic regulation in human cutaneous melanoma and peritumoral skin. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2014, 35, 8225–8233. [Google Scholar] [CrossRef]
- Terranova, C.; Tang, M.; Maitituoheti, M.; Raman, A.T.; Schulz, J.; Amin, S.B.; Orouji, E.; Tomczak, K.; Sarkar, S.; Oba, J.; et al. Bivalent and broad chromatin domains regulate pro-metastatic drivers in melanoma. bioRxiv 2019, 721480. [Google Scholar] [CrossRef]
- Neagu, M.; Constantin, C.; Bostan, M.; Caruntu, C.; Ignat, S.R.; Dinescu, S.; Costache, M. Proteomic Technology “Lens” for Epithelial-Mesenchymal Transition Process Identification in Oncology. Anal. Cell. Pathol. 2019, 2019, 3565970. [Google Scholar] [CrossRef]
- Hoffmann, F.; Niebel, D.; Aymans, P.; Ferring-Schmitt, S.; Dietrich, D.; Landsberg, J. H3K27me3 and EZH2 expression in melanoma: Relevance for melanoma progression and response to immune checkpoint blockade. Clin. Epigenetics 2020, 12, 24. [Google Scholar] [CrossRef] [PubMed]
- McHugh, J.B.; Fullen, D.R.; Ma, L.; Kleer, C.G.; Su, L.D. Expression of polycomb group protein EZH2 in nevi and melanoma. J. Cutan. Pathol. 2007, 34, 597–600. [Google Scholar] [CrossRef]
- Bachmann, I.M.; Halvorsen, O.J.; Collett, K.; Stefansson, I.M.; Straume, O.; Haukaas, S.A.; Salvesen, H.B.; Otte, A.P.; Akslen, L.A. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2006, 24, 268–273. [Google Scholar] [CrossRef] [PubMed]
- Fan, T.; Jiang, S.; Chung, N.; Alikhan, A.; Ni, C.; Lee, C.-C.R.; Hornyak, T.J. EZH2-dependent suppression of a cellular senescence phenotype in melanoma cells by inhibition of p21/CDKN1A expression. Mol. Cancer Res. 2011, 9, 418–429. [Google Scholar] [CrossRef] [PubMed]
- De Donatis, G.M.; Le Pape, E.; Pierron, A.; Cheli, Y.; Hofman, V.; Hofman, P.; Allegra, M.; Zahaf, K.; Bahadoran, P.; Rocchi, S.; et al. NF-kB2 induces senescence bypass in melanoma via a direct transcriptional activation of EZH2. Oncogene 2016, 35, 2735–2745. [Google Scholar] [CrossRef] [PubMed]
- Barsotti, A.M.; Ryskin, M.; Zhong, W.; Zhang, W.-G.; Giannakou, A.; Loreth, C.; Diesl, V.; Follettie, M.; Golas, J.; Lee, M.; et al. Epigenetic reprogramming by tumor-derived EZH2 gain-of-function mutations promotes aggressive 3D cell morphologies and enhances melanoma tumor growth. Oncotarget 2015, 6, 2928–2938. [Google Scholar] [CrossRef] [PubMed]
- Ceol, C.J.; Houvras, Y.; Jane-Valbuena, J.; Bilodeau, S.; Orlando, D.A.; Battisti, V.; Fritsch, L.; Lin, W.M.; Hollmann, T.J.; Ferré, F.; et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature 2011, 471, 513–517. [Google Scholar] [CrossRef] [PubMed]
- Schultz, D.C.; Ayyanathan, K.; Negorev, D.; Maul, G.G.; Rauscher, F.J. 3rd SETDB1: A novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002, 16, 919–932. [Google Scholar] [CrossRef] [PubMed]
- Orouji, E.; Utikal, J. Tackling malignant melanoma epigenetically: Histone lysine methylation. Clin. Epigenetics 2018, 10, 145. [Google Scholar] [CrossRef]
- Kostaki, M.; Manona, A.D.; Stavraka, I.; Korkolopoulou, P.; Levidou, G.; Trigka, E.-A.; Christofidou, E.; Champsas, G.; Stratigos, A.J.; Katsambas, A.; et al. High-frequency p16(INK) (4A) promoter methylation is associated with histone methyltransferase SETDB1 expression in sporadic cutaneous melanoma. Exp. Dermatol. 2014, 23, 332–338. [Google Scholar] [CrossRef]
- Orouji, E.; Federico, A.; Larribère, L.; Novak, D.; Lipka, D.B.; Assenov, Y.; Sachindra, S.; Hüser, L.; Granados, K.; Gebhardt, C.; et al. Histone methyltransferase SETDB1 contributes to melanoma tumorigenesis and serves as a new potential therapeutic target. Int. J. Cancer 2019, 145, 3462–3477. [Google Scholar] [CrossRef]
- Trivedi, A.; Mehrotra, A.; Baum, C.E.; Lewis, B.; Basuroy, T.; Blomquist, T.; Trumbly, R.; Filipp, F.V.; Setaluri, V.; De La Serna, I.L. Bromodomain and extra-terminal domain (BET) proteins regulate melanocyte differentiation. Epigenetics Chromatin 2020, 13, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Denis, G.V.; McComb, M.E.; Faller, D.V.; Sinha, A.; Romesser, P.B.; Costello, C.E. Identification of transcription complexes that contain the double bromodomain protein Brd2 and chromatin remodeling machines. J. Proteome Res. 2006, 5, 502–511. [Google Scholar] [CrossRef] [PubMed]
- Segura, M.F.; Fontanals-Cirera, B.; Gaziel-Sovran, A.; Guijarro, M.V.; Hanniford, D.; Zhang, G.; González-Gomez, P.; Morante, M.; Jubierre, L.; Zhang, W.; et al. BRD4 sustains melanoma proliferation and represents a new target for epigenetic therapy. Cancer Res. 2013, 73, 6264–6276. [Google Scholar] [CrossRef]
- Li, Y.; Seto, E. HDACs and HDAC Inhibitors in Cancer Development and Therapy. Cold Spring Harb. Perspect. Med. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Fiziev, P.; Akdemir, K.C.; Miller, J.P.; Keung, E.Z.; Samant, N.S.; Sharma, S.; Natale, C.A.; Terranova, C.J.; Maitituoheti, M.; Amin, S.B.; et al. Systematic Epigenomic Analysis Reveals Chromatin States Associated with Melanoma Progression. Cell Rep. 2017, 19, 875–889. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Rong, Y.; Li, S.; Qu, S.; Huang, S. Upregulated Histone Deacetylase 6 Associates with Malignant Progression of Melanoma and Predicts the Prognosis of Patients. Cancer Manag. Res. 2020, 12, 12993–13001. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Gu, J.; Feng, Z.; Yang, Y.; Zhu, N.; Lu, W.; Qi, F. Both HDAC5 and HDAC6 are required for the proliferation and metastasis of melanoma cells. J. Transl. Med. 2016, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Wilking, M.J.; Singh, C.; Nihal, M.; Zhong, W.; Ahmad, N. SIRT1 deacetylase is overexpressed in human melanoma and its small molecule inhibition imparts anti-proliferative response via p53 activation. Arch. Biochem. Biophys. 2014, 563, 94–100. [Google Scholar] [CrossRef]
- Sun, T.; Jiao, L.; Wang, Y.; Yu, Y.; Ming, L. SIRT1 induces epithelial-mesenchymal transition by promoting autophagic degradation of E-cadherin in melanoma cells article. Cell Death Dis. 2018, 9, 1–10. [Google Scholar] [CrossRef]
- George, J.; Nihal, M.; Singh, C.K.; Zhong, W.; Liu, X.; Ahmad, N. Pro-Proliferative Function of Mitochondrial Sirtuin Deacetylase SIRT3 in Human Melanoma. J. Invest. Dermatol. 2016, 136, 809–818. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Peterson, L.M.; Ndiaye, M.A.; Singh, C.K.; Chhabra, G.; Huang, W.; Ahmad, N. SIRT6 histone deacetylase functions as a potential oncogene in human melanoma. Genes Cancer 2017, 8, 701–712. [Google Scholar] [CrossRef]
- George, J.; Nihal, M.; Singh, C.K.; Ahmad, N. 4′-Bromo-resveratrol, a dual Sirtuin-1 and Sirtuin-3 inhibitor, inhibits melanoma cell growth through mitochondrial metabolic reprogramming. Mol. Carcinog. 2019, 58, 1876–1885. [Google Scholar] [CrossRef] [PubMed]
- Kuźbicki, L.; Lange, D.; Strączyńska-Niemiec, A.; Chwirot, B.W. JARID1B expression in human melanoma and benign melanocytic skin lesions. Melanoma Res. 2013, 23, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Roesch, A.; Becker, B.; Meyer, S.; Wild, P.; Hafner, C.; Landthaler, M.; Vogt, T. Retinoblastoma-binding protein 2-homolog 1: A retinoblastoma-binding protein downregulated in malignant melanomas. Mod. Pathol. Off. J. USA Can. Acad. Pathol. Inc 2005, 18, 1249–1257. [Google Scholar] [CrossRef]
- Chauvistré, H.; Daignault, S.; Shannan, B.; Ju, R.; Picard, D.; Vogel, F.; Egetemaier, S.; Krepler, C.; Rebecca, V.; Sechi, A.; et al. The Janus-faced role of KDM5B heterogeneity in melanoma: Differentiation as a situational driver of both growth arrest and drug-resistance. 2020; preprint. [Google Scholar] [CrossRef]
- Roesch, A.; Fukunaga-Kalabis, M.; Schmidt, E.C.; Zabierowski, S.E.; Brafford, P.A.; Vultur, A.; Basu, D.; Gimotty, P.; Vogt, T.; Herlyn, M. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell 2010, 141, 583–594. [Google Scholar] [CrossRef] [PubMed]
- Flaherty, K.T.; Robert, C.; Hersey, P.; Nathan, P.; Garbe, C.; Milhem, M.; Demidov, L.V.; Hassel, J.C.; Rutkowski, P.; Mohr, P.; et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N. Engl. J. Med. 2012, 367, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Flaherty, K.T.; Infante, J.R.; Daud, A.; Gonzalez, R.; Kefford, R.F.; Sosman, J.; Hamid, O.; Schuchter, L.; Cebon, J.; Ibrahim, N.; et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N. Engl. J. Med. 2012, 367, 1694–1703. [Google Scholar] [CrossRef]
- Shi, H.; Hugo, W.; Kong, X.; Hong, A.; Koya, R.C.; Moriceau, G.; Chodon, T.; Guo, R.; Johnson, D.B.; Dahlman, K.B.; et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov. 2014, 4, 80–93. [Google Scholar] [CrossRef]
- Clark, M.E.; Rizos, H.; Pereira, M.R.; McEvoy, A.C.; Marsavela, G.; Calapre, L.; Meehan, K.; Ruhen, O.; Khattak, M.A.; Meniawy, T.M.; et al. Detection of BRAF splicing variants in plasma-derived cell-free nucleic acids and extracellular vesicles of melanoma patients failing targeted therapy therapies. Oncotarget 2020, 11, 4016–4027. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Moriceau, G.; Kong, X.; Lee, M.-K.; Lee, H.; Koya, R.C.; Ng, C.; Chodon, T.; Scolyer, R.A.; Dahlman, K.B.; et al. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat. Commun. 2012, 3, 724. [Google Scholar] [CrossRef] [PubMed]
- Perna, D.; Karreth, F.A.; Rust, A.G.; Perez-Mancera, P.A.; Rashid, M.; Iorio, F.; Alifrangis, C.; Arends, M.J.; Bosenberg, M.W.; Bollag, G.; et al. BRAF inhibitor resistance mediated by the AKT pathway in an oncogenic BRAF mouse melanoma model. Proc. Natl. Acad. Sci. USA 2015, 112, E536–E545. [Google Scholar] [CrossRef]
- Basile, K.J.; Aplin, A.E. Downregulation of Noxa by RAF/MEK inhibition counteracts cell death response in mutant B-RAF melanoma cells. Am. J. Cancer Res. 2012, 2, 726–735. [Google Scholar] [PubMed]
- Nazarian, R.; Shi, H.; Wang, Q.; Kong, X.; Koya, R.C.; Lee, H.; Chen, Z.; Lee, M.-K.; Attar, N.; Sazegar, H.; et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 2010, 468, 973–977. [Google Scholar] [CrossRef]
- Dugo, M.; Nicolini, G.; Tragni, G.; Bersani, I.; Tomassetti, A.; Colonna, V.; Del Vecchio, M.; De Braud, F.; Canevari, S.; Anichini, A.; et al. A melanoma subtype with intrinsic resistance to BRAF inhibition identified by receptor tyrosine kinases gene-driven classification. Oncotarget 2015, 6, 5118–5133. [Google Scholar] [CrossRef]
- Su, Y.; Lu, X.; Li, G.; Liu, C.; Kong, Y.; Lee, J.W.; Ng, R.; Wong, S.; Robert, L.; Warden, C.; et al. Kinetic Inference Resolves Epigenetic Mechanism of Drug Resistance in Melanoma. Cell 2019. preprint. [Google Scholar] [CrossRef]
- Wilting, R.H.; Dannenberg, J.H. Epigenetic mechanisms in tumorigenesis, tumor cell heterogeneity and drug resistance. Drug Resist. Updat. 2012, 15, 21–38. [Google Scholar] [CrossRef] [PubMed]
- Al Emran, A.; Marzese, D.M.; Menon, D.R.; Stark, M.S.; Torrano, J.; Hammerlindl, H.; Zhang, G.; Brafford, P.; Salomon, M.P.; Nelson, N.; et al. Distinct histone modifications denote early stress-induced drug tolerance in cancer. Oncotarget 2018, 9, 8206–8222. [Google Scholar] [CrossRef]
- Hugo, W.; Shi, H.; Sun, L.; Piva, M.; Song, C.; Kong, X.; Moriceau, G.; Hong, A.; Dahlman, K.B.; Johnson, D.B.; et al. Non-genomic and Immune Evolution of Melanoma Acquiring MAPKi Resistance. Cell 2015, 162, 1271–1285. [Google Scholar] [CrossRef] [PubMed]
- Seghers, A.C.; Wilgenhof, S.; Lebbé, C.; Neyns, B. Successful rechallenge in two patients with BRAF-V600-mutant melanoma who experienced previous progression during treatment with a selective BRAF inhibitor. Melanoma Res. 2012, 22, 466–472. [Google Scholar] [CrossRef]
- Atkinson, V.; Batty, K.; Long, G.V.; Carlino, M.S.; Peters, G.D.; Bhave, P.; Moore, M.A.; Xu, W.; Brown, L.J.; Arneil, M.; et al. Activity and safety of third-line BRAF-targeted therapy (TT) following first-line TT and second-line immunotherapy (IT) in advanced melanoma. J. Clin. Oncol. 2020, 38, 10049. [Google Scholar] [CrossRef]
- Cybulska-Stopa, B.; Rogala, P.; Czarnecka, A.M.; Galus, Ł.; Dziura, R.; Rajczykowski, M.; Kubiatowski, T.; Wiśniewska, M.; Gęga-Czarnota, A.; Teterycz, P.; et al. BRAF and MEK inhibitors rechallenge as effective treatment for patients with metastatic melanoma. Melanoma Res. 2020, 30, 465–471. [Google Scholar] [CrossRef]
- Sharma, S.V.; Lee, D.Y.; Li, B.; Quinlan, M.P.; Takahashi, F.; Maheswaran, S.; McDermott, U.; Azizian, N.; Zou, L.; Fischbach, M.A.; et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 2010, 141, 69–80. [Google Scholar] [CrossRef]
- Roesch, A.; Vultur, A.; Bogeski, I.; Wang, H.; Zimmermann, K.M.; Speicher, D.; Körbel, C.; Laschke, M.W.; Gimotty, P.A.; Philipp, S.E.; et al. Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B(high) cells. Cancer Cell 2013, 23, 811–825. [Google Scholar] [CrossRef]
- Ravindran Menon, D.; Das, S.; Krepler, C.; Vultur, A.; Rinner, B.; Schauer, S.; Kashofer, K.; Wagner, K.; Zhang, G.; Bonyadi Rad, E.; et al. A stress-induced early innate response causes multidrug tolerance in melanoma. Oncogene 2015, 34, 4448–4459. [Google Scholar] [CrossRef] [PubMed]
- Cartlidge, R.A.; Thomas, G.R.; Cagnol, S.; Jong, K.A.; Molton, S.A.; Finch, A.J.; McMahon, M. Oncogenic BRAF(V600E) inhibits BIM expression to promote melanoma cell survival. Pigment Cell Melanoma Res. 2008, 21, 534–544. [Google Scholar] [CrossRef] [PubMed]
- Tsai, J.; Lee, J.T.; Wang, W.; Zhang, J.; Cho, H.; Mamo, S.; Bremer, R.; Gillette, S.; Kong, J.; Haass, N.K.; et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc. Natl. Acad. Sci. USA 2008, 105, 3041–3046. [Google Scholar] [CrossRef] [PubMed]
- Mohana-Kumaran, N.; Hill, D.S.; Allen, J.D.; Haass, N.K. Targeting the intrinsic apoptosis pathway as a strategy for melanoma therapy. Pigment Cell Melanoma Res. 2014, 27, 525–539. [Google Scholar] [CrossRef]
- Tiffen, J.C.; Gunatilake, D.; Gallagher, S.J.; Gowrishankar, K.; Heinemann, A.; Cullinane, C.; Dutton-Regester, K.; Pupo, G.M.; Strbenac, D.; Yang, J.Y.; et al. Targeting activating mutations of EZH2 leads to potent cell growth inhibition in human melanoma by derepression of tumor suppressor genes. Oncotarget 2015, 6, 27023–27036. [Google Scholar] [CrossRef]
- Gallagher, S.J.; Mijatov, B.; Gunatilake, D.; Tiffen, J.C.; Gowrishankar, K.; Jin, L.; Pupo, G.M.; Cullinane, C.; Prinjha, R.K.; Smithers, N.; et al. The epigenetic regulator I-BET151 induces BIM-dependent apoptosis and cell cycle arrest of human melanoma cells. J. Invest. Dermatol. 2014, 134, 2795–2805. [Google Scholar] [CrossRef]
- Heinemann, A.; Cullinane, C.; De Paoli-Iseppi, R.; Wilmott, J.S.; Gunatilake, D.; Madore, J.; Strbenac, D.; Yang, J.Y.; Gowrishankar, K.; Tiffen, J.C.; et al. Combining BET and HDAC inhibitors synergistically induces apoptosis of melanoma and suppresses AKT and YAP signaling. Oncotarget 2015, 6, 21507–21521. [Google Scholar] [CrossRef]
- Neagu, M. Metabolic Traits in Cutaneous Melanoma. Front. Oncol. 2020, 10, 851. [Google Scholar] [CrossRef]
- Witz, I.P. Tumor-microenvironment interactions: Dangerous liaisons. Adv. Cancer Res. 2008, 100, 203–229. [Google Scholar] [CrossRef] [PubMed]
- Falcone, I.; Conciatori, F.; Bazzichetto, C.; Ferretti, G.; Cognetti, F.; Ciuffreda, L.; Milella, M. Tumor Microenvironment: Implications in Melanoma Resistance to Targeted Therapy and Immunotherapy. Cancers 2020, 12, 2870. [Google Scholar] [CrossRef] [PubMed]
- Mazurkiewicz, J.; Simiczyjew, A.; Dratkiewicz, E.; Ziętek, M.; Matkowski, R.; Nowak, D. Stromal Cells Present in the Melanoma Niche Affect Tumor Invasiveness and Its Resistance to Therapy. Int. J. Mol. Sci. 2021, 22, 529. [Google Scholar] [CrossRef] [PubMed]
- Passarelli, A.; Mannavola, F.; Stucci, L.S.; Tucci, M.; Silvestris, F. Immune system and melanoma biology: A balance between immunosurveillance and immune escape. Oncotarget 2017, 8, 106132–106142. [Google Scholar] [CrossRef]
- Farhood, B.; Najafi, M.; Mortezaee, K. CD8(+) cytotoxic T lymphocytes in cancer immunotherapy: A review. J. Cell. Physiol. 2019, 234, 8509–8521. [Google Scholar] [CrossRef]
- Liu, Y.; Guo, J.; Huang, L. Modulation of tumor microenvironment for immunotherapy: Focus on nanomaterial-based strategies. Theranostics 2020, 10, 3099–3117. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Xu, L.; Yi, M.; Yu, S.; Wu, K.; Luo, S. Novel immune checkpoint targets: Moving beyond PD-1 and CTLA-4. Mol. Cancer 2019, 18, 155. [Google Scholar] [CrossRef] [PubMed]
- Turnis, M.E.; Andrews, L.P.; Vignali, D.A.A. Inhibitory receptors as targets for cancer immunotherapy. Eur. J. Immunol. 2015, 45, 1892–1905. [Google Scholar] [CrossRef]
- Yervoy (ipilimumab) FDA Approval History Drugs.com. Available online: https://www.drugs.com/history/yervoy.html (accessed on 17 February 2021).
- Opdivo (nivolumab) FDA Approval History Drugs.com. Available online: https://www.drugs.com/history/opdivo.html (accessed on 17 February 2021).
- Asher, N.; Ben-Betzalel, G.; Lev-Ari, S.; Shapira-Frommer, R.; Steinberg-Silman, Y.; Gochman, N.; Schachter, J.; Meirson, T.; Markel, G. Real World Outcomes of Ipilimumab and Nivolumab in Patients with Metastatic Melanoma. Cancers 2020, 12, 2329. [Google Scholar] [CrossRef]
- Nobel Prize Awarded to Cancer Immunotherapy Researchers. Available online: https://www.cancer.org/latest-news/nobel-prize-awarded-to-cancer-immunotherapy-researchers.html (accessed on 17 February 2021).
- Lee, P.P.; Fitzpatrick, D.R.; Beard, C.; Jessup, H.K.; Lehar, S.; Makar, K.W.; Pérez-Melgosa, M.; Sweetser, M.T.; Schlissel, M.S.; Nguyen, S.; et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 2001, 15, 763–774. [Google Scholar] [CrossRef]
- Scanlon, S. DNA methylation makes for tired T cells. Science. 2017, 357, 367–368. [Google Scholar] [CrossRef]
- Xiao, Q.; Nobre, A.; Piñeiro, P.; Berciano-Guerrero, M.-Á.; Alba, E.; Cobo, M.; Lauschke, V.M.; Barragán, I. Genetic and Epigenetic Biomarkers of Immune Checkpoint Blockade Response. J. Clin. Med. 2020, 9, 286. [Google Scholar] [CrossRef] [PubMed]
- Fietz, S.; Zarbl, R.; Niebel, D.; Posch, C.; Brossart, P.; Gielen, G.H.; Strieth, S.; Pietsch, T.; Kristiansen, G.; Bootz, F.; et al. CTLA4 promoter methylation predicts response and progression-free survival in stage IV melanoma treated with anti-CTLA-4 immunotherapy (ipilimumab). Cancer Immunol. Immunother. 2020. [Google Scholar] [CrossRef] [PubMed]
- Madore, J.; Vilain, R.E.; Menzies, A.M.; Kakavand, H.; Wilmott, J.S.; Hyman, J.; Yearley, J.H.; Kefford, R.F.; Thompson, J.F.; Long, G.V.; et al. PD-L1 expression in melanoma shows marked heterogeneity within and between patients: Implications for anti-PD-1/PD-L1 clinical trials. Pigment Cell Melanoma Res. 2015, 28, 245–253. [Google Scholar] [CrossRef]
- Sznol, M.; Chen, L. Antagonist antibodies to PD-1 and B7-H1 (PD-L1) in the treatment of advanced human cancer. Clin. cancer Res. an Off. J. Am. Assoc. Cancer Res. 2013, 19, 1021–1034. [Google Scholar] [CrossRef] [PubMed]
- Eddy, K.; Chen, S. Overcoming Immune Evasion in Melanoma. Int. J. Mol. Sci. 2020, 21, 8984. [Google Scholar] [CrossRef] [PubMed]
- Madore, J.; Strbenac, D.; Vilain, R.; Menzies, A.M.; Yang, J.Y.H.; Thompson, J.F.; Long, G.V.; Mann, G.J.; Scolyer, R.A.; Wilmott, J.S. PD-L1 Negative Status is Associated with Lower Mutation Burden, Differential Expression of Immune-Related Genes, and Worse Survival in Stage III Melanoma. Clin. cancer Res. an Off. J. Am. Assoc. Cancer Res. 2016, 22, 3915–3923. [Google Scholar] [CrossRef]
- Micevic, G.; Thakral, D.; McGeary, M.; Bosenberg, M.W. PD-L1 methylation regulates PD-L1 expression and is associated with melanoma survival. Pigment Cell Melanoma Res. 2019, 32, 435–440. [Google Scholar] [CrossRef]
- Fröhlich, A.; Sirokay, J.; Fietz, S.; Vogt, T.J.; Dietrich, J.; Zarbl, R.; Florin, M.; Kuster, P.; Saavedra, G.; Valladolid, S.R.; et al. Molecular, clinicopathological, and immune correlates of LAG3 promoter DNA methylation in melanoma. EBioMedicine 2020, 59, 102962. [Google Scholar] [CrossRef]
- Chiappinelli, K.B.; Strissel, P.L.; Desrichard, A.; Li, H.; Henke, C.; Akman, B.; Hein, A.; Rote, N.S.; Cope, L.M.; Snyder, A.; et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell 2015, 162, 974–986. [Google Scholar] [CrossRef]
- Zingg, D.; Arenas-Ramirez, N.; Sahin, D.; Rosalia, R.A.; Antunes, A.T.; Haeusel, J.; Sommer, L.; Boyman, O. The Histone Methyltransferase Ezh2 Controls Mechanisms of Adaptive Resistance to Tumor Immunotherapy. Cell Rep. 2017, 20, 854–867. [Google Scholar] [CrossRef] [PubMed]
- Tiffen, J.; Wilson, S.; Gallagher, S.J.; Hersey, P.; Filipp, F. V Somatic Copy Number Amplification and Hyperactivating Somatic Mutations of EZH2 Correlate With DNA Methylation and Drive Epigenetic Silencing of Genes Involved in Tumor Suppression and Immune Responses in Melanoma. Neoplasia 2016, 18, 121–132. [Google Scholar] [CrossRef]
- Chen, B.-F.; Chan, W.-Y. The de novo DNA methyltransferase DNMT3A in development and cancer. Epigenetics 2014, 9, 669–677. [Google Scholar] [CrossRef]
- Badeaux, A.I.; Shi, Y. Emerging roles for chromatin as a signal integration and storage platform. Nat. Rev. Mol. Cell Biol. 2013, 14, 211–224. [Google Scholar] [CrossRef] [PubMed]
- Schlesinger, Y.; Straussman, R.; Keshet, I.; Farkash, S.; Hecht, M.; Zimmerman, J.; Eden, E.; Yakhini, Z.; Ben-Shushan, E.; Reubinoff, B.E.; et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet. 2007, 39, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Montalvo-Casimiro, M.; González-Barrios, R.; Meraz-Rodriguez, M.A.; Juárez-González, V.T.; Arriaga-Canon, C.; Herrera, L.A. Epidrug Repurposing: Discovering New Faces of Old Acquaintances in Cancer Therapy. Front. Oncol. 2020, 10, 605386. [Google Scholar] [CrossRef]
- Majchrzak-Celińska, A.; Warych, A.; Szoszkiewicz, M. Novel Approaches to Epigenetic Therapies: From Drug Combinations to Epigenetic Editing. Genes 2021, 12. [Google Scholar]
- Bioequivalence & Food Effect Study in Patients With Solid Tumor or Hematologic Malignancies Full Text View ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/NCT02223052?term=azacitidine&cond=Melanoma&draw=6&rank=6 (accessed on 9 August 2021).
- Hussein Tawbi PHD, M.D. Study of Oral Azacitidine (CC-486) in Combination With Pembrolizumab (MK-3475) in Patients With Metastatic Melanoma. Available online: https://clinicaltrials.gov/ct2/show/NCT02816021?term=azacitidine&cond=Melanoma&draw=2&rank=1 (accessed on 9 August 2021).
- Van Der Westhuizen, A.; Graves, M.; Levy, R.; Majid, A.; Vilain, R.; Bowden, N. PRIME002: Early phase II study of azacitidine and carboplatin priming for avelumab in patients with advanced melanoma who are resistant to immunotherapy. In Abstracts B, Proceedings of the 44th ESMO Congress (ESMO 2019), Barcelona, Spain, 27 September—1 October 2019; European Society for Medical Oncology: Lugano, Switzerland, 2019; Volume 561, p. 30. [Google Scholar] [CrossRef]
- Decitabine in Treating Patients With Melanoma or Other Advanced Cancer Full Text View ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/NCT00002980?term=azacitidine&cond=Melanoma&draw=2&rank=9 (accessed on 9 August 2021).
- Gene-Modified T Cells With or Without Decitabine in Treating Patients With Advanced Malignancies Expressing NY-ESO-1 Full Text View ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/NCT02650986?term=azacitidine&cond=Melanoma&draw=5&rank=11 (accessed on 9 August 2021).
- Xia, C.; Leon-Ferre, R.; Laux, D.; Deutsch, J.; Smith, B.J.; Frees, M.; Milhem, M. Treatment of resistant metastatic melanoma using sequential epigenetic therapy (decitabine and panobinostat) combined with chemotherapy (temozolomide). Cancer Chemother. Pharmacol. 2014, 74, 691–697. [Google Scholar] [CrossRef] [PubMed]
- Combination of Decitabine and Temozolomide in the Treatment of Patients With Metastatic Melanoma Full Text View ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/NCT00715793?term=decitabine&cond=Melanoma&draw=4&rank=3 (accessed on 9 August 2021).
- Phadke, S.D.; Engelman, E.S.; Leon-Ferre, R.A.; Abushahin, L.I.; Mott, S.C.; Zakharia, Y.; Milhem, M.M. A phase I study of vemurafenib and decitabine in metastatic melanoma. J. Clin. Oncol. 2015, 33, 9056. [Google Scholar] [CrossRef]
- Zakharia, Y.; Monga, V.; Swami, U.; Bossler, A.D.; Freesmeier, M.; Frees, M.; Khan, M.; Frydenlund, N.; Srikantha, R.; Vanneste, M.; et al. Targeting epigenetics for treatment of BRAF mutated metastatic melanoma with decitabine in combination with vemurafenib: A phase lb study. Oncotarget 2017, 8, 89182–89193. [Google Scholar] [CrossRef]
- A Study Investigating SGI-110 in Combination With Ipilimumab in Unresectable or Metastatic Melanoma Patients Full Text View ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/NCT02608437?term=Guadecitabine&cond=Melanoma&draw=2&rank=1 (accessed on 3 August 2021).
- Giacomo, A.M.D.; Covre, A.; Finotello, F.; Rieder, D.; Danielli, R.; Sigalotti, L.; Giannarelli, D.; Petitprez, F.; Lacroix, L.; Valente, M.; et al. Guadecitabine Plus Ipilimumab in Unresectable Melanoma: The NIBIT-M4 Clinical Trial. Clin. Cancer Res. 2019, 25, 7351–7362. [Google Scholar] [CrossRef]
- A Study of NIVO Plus IPI and Guadecitabine or NIVO Plus IPI in Melanoma and NSCLC Resistant to Anti-PD1/PDL1 Full Text View ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/NCT04250246?term=Guadecitabine&cond=Melanoma&draw=2&rank=2 (accessed on 3 August 2021).
- Hellebrekers, D.M.E.I.; Castermans, K.; Viré, E.; Dings, R.P.M.; Hoebers, N.T.H.; Mayo, K.H.; Egbrink, M.G.A.; Molema, G.; Fuks, F.; Engeland, M.; et al. Epigenetic Regulation of Tumor Endothelial Cell Anergy: Silencing of Intercellular Adhesion Molecule-1 by Histone Modifications. Cancer Res. 2006, 66, 10770–10777. [Google Scholar] [CrossRef]
- Disulfiram in Patients With Metastatic Melanoma Full Text View ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/NCT00256230?term=disulfiram&cond=Melanoma&draw=2&rank=2 (accessed on 9 August 2021).
- Disulfiram Plus Arsenic Trioxide In Patients With Metastatic Melanoma and at Least One Prior Systemic Therapy Full Text View ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/NCT00571116?term=disulfiram&cond=Melanoma&draw=2&rank=1 (accessed on 9 August 2021).
- Huijberts, S.; Wang, L.; de Oliveira, R.L.; Rosing, H.; Nuijen, B.; Beijnen, J.; Bernards, R.; Schellens, J.; Wilgenhof, S. Vorinostat in patients with resistant BRAF(V600E) mutated advanced melanoma: A proof of concept study. Future Oncol. 2020, 16, 619–629. [Google Scholar] [CrossRef] [PubMed]
- Reijers, I.L.M.; Dimitriadis, P.; Rozeman, E.A.; Versluis, J.M.; Broeks, A.; Bosch, L.J.W.; Bouwman, J.; Cornelissen, S.; Krijgsman, O.; Gonzalez, M.; et al. Personalized combination of neoadjuvant domatinostat, nivolumab and ipilimumab in macroscopic stage III melanoma patients stratified according to the interferon-gamma signature: The DONIMI study. J. Clin. Oncol. 2020, 38, TPS10087. [Google Scholar] [CrossRef]
- Panobinostat (LBH589) in Patients With Metastatic Melanoma Full Text View ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/NCT01065467 (accessed on 9 August 2021).
- Khushalani, N.I.; Markowitz, J.; Eroglu, Z.; Giuroiu, I.; Ladanova, V.; Reiersen, P.; Rich, J.; Thapa, R.; Schell, M.J.; Sotomayor, E.M.; et al. A phase I trial of panobinostat with ipilimumab in advanced melanoma. J. Clin. Oncol. 2017, 35, 9547. [Google Scholar] [CrossRef]
- FR901228 in Treating Patients With Unresectable Stage III or Stage IV Malignant Melanoma Full Text View ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/NCT00104884 (accessed on 9 August 2021).
- Johnson, M.L.; Gonzalez, R.; Opyrchal, M.; Gabrilovich, D.; Ordentlich, P.; Brouwer, S.; Sankoh, S.; Schmidt, E.V.; Meyers, M.L.; Agarwala, S.S. ENCORE 601: A phase II study of entinostat (ENT) in combination with pembrolizumab (PEMBRO) in patients with melanoma. J. Clin. Oncol. 2017, 35, 9529. [Google Scholar] [CrossRef]
- Woods, D.M.; Laino, A.S.; Vassallo, M.; Weber, J. Abstract PO-007: The Class I/IV HDAC Inhibitor Mocetinostat Augments Anti-Tumor Immune Responses in Melanoma Patients. Cancer Res. 2020, 80, PO-007. [Google Scholar] [CrossRef]
- Tinostamustine and Nivolumab in Advanced Melanoma Full Text View ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/NCT03903458 (accessed on 9 August 2021).
- Rocca, A.; Minucci, S.; Tosti, G.; Croci, D.; Contegno, F.; Ballarini, M.; Nolè, F.; Munzone, E.; Salmaggi, A.; Goldhirsch, A.; et al. A phase I–II study of the histone deacetylase inhibitor valproic acid plus chemoimmunotherapy in patients with advanced melanoma. Br. J. Cancer 2009, 100, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Tahata, S.; Singh, S.V.; Lin, Y.; Hahm, E.-R.; Beumer, J.H.; Christner, S.M.; Rao, U.N.; Sander, C.; Tarhini, A.A.; Tawbi, H.; et al. Evaluation of Biodistribution of Sulforaphane after Administration of Oral Broccoli Sprout Extract in Melanoma Patients with Multiple Atypical Nevi. Cancer Prev. Res. 2018, 11, 429–438. [Google Scholar] [CrossRef]
- Garmpis, N.; Damaskos, C.; Garmpi, A.; Dimitroulis, D.; Spartalis, E.; Margonis, G.A.; Schizas, D.; Deskou, I.; Doula, C.; Magkouti, E.; et al. Targeting histone deacetylases in malignant melanoma: A future therapeutic agent or just great expectations? Anticancer Res. 2017, 37, 5355–5362. [Google Scholar] [PubMed]
- Testing the Addition of the Anti-cancer Drug, Tazemetostat, to the Usual Treatment (Dabrafenib and Trametinib) for Metastatic Melanoma That Has Progressed on the Usual Treatment Full Text View ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/NCT04557956 (accessed on 9 August 2021).
- ORIOn-E: A Study Evaluating CPI-1205 in Patients With Advanced Solid Tumors Full Text View ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/NCT03525795 (accessed on 9 August 2021).
- Ameratunga, M.; Braña, I.; Bono, P.; Postel-Vinay, S.; Plummer, R.; Aspegren, J.; Korjamo, T.; Snapir, A.; de Bono, J.S. First-in-human Phase 1 open label study of the BET inhibitor ODM-207 in patients with selected solid tumours. Br. J. Cancer 2020, 123, 1730–1736. [Google Scholar] [CrossRef] [PubMed]
- Yin, M.; Guo, Y.; Hu, R.; Cai, W.L.; Li, Y.; Pei, S.; Sun, H.; Peng, C.; Li, J.; Ye, R.; et al. Potent BRD4 inhibitor suppresses cancer cell-macrophage interaction. Nat. Commun. 2020, 11, 1833. [Google Scholar] [CrossRef] [PubMed]
- Erkes, D.A.; Rosenbaum, S.R.; Field, C.O.; Chervoneva, I.; Villanueva, J.; Aplin, A.E. PLX3397 inhibits the accumulation of intra-tumoral macrophages and improves bromodomain and extra-terminal inhibitor efficacy in melanoma. Pigment Cell Melanoma Res. 2020, 33, 372–377. [Google Scholar] [CrossRef]
- Rollins, R.A.; Kim, K.H.; Tsao, C.-C. The Emerging Epigenetic Landscape in Melanoma. In Human Skin Cancer, Potential Biomarkers and Therapeutic Targets; InTech: London, UK, 2016. [Google Scholar]
- Jin, N.; George, T.L.; Otterson, G.A.; Verschraegen, C.; Wen, H.; Carbone, D.; Herman, J.; Bertino, E.M.; He, K. Advances in epigenetic therapeutics with focus on solid tumors. Clin. Epigenetics 2021, 13, 83. [Google Scholar] [CrossRef]
- Ganesan, A.; Arimondo, P.B.; Rots, M.G.; Jeronimo, C.; Berdasco, M. The timeline of epigenetic drug discovery: From reality to dreams. Clin. Epigenetics 2019, 11, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Cartron, P.-F.; Cheray, M.; Bretaudeau, L. Epigenetic protein complexes: The adequate candidates for the use of a new generation of epidrugs in personalized and precision medicine in cancer. Epigenomics 2020, 12, 171–177. [Google Scholar] [CrossRef]
- Lu, Y.; Chan, Y.-T.; Tan, H.-Y.; Li, S.; Wang, N.; Feng, Y. Epigenetic regulation in human cancer: The potential role of epi-drug in cancer therapy. Mol. Cancer 2020, 19, 79. [Google Scholar] [CrossRef]
- Martinez-Useros, J.; Martin-Galan, M.; Florez-Cespedes, M.; Garcia-Foncillas, J. Epigenetics of Most Aggressive Solid Tumors: Pathways, Targets and Treatments. Cancers 2021, 13, 3209. [Google Scholar] [CrossRef]
- Chu, B.F.; Karpenko, M.J.; Liu, Z.; Aimiuwu, J.; Villalona-Calero, M.A.; Chan, K.K.; Grever, M.R.; Otterson, G.A. Phase I study of 5-aza-2′-deoxycytidine in combination with valproic acid in non-small-cell lung cancer. Cancer Chemother. Pharmacol. 2013, 71, 115–121. [Google Scholar] [CrossRef]
- Jansen, Y.J.L.; Verset, G.; Schats, K.; Van Dam, P.-J.; Seremet, T.; Kockx, M.; Van Laethem, J.-L.B.; Neyns, B. Phase I clinical trial of decitabine (5-aza-2′-deoxycytidine) administered by hepatic arterial infusion in patients with unresectable liver-predominant metastases. ESMO Open 2019, 4, e000464. [Google Scholar] [CrossRef]
- Jones, P.A.; Ohtani, H.; Chakravarthy, A.; De Carvalho, D.D. Epigenetic therapy in immune-oncology. Nat. Rev. Cancer 2019, 19, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Tawbi, H.A.; Beumer, J.H.; Tarhini, A.A.; Moschos, S.J.; Egorin, M.J.; Buch, S.C.; Lin, Y.; Kirkwood, J.M. Phase I/II study of the combination of decitabine (DAC) and temozolomide (TMZ) in patients (pts) with metastatic melanoma (MM). J. Clin. Oncol. 2010, 28, 8533. [Google Scholar] [CrossRef]
- Patnaik, S. Anupriya Drugs Targeting Epigenetic Modifications and Plausible Therapeutic Strategies Against Colorectal Cancer. Front. Pharmacol. 2019, 10, 588. [Google Scholar] [CrossRef] [PubMed]
- Meraz-Torres, F.; Plöger, S.; Garbe, C.; Niessner, H.; Sinnberg, T. Disulfiram as a Therapeutic Agent for Metastatic Malignant Melanoma—Old Myth or New Logos? Cancers 2020, 12, 3538. [Google Scholar] [CrossRef]
- Hull, E.E.; Montgomery, M.R.; Leyva, K.J. HDAC Inhibitors as Epigenetic Regulators of the Immune System: Impacts on Cancer Therapy and Inflammatory Diseases. Biomed Res. Int. 2016, 2016, 8797206. [Google Scholar] [CrossRef]
- Mrakovcic, M.; Fröhlich, L.F. Molecular Determinants of Cancer Therapy Resistance to HDAC Inhibitor-Induced Autophagy. Cancers 2019, 12, 109. [Google Scholar] [CrossRef]
- Ibrahim, N.; Buchbinder, E.I.; Granter, S.R.; Rodig, S.J.; Giobbie-Hurder, A.; Becerra, C.; Tsiaras, A.; Gjini, E.; Fisher, D.E.; Hodi, F.S. A phase I trial of panobinostat (LBH589) in patients with metastatic melanoma. Cancer Med. 2016, 5, 3041–3050. [Google Scholar] [CrossRef]
- Hauschild, A.; Trefzer, U.; Garbe, C.; Kaehler, K.; Ugurel, S.; Kiecker, F.; Eigentler, T.; Krissel, H.; Schadendorf, D. A phase II multicenter study on the histone deacetylase (HDAC) inhibitor MS-275, comparing two dosage schedules in metastatic melanoma. J. Clin. Oncol. 2006, 24, 8044. [Google Scholar] [CrossRef]
- Haas, N.B.; Quirt, I.; Hotte, S.; McWhirter, E.; Polintan, R.; Litwin, S.; Adams, P.D.; McBryan, T.; Wang, L.; Martin, L.P.; et al. Phase II trial of vorinostat in advanced melanoma. Invest. New Drugs 2014, 32, 526–534. [Google Scholar] [CrossRef]
- Venugopal, B.; Baird, R.; Kristeleit, R.S.; Plummer, R.; Cowan, R.; Stewart, A.; Fourneau, N.; Hellemans, P.; Elsayed, Y.; McClue, S.; et al. A phase I study of quisinostat (JNJ-26481585), an oral hydroxamate histone deacetylase inhibitor with evidence of target modulation and antitumor activity, in patients with advanced solid tumors. Clin. cancer Res. Off. J. Am. Assoc. Cancer Res. 2013, 19, 4262–4272. [Google Scholar] [CrossRef] [PubMed]
- Dinkova-Kostova, A.T.; Jenkins, S.N.; Fahey, J.W.; Ye, L.; Wehage, S.L.; Liby, K.T.; Stephenson, K.K.; Wade, K.L.; Talalay, P. Protection against UV-light-induced skin carcinogenesis in SKH-1 high-risk mice by sulforaphane-containing broccoli sprout extracts. Cancer Lett. 2006, 240, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Shibata, A.; Nakagawa, K.; Yamanoi, H.; Tsuduki, T.; Sookwong, P.; Higuchi, O.; Kimura, F.; Miyazawa, T. Sulforaphane suppresses ultraviolet B-induced inflammation in HaCaT keratinocytes and HR-1 hairless mice. J. Nutr. Biochem. 2010, 21, 702–709. [Google Scholar] [CrossRef] [PubMed]
- Arcidiacono, P.; Ragonese, F.; Stabile, A.; Pistilli, A.; Kuligina, E.; Rende, M.; Bottoni, U.; Calvieri, S.; Crisanti, A.; Spaccapelo, R. Antitumor activity and expression profiles of genes induced by sulforaphane in human melanoma cells. Eur. J. Nutr. 2018, 57, 2547–2569. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, S.J.; Mishra, R.; Sharma, P.; Kundu, G.C. Quercetin and sulforaphane in combination suppress the progression of melanoma through the down-regulation of matrix metalloproteinase-9. Exp. Ther. Med. 2010, 1, 915–920. [Google Scholar] [CrossRef]
- Alyoussef, A.; Taha, M. Antitumor activity of sulforaphane in mice model of skin cancer via blocking sulfatase-2. Exp. Dermatol. 2019, 28, 28–34. [Google Scholar] [CrossRef]
- Duan, R.; Du, W.; Guo, W. EZH2: A novel target for cancer treatment. J. Hematol. Oncol. 2020, 13, 104. [Google Scholar] [CrossRef] [PubMed]
- Yap, T.A.; Winter, J.N.; Giulino-Roth, L.; Longley, J.; Lopez, J.; Michot, J.-M.; Leonard, J.P.; Ribrag, V.; McCabe, M.T.; Creasy, C.L.; et al. Phase I Study of the Novel Enhancer of Zeste Homolog 2 (EZH2) Inhibitor GSK2816126 in Patients with Advanced Hematologic and Solid Tumors. Clin. Cancer Res. 2019, 25, 7331–7339. [Google Scholar] [CrossRef]
- Doroshow, D.B.; Eder, J.P.; LoRusso, P.M. BET inhibitors: A novel epigenetic approach. Ann. Oncol. 2017, 28, 1776–1787. [Google Scholar] [CrossRef]
- Shorstova, T.; Foulkes, W.D.; Witcher, M. Achieving clinical success with BET inhibitors as anti-cancer agents. Br. J. Cancer 2021, 124, 1478–1490. [Google Scholar] [CrossRef] [PubMed]
- Deng, G.; Zeng, F.; Su, J.; Zhao, S.; Hu, R.; Zhu, W.; Hu, S.; Chen, X.; Yin, M. BET inhibitor suppresses melanoma progression via the noncanonical NF-κB/SPP1 pathway. Theranostics 2020, 10, 11428–11443. [Google Scholar] [CrossRef]
- Gallagher, S.J.; Mijatov, B.; Gunatilake, D.; Gowrishankar, K.; Tiffen, J.; James, W.; Jin, L.; Pupo, G.; Cullinane, C.; McArthur, G.A.; et al. Control of NF-kB activity in human melanoma by bromodomain and extra-terminal protein inhibitor I-BET151. Pigment Cell Melanoma Res. 2014, 27, 1126–1137. [Google Scholar] [CrossRef]
- Erkes, D.A.; Field, C.O.; Capparelli, C.; Tiago, M.; Purwin, T.J.; Chervoneva, I.; Berger, A.C.; Hartsough, E.J.; Villanueva, J.; Aplin, A.E. The next-generation BET inhibitor, PLX51107, delays melanoma growth in a CD8-mediated manner. Pigment Cell Melanoma Res. 2019, 32, 687–696. [Google Scholar] [CrossRef]
- Punnia-Moorthy, G.; Hersey, P.; Emran, A.A.; Tiffen, J. Lysine Demethylases: Promising Drug Targets in Melanoma and Other Cancers. Front. Genet. 2021, 12, 680633. [Google Scholar] [CrossRef] [PubMed]
- Vogel, F.C.E.; Bordag, N.; Zügner, E.; Trajkovic-Arsic, M.; Chauvistré, H.; Shannan, B.; Váraljai, R.; Horn, S.; Magnes, C.; Thomas Siveke, J.; et al. Targeting the H3K4 Demethylase KDM5B Reprograms the Metabolome and Phenotype of Melanoma Cells. J. Invest. Dermatol. 2019, 139, 2506–2516.e10. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Cheng, X.; Zhou, X. The BET-bromodomain inhibitor JQ1 mitigates vemurafenib drug resistance in melanoma. Melanoma Res. 2018, 28, 521–526. [Google Scholar] [CrossRef]
- Gallagher, S.J.; Gunatilake, D.; Beaumont, K.A.; Sharp, D.M.; Tiffen, J.C.; Heinemann, A.; Weninger, W.; Haass, N.K.; Wilmott, J.S.; Madore, J.; et al. HDAC inhibitors restore BRAF-inhibitor sensitivity by altering PI3K and survival signalling in a subset of melanoma. Int. J. cancer 2018, 142, 1926–1937. [Google Scholar] [CrossRef]
- Héninger, E.; Krueger, T.E.G.; Lang, J.M. Augmenting antitumor immune responses with epigenetic modifying agents. Front. Immunol. 2015, 6, 29. [Google Scholar] [CrossRef] [PubMed]
- Dan, H.; Zhang, S.; Zhou, Y.; Guan, Q. DNA Methyltransferase Inhibitors: Catalysts For Antitumour Immune Responses. Onco. Targets. Ther. 2019, 12, 10903–10916. [Google Scholar] [CrossRef]
- Serrano, A.; Tanzarella, S.; Lionello, I.; Mendez, R.; Traversari, C.; Ruiz-Cabello, F.; Garrido, F. Rexpression of HLA class I antigens and restoration of antigen-specific CTL response in melanoma cells following 5-aza-2′-deoxycytidine treatment. Int. J. Cancer 2001, 94, 243–251. [Google Scholar] [CrossRef]
- Jansz, N.; Faulkner, G.J. Endogenous retroviruses in the origins and treatment of cancer. Genome Biol. 2021, 22, 147. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-F.; Ren, P.; Shen, W.-Z.; Jin, X.; Zhang, J. The expression, modulation and use of cancer-testis antigens as potential biomarkers for cancer immunotherapy. Am. J. Transl. Res. 2020, 12, 7002–7019. [Google Scholar]
- Burton, E.M.; Woody, T.; Glitza, I.C.; Amaria, R.N.; Keung, E.Z.-Y.; Diab, A.; Patel, S.P.; Wong, M.K.K.; Yee, C.; Hwu, P.; et al. A phase II study of oral azacitidine (CC-486) in combination with pembrolizumab (PEMBRO) in patients (pts) with metastatic melanoma (MM). J. Clin. Oncol. 2019, 37, 9560. [Google Scholar] [CrossRef]
- Woods, D.M.; Sodré, A.L.; Villagra, A.; Sarnaik, A.; Sotomayor, E.M.; Weber, J. HDAC Inhibition Upregulates PD-1 Ligands in Melanoma and Augments Immunotherapy with PD-1 Blockade. Cancer Immunol. Res. 2015, 3, 1375–1385. [Google Scholar] [CrossRef] [PubMed]
- Banik, D.; Moufarrij, S.; Villagra, A. Immunoepigenetics Combination Therapies: An Overview of the Role of HDACs in Cancer Immunotherapy. Int. J. Mol. Sci. 2019, 20, 2241. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-F.; Ning, F.; Liu, Z.-C.; Wu, L.; Li, Z.-Q.; Qi, Y.-F.; Zhang, G.; Wang, H.-S.; Cai, S.-H.; Du, J. Histone deacetylase inhibitors deplete myeloid-derived suppressor cells induced by 4T1 mammary tumors in vivo and in vitro. Cancer Immunol. Immunother. 2017, 66, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Knox, T.; Sahakian, E.; Banik, D.; Hadley, M.; Palmer, E.; Noonepalle, S.; Kim, J.; Powers, J.; Gracia-Hernandez, M.; Oliveira, V.; et al. Selective HDAC6 inhibitors improve anti-PD-1 immune checkpoint blockade therapy by decreasing the anti-inflammatory phenotype of macrophages and down-regulation of immunosuppressive proteins in tumor cells. Sci. Rep. 2019, 9, 6136. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Skora, A.D.; Li, Z.; Liu, Q.; Tam, A.J.; Blosser, R.L.; Diaz, L.A.J.; Papadopoulos, N.; Kinzler, K.W.; Vogelstein, B.; et al. Eradication of metastatic mouse cancers resistant to immune checkpoint blockade by suppression of myeloid-derived cells. Proc. Natl. Acad. Sci. USA 2014, 111, 11774–11779. [Google Scholar] [CrossRef]
- Winder, M.; Virós, A. Mechanisms of Drug Resistance in Melanoma. Handb. Exp. Pharmacol. 2018, 249, 91–108. [Google Scholar] [CrossRef]
- Signetti, L.; Elizarov, N.; Simsir, M.; Paquet, A.; Douguet, D.; Labbal, F.; Debayle, D.; Di Giorgio, A.; Biou, V.; Girard, C.; et al. Inhibition of Patched Drug Efflux Increases Vemurafenib Effectiveness against Resistant Braf(V600E) Melanoma. Cancers 2020, 12, 1500. [Google Scholar] [CrossRef]
- Zhang, J.; Shih, D.J.H.; Lin, S.-Y. Role of DNA repair defects in predicting immunotherapy response. Biomark. Res. 2020, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Czerwinska, P.; Jaworska, A.M.; Wlodarczyk, N.A.; Mackiewicz, A.A. Melanoma Stem Cell-Like Phenotype and Significant Suppression of Immune Response within a Tumor Are Regulated by TRIM28 Protein. Cancers 2020, 12, 2998. [Google Scholar] [CrossRef]
- Mancarella, D.; Plass, C. Epigenetic signatures in cancer: Proper controls, current challenges and the potential for clinical translation. Genome Med. 2021, 13, 23. [Google Scholar] [CrossRef] [PubMed]
- Wouters, J.; Vizoso, M.; Martinez-Cardus, A.; Carmona, F.J.; Govaere, O.; Laguna, T.; Joseph, J.; Dynoodt, P.; Aura, C.; Foth, M.; et al. Comprehensive DNA methylation study identifies novel progression-related and prognostic markers for cutaneous melanoma. BMC Med. 2017, 15, 101. [Google Scholar] [CrossRef] [PubMed]
- Stacchiotti, S.; Schoffski, P.; Jones, R.; Agulnik, M.; Villalobos, V.M.; Jahan, T.M.; Chen, T.W.-W.; Italiano, A.; Demetri, G.D.; Cote, G.M.; et al. Safety and efficacy of tazemetostat, a first-in-class EZH2 inhibitor, in patients (pts) with epithelioid sarcoma (ES) (NCT02601950). J. Clin. Oncol. 2019, 37, 11003. [Google Scholar] [CrossRef]
- Lewin, J.; Soria, J.-C.; Stathis, A.; Delord, J.-P.; Peters, S.; Awada, A.; Aftimos, P.G.; Bekradda, M.; Rezai, K.; Zeng, Z.; et al. Phase Ib Trial With Birabresib, a Small-Molecule Inhibitor of Bromodomain and Extraterminal Proteins, in Patients With Selected Advanced Solid Tumors. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2018, 36, 3007–3014. [Google Scholar] [CrossRef] [PubMed]
- Bolden, J.E.; Tasdemir, N.; Dow, L.E.; van Es, J.H.; Wilkinson, J.E.; Zhao, Z.; Clevers, H.; Lowe, S.W. Inducible in vivo silencing of Brd4 identifies potential toxicities of sustained BET protein inhibition. Cell Rep. 2014, 8, 1919–1929. [Google Scholar] [CrossRef]
| Class | Agent | Mechanism of Action | Development Stage | Combination | References |
|---|---|---|---|---|---|
| DNMTi | Azacitidine (Vidaza©) | Incorporation into DNA, covalently linking the DNMTs, leading to DNMTs exhaustion and DNA damage | Phase I/II clinical trials | - (NCT02223052) | [204] |
| Pembrolizumab (NCT02816021) | [205] | ||||
| Carboplatin + Avelumab (ACTRN12618000053224) | [206] | ||||
| Decitabine (Dacogen©) | Phase I/II clinical trials | - (NCT00002980) | [207] | ||
| TCR-engineered T-cell immunotherapy (NCT02650986) | [208] | ||||
| Decitabine, Temozolomide and Panobinostat (NCT00925132) | [209] | ||||
| Temozolomide (NCT00715793) | [210] | ||||
| Vemurafenib (NCT01876641) | [211] | ||||
| Vemurafenib + Cobimetinib (NCT01876641) | [212] | ||||
| Guadecitabine (SGI-110) | Phase I/II clinical trials | Ipilimumab (NCT02608437) | [213,214] | ||
| Nivolumab + ipilimumab (NCT04250246) | [215] | ||||
| Zebularine | Preclinical | - | [216] | ||
| Disulfiram (Antabuse©) | Prevent the interaction of DNMTs with their target sequences either by binding to the catalytic site of DNMTs or by binding to CpG-enriched sequences | Phase I clinical trials | - (NCT00256230) | [217] | |
| Arsenic Trioxide (NCT00571116) | [218] | ||||
| HDACi | Vorinostat/Suberoylanilide hydroxamic acid (SAHA/Zolinza©) | Targeting class I, II and IV HDACs | Phase I/II clinical trials | - (NCT02836548) | [219] |
| Domatinostat (4SC-202) | Targeting class I HDACs | Phase I clinical trials | Nivolumab + Ipilimumab (NCT04133948) | [220] | |
| Panobinostat (LBH589) | Inhibition of class I, II, and IV enzymes | Phase I clinical trials | - (NCT01065467) | [221] | |
| Ipilimumab (NCT02032810) | [222] | ||||
| Romidepsin (Despipeptide/FR901228) | Targeting class I HDACs | Phase II clinical trials | - (NCT00104884) | [223] | |
| Entinostat (SNDX-275/MS-275) | Inhibition of class I HDACs | Phase II clinical trials | Pembrolizumab (NCT02437136) | [224] | |
| Mocetinostat (DB11830) | Targeting class I HDACs | Phase I clinical trials | Ipilimumab (NCT03565406) | [225] | |
| Tinostamustine | Targeting all the classical HDACs | Phase I clinical trials | Nivolumab (NCT03903458) | [226] | |
| Valproic acid (Depakote©) | Inhibition of class I and II enzymes | Phase I clinical trials | Chemoimmunotherapy | [227] | |
| Sulforaphane | Regulation of inflammatory and cell survival pathways | Pilot clinical studies | - (NCT01568996) | [228] | |
| Trichostatin A (TSA) | Induction of cell cycle arrest and apoptotic pathways | Preclinical | - | [229] | |
| Dacinostat (LAQ824) | Regulation of cell cycle and apoptosis | Preclinical | - | [229] | |
| Suberic bishydroxamate (SBHA) | Induction of apoptosis | Preclinical | - | [229] | |
| EZH2i | Tazemetostat | Targeting EZH2 activity | Phase I/II clinical studies | Dabrafenib trametinib (NCT04557956) | [230] |
| CPI-1205 | Targeting EZH2 activity | Phase I clinical studies | Ipilimumab (NCT03525795) | [231] | |
| BETi | ODM-207 | Preventing BETs-acetylated histones interaction | Phase I clinical studies | - (NCT03035591) | [232] |
| NHWD-870 | Preventing BETs-acetylated histones interaction | Preclinical | - | [233] | |
| PLX51107 | Preventing BETs-acetylated histones interaction, regulation of TME | Preclinical | PLX3397 | [234] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dobre, E.-G.; Constantin, C.; Costache, M.; Neagu, M. Interrogating Epigenome toward Personalized Approach in Cutaneous Melanoma. J. Pers. Med. 2021, 11, 901. https://doi.org/10.3390/jpm11090901
Dobre E-G, Constantin C, Costache M, Neagu M. Interrogating Epigenome toward Personalized Approach in Cutaneous Melanoma. Journal of Personalized Medicine. 2021; 11(9):901. https://doi.org/10.3390/jpm11090901
Chicago/Turabian StyleDobre, Elena-Georgiana, Carolina Constantin, Marieta Costache, and Monica Neagu. 2021. "Interrogating Epigenome toward Personalized Approach in Cutaneous Melanoma" Journal of Personalized Medicine 11, no. 9: 901. https://doi.org/10.3390/jpm11090901
APA StyleDobre, E.-G., Constantin, C., Costache, M., & Neagu, M. (2021). Interrogating Epigenome toward Personalized Approach in Cutaneous Melanoma. Journal of Personalized Medicine, 11(9), 901. https://doi.org/10.3390/jpm11090901









