Identification of Potential Repurposable Drugs in Alzheimer’s Disease Exploiting a Bioinformatics Analysis
Abstract
1. Introduction
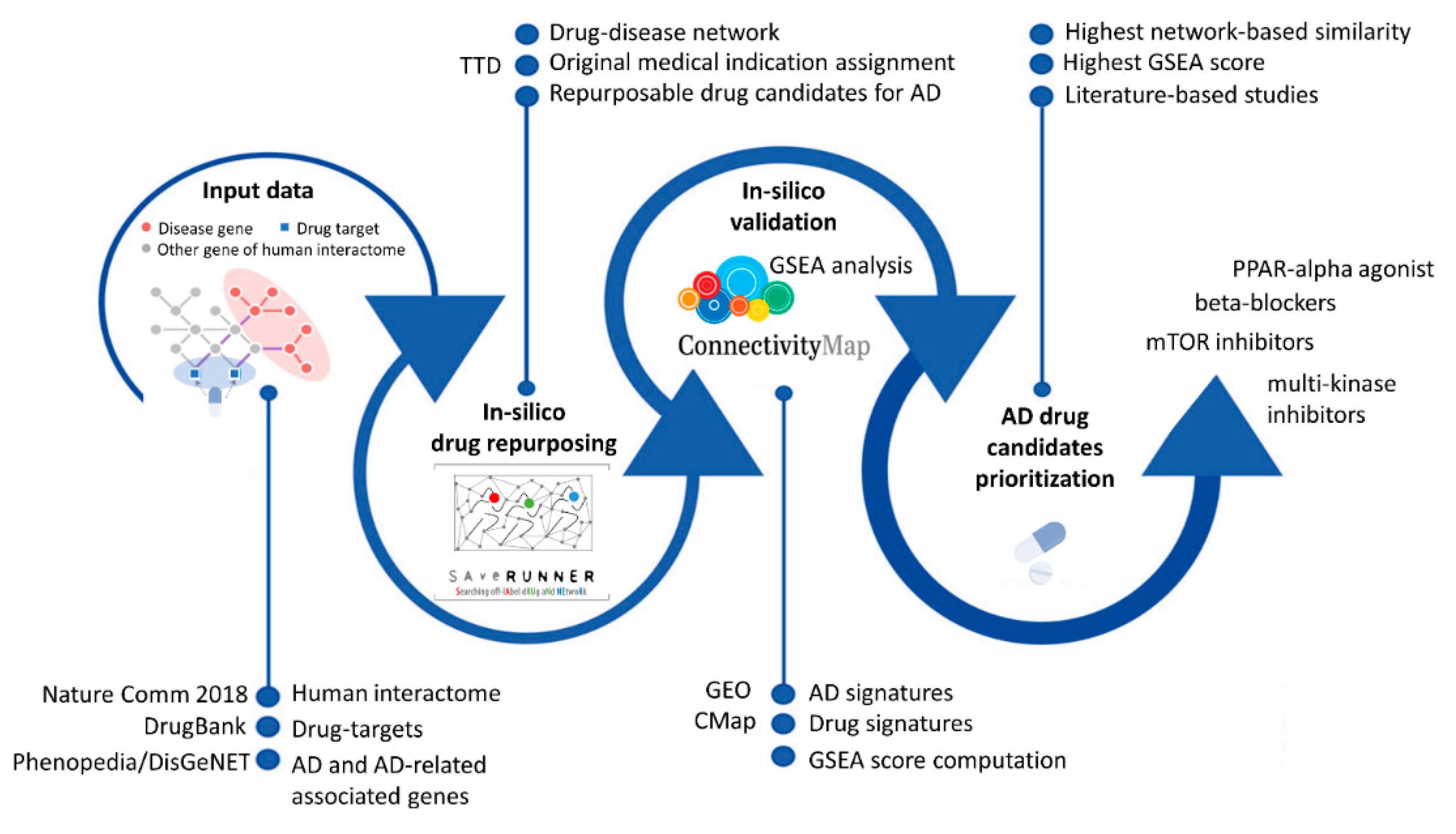
2. Materials and Methods
2.1. Data Retrieval
2.1.1. Human Interactome
2.1.2. Drug–Target Interactions
2.1.3. Drug Medical Indications
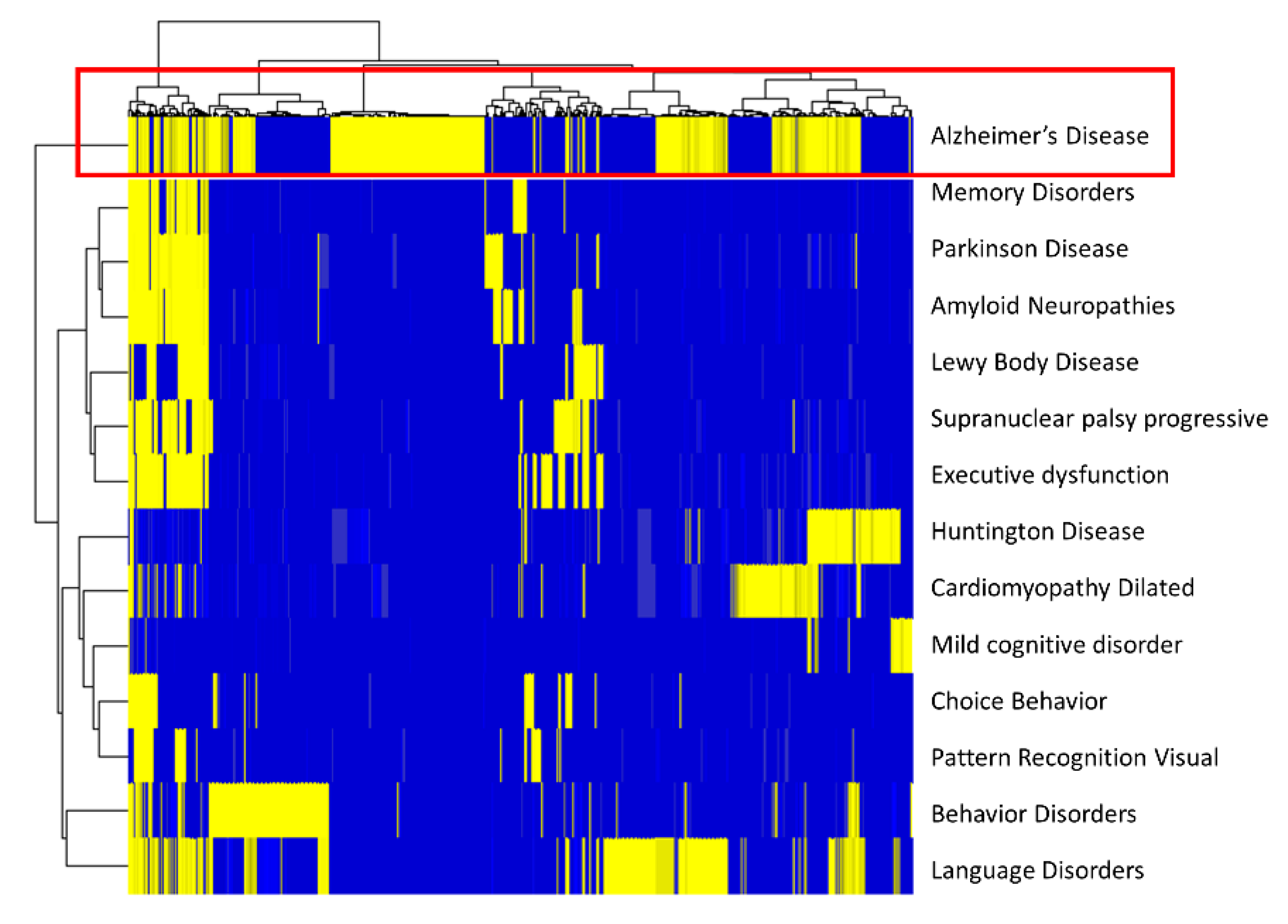
2.1.4. Disease Gene Associations
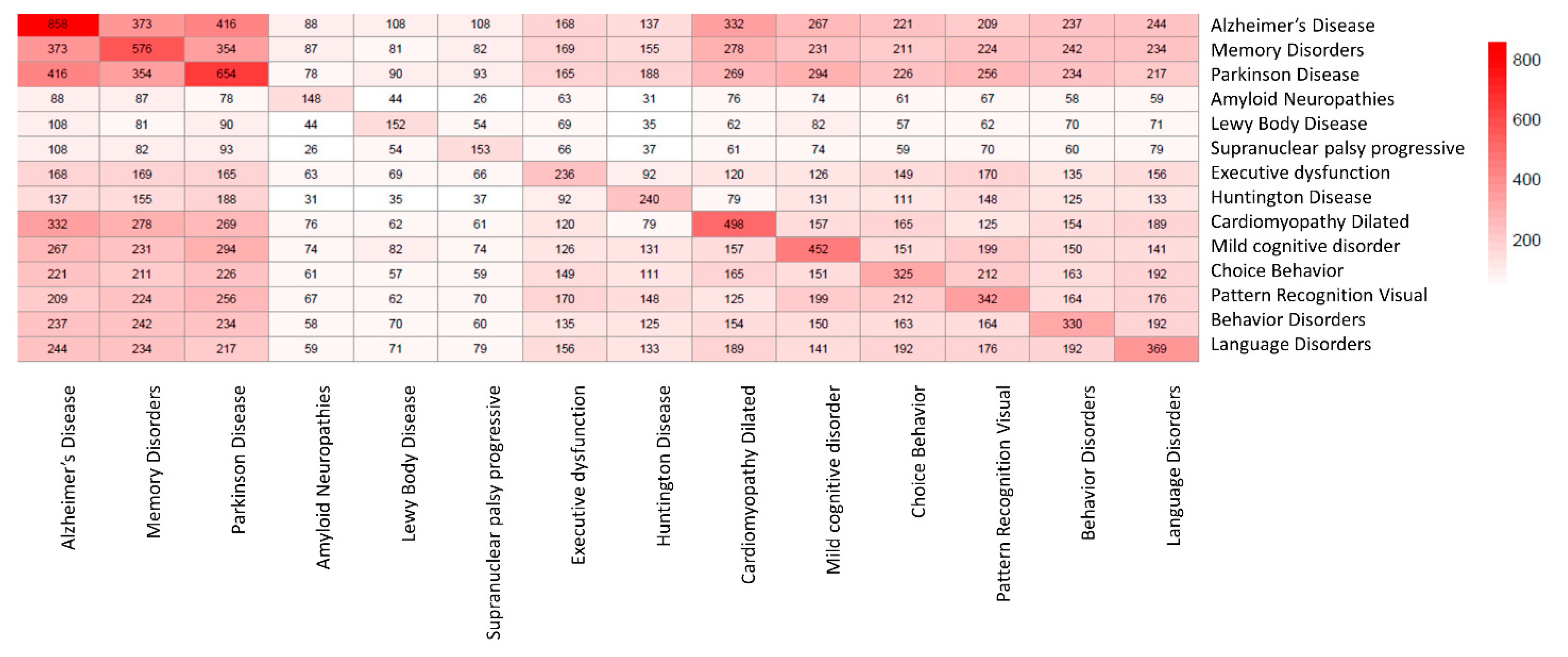
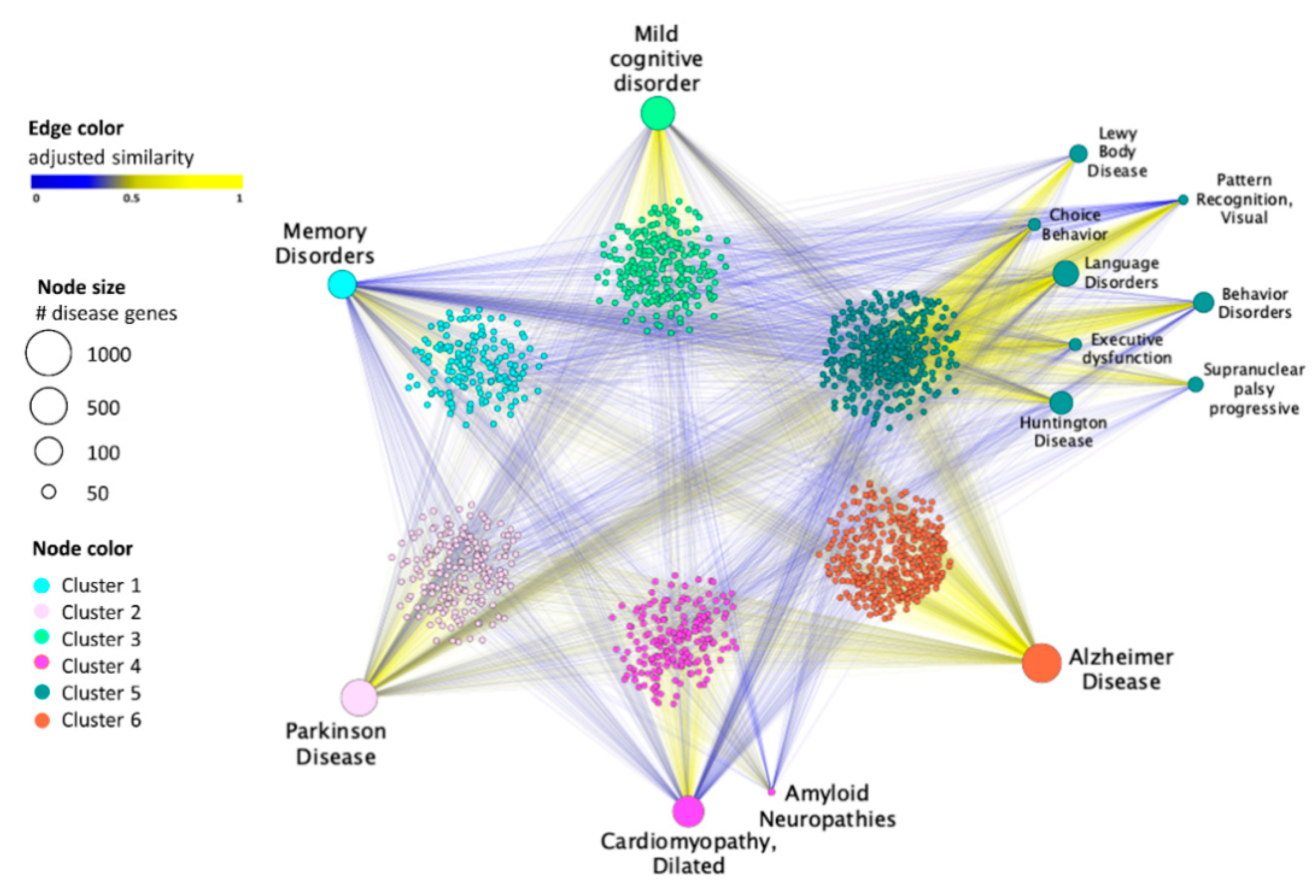
2.2. SAveRUNNER Algorithm
2.3. In Silico Validation: Gene Set Enrichment Analysis
2.3.1. Disease Signature
- (i)
- Expression profiling by an array of human frozen hippocampal tissue blocks containing both gray and white matter from a total of 30 subjects, including 22 patients with AD and 8 control samples (GSE28146 [29]).
- (ii)
- Expression profiling by an array of entorhinal cortex neurons from 19 AD patients and 14 non-demented controls (GSE4757 [30]).
- (iii)
- Expression profiling by array related to AD and control samples originating from the EU funded AddNeuroMed Cohort [31] and available from the GEO repository via the following accession numbers: GSE63060—batch 1 and GSE63061—batch 2 [32]. In particular, batch 1 (GSE63060) has a total of 249 samples, including 145 AD and 104 control samples, whereas batch 2 (GSE63061) has a total of 273 samples, including 139 AD and 134 control samples. The probe-sets were mapped to official gene symbols using the relative platform (GPL6947-13512 for GSE63060 and GPL10558-50081 for GSE63061). Multiple probe measurements of a given gene were collapsed into a single gene measurement by considering the mean. By matching genes based on gene symbols, we created a single merged dataset with both batches. We ran Combat function from R/Bioconductor package SVA to correct for batch-specific effects. Finally, we obtained a data matrix of 19,460 gene symbols (rows) and 522 samples (columns) including 284 AD and 238 control samples.
2.3.2. Drug Signature
2.3.3. GSEA Score Computation
3. Results and Discussion
3.1. Drug–Disease Network
3.2. In Silico Validation: GSEA Analysis
3.2.1. GSEA Score 3
3.2.2. GSEA Score 2
3.2.3. GSEA Score 1
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Knopman, D.S.; Amieva, H.; Petersen, R.C.; Chételat, G.; Holtzman, D.M.; Hyman, B.T.; Nixon, R.A.; Jones, D.T. Alzheimer Disease. Nat. Rev. Dis. Primer 2021, 7, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Haque, R.U.; Levey, A.I. Alzheimer’s Disease: A Clinical Perspective and Future Nonhuman Primate Research Opportunities. Proc. Natl. Acad. Sci. USA 2019, 116, 26224–26229. [Google Scholar] [CrossRef] [PubMed]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug Repurposing: Progress, Challenges and Recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Kukharsky, M.S.; Skvortsova, V.I.; Bachurin, S.O.; Buchman, V.L. In a Search for Efficient Treatment for Amyotrophic Lateral Sclerosis: Old Drugs for New Approaches. Med. Res. Rev. 2020, 41, 2804–2822. [Google Scholar] [CrossRef] [PubMed]
- Roessler, H.I.; Knoers, N.V.A.M.; van Haelst, M.M.; van Haaften, G. Drug Repurposing for Rare Diseases. Trends Pharmacol. Sci. 2021, 42, 255–267. [Google Scholar] [CrossRef]
- Cheng, F.; Desai, R.J.; Handy, D.E.; Wang, R.; Schneeweiss, S.; Barabási, A.-L.; Loscalzo, J. Network-Based Approach to Prediction and Population-Based Validation of in Silico Drug Repurposing. Nat. Commun. 2018, 9, 2691. [Google Scholar] [CrossRef] [PubMed]
- Barabási, A.-L.; Gulbahce, N.; Loscalzo, J. Network Medicine: A Network-Based Approach to Human Disease. Nat. Rev. Genet. 2011, 12, 56–68. [Google Scholar] [CrossRef]
- Gysi, D.M.; Do Valle, Í.; Zitnik, M.; Ameli, A.; Gan, X.; Varol, O.; Ghiassian, S.D.; Patten, J.J.; Davey, R.A.; Loscalzo, J.; et al. Network Medicine Framework for Identifying Drug-Repurposing Opportunities for COVID-19. Proc. Natl. Acad. Sci. USA 2021, 118, e2025581118. [Google Scholar] [CrossRef]
- Fiscon, G.; Conte, F.; Farina, L.; Paci, P. SAveRUNNER: A Network-Based Algorithm for Drug Repurposing and Its Application to COVID-19. PLOS Comput. Biol. 2021, 17, e1008686. [Google Scholar] [CrossRef]
- Fiscon, G.; Paci, P. SAveRUNNER: An R-Based Tool for Drug Repurposing. BMC Bioinform. 2021, 22, 150. [Google Scholar] [CrossRef]
- Fiscon, G.; Conte, F.; Amadio, S.; Volonté, C.; Paci, P. Drug Repurposing: A Network-Based Approach to Amyotrophic Lateral Sclerosis. Neurotherapeutics 2021, 18, 1678–1691. [Google Scholar] [CrossRef] [PubMed]
- Amadio, S.; Conte, F.; Esposito, G.; Fiscon, G.; Paci, P.; Volonté, C. Repurposing Histaminergic Drugs in Multiple Sclerosis. Int. J. Mol. Sci. 2022, 23, 6347. [Google Scholar] [CrossRef] [PubMed]
- Beaman, E.E.; Bonde, A.N.; Ulv Larsen, S.M.; Ozenne, B.; Lohela, T.J.; Nedergaard, M.; Gíslason, G.H.; Knudsen, G.M.; Holst, S.C. Blood–Brain Barrier Permeable β-Blockers Linked to Lower Risk of Alzheimer’s Disease in Hypertension. Brain 2022, awac076. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.X.; Correia, S.C.; Cardoso, S.; Carvalho, C.; Santos, M.S.; Moreira, P.I. Effects of Rapamycin and TOR on Aging and Memory: Implications for Alzheimer’s Disease. J. Neurochem. 2011, 117, 927–936. [Google Scholar] [CrossRef] [PubMed]
- Richardson, A.; Galvan, V.; Lin, A.-L.; Oddo, S. How Longevity Research Can Lead to Therapies for Alzheimer’s Disease: The Rapamycin Story. Exp. Gerontol. 2015, 68, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Rong, X.; Li, H.; Deng, Z.; Wang, J.; Liu, X.; He, L.; Xu, Y.; Tang, Y. Anti-Platelet Therapy Is Associated With Lower Risk of Dementia in Patients With Cerebral Small Vessel Disease. Front. Aging Neurosci. 2022, 14, 788407. [Google Scholar] [CrossRef]
- Khalaf, N.E.A.; El Banna, F.M.; Youssef, M.Y.; Mosaad, Y.M.; Daba, M.-H.Y.; Ashour, R.H. Clopidogrel Combats Neuroinflammation and Enhances Learning Behavior and Memory in a Rat Model of Alzheimer’s Disease. Pharmacol. Biochem. Behav. 2020, 195, 172956. [Google Scholar] [CrossRef]
- Han, K.-M.; Kang, R.J.; Jeon, H.; Lee, H.; Lee, J.-S.; Park, H.; Gak Jeon, S.; Suk, K.; Seo, J.; Hoe, H.-S. Regorafenib Regulates AD Pathology, Neuroinflammation, and Dendritic Spinogenesis in Cells and a Mouse Model of AD. Cells 2020, 9, 1655. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A Major Update to the DrugBank Database for 2018. Nucleic Acids Res. 2018, 46, D1074. [Google Scholar] [CrossRef]
- Yu, W.; Clyne, M.; Khoury, M.J.; Gwinn, M. Phenopedia and Genopedia: Disease-Centered and Gene-Centered Views of the Evolving Knowledge of Human Genetic Associations. Bioinformatics 2010, 26, 145–146. [Google Scholar] [CrossRef]
- Piñero, J.; Bravo, À.; Queralt-Rosinach, N.; Gutiérrez-Sacristán, A.; Deu-Pons, J.; Centeno, E.; García-García, J.; Sanz, F.; Furlong, L.I. DisGeNET: A Comprehensive Platform Integrating Information on Human Disease-Associated Genes and Variants. Nucleic Acids Res. 2017, 45, D833–D839. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K.; et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017, 171, 1437–1452.e17. [Google Scholar] [CrossRef] [PubMed]
- Lamb, J.; Crawford, E.D.; Peck, D.; Modell, J.W.; Blat, I.C.; Wrobel, M.J.; Lerner, J.; Brunet, J.-P.; Subramanian, A.; Ross, K.N.; et al. The Connectivity Map: Using Gene-Expression Signatures to Connect Small Molecules, Genes, and Disease. Science 2006, 313, 1929–1935. [Google Scholar] [CrossRef]
- Wang, Z.; Monteiro, C.D.; Jagodnik, K.M.; Fernandez, N.F.; Gundersen, G.W.; Rouillard, A.D.; Jenkins, S.L.; Feldmann, A.S.; Hu, K.S.; McDermott, M.G.; et al. Extraction and Analysis of Signatures from the Gene Expression Omnibus by the Crowd. Nat. Commun. 2016, 7, 12846. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA Circles Function as Efficient MicroRNA Sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Licata, L.; Briganti, L.; Peluso, D.; Perfetto, L.; Iannuccelli, M.; Galeota, E.; Sacco, F.; Palma, A.; Nardozza, A.P.; Santonico, E.; et al. MINT, the Molecular Interaction Database: 2012 Update. Nucleic Acids Res. 2012, 40, D857–D861. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, S.; Li, F.; Zhou, Y.; Zhang, Y.; Wang, Z.; Zhang, R.; Zhu, J.; Ren, Y.; Tan, Y.; et al. Therapeutic Target Database 2020: Enriched Resource for Facilitating Research and Early Development of Targeted Therapeutics. Nucleic Acids Res. 2020, 48, D1031–D1041. [Google Scholar] [CrossRef]
- Yu, W.; Gwinn, M.; Clyne, M.; Yesupriya, A.; Khoury, M.J. A Navigator for Human Genome Epidemiology. Nat. Genet. 2008, 40, 124–125. [Google Scholar] [CrossRef]
- Blalock, E.M.; Buechel, H.M.; Popovic, J.; Geddes, J.W.; Landfield, P.W. Microarray Analyses of Laser-Captured Hippocampus Reveal Distinct Gray and White Matter Signatures Associated with Incipient Alzheimer’s Disease. J. Chem. Neuroanat. 2011, 42, 118–126. [Google Scholar] [CrossRef]
- Dunckley, T.; Beach, T.G.; Ramsey, K.E.; Grover, A.; Mastroeni, D.; Walker, D.G.; LaFleur, B.J.; Coon, K.D.; Brown, K.M.; Caselli, R.; et al. Gene Expression Correlates of Neurofibrillary Tangles in Alzheimer’s Disease. Neurobiol. Aging 2006, 27, 1359–1371. [Google Scholar] [CrossRef]
- Lovestone, S.; Francis, P.; Kloszewska, I.; Mecocci, P.; Simmons, A.; Soininen, H.; Spenger, C.; Tsolaki, M.; Vellas, B.; Wahlund, L.-O.; et al. AddNeuroMed—the European Collaboration for the Discovery of Novel Biomarkers for Alzheimer’s Disease. Ann. N. Y. Acad. Sci. 2009, 1180, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Sood, S.; Gallagher, I.J.; Lunnon, K.; Rullman, E.; Keohane, A.; Crossland, H.; Phillips, B.E.; Cederholm, T.; Jensen, T.; van Loon, L.J.C.; et al. A Novel Multi-Tissue RNA Diagnostic of Healthy Ageing Relates to Cognitive Health Status. Genome Biol. 2015, 16, 185. [Google Scholar] [CrossRef] [PubMed]
- Sirota, M.; Dudley, J.T.; Kim, J.; Chiang, A.P.; Morgan, A.A.; Sweet-Cordero, A.; Sage, J.; Butte, A.J. Discovery and Preclinical Validation of Drug Indications Using Compendia of Public Gene Expression Data. Sci. Transl. Med. 2011, 3, 96ra77. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Clauset, A.; Newman, M.E.J.; Moore, C. Finding Community Structure in Very Large Networks. Phys. Rev. E 2004, 70, 066111. [Google Scholar] [CrossRef]
- Ferrero, J.; Williams, L.; Stella, H.; Leitermann, K.; Mikulskis, A.; O’Gorman, J.; Sevigny, J. First-in-Human, Double-Blind, Placebo-Controlled, Single-Dose Escalation Study of Aducanumab (BIIB037) in Mild-to-Moderate Alzheimer’s Disease. Alzheimers Dement. N. Y. 2016, 2, 169–176. [Google Scholar] [CrossRef]
- Branigan, G.L.; Soto, M.; Neumayer, L.; Rodgers, K.; Brinton, R.D. Association Between Hormone-Modulating Breast Cancer Therapies and Incidence of Neurodegenerative Outcomes for Women With Breast Cancer. JAMA Netw. Open 2020, 3, e201541. [Google Scholar] [CrossRef]
- Hui, Z.; Zhijun, Y.; Yushan, Y.; Liping, C.; Yiying, Z.; Difan, Z.; Chunglit, C.T.; Wei, C. The Combination of Acyclovir and Dexamethasone Protects against Alzheimer’s Disease-Related Cognitive Impairments in Mice. Psychopharmacology 2020, 237, 1851–1860. [Google Scholar] [CrossRef]
- Chandra, S.; Pahan, K. Gemfibrozil, a Lipid-Lowering Drug, Lowers Amyloid Plaque Pathology and Enhances Memory in a Mouse Model of Alzheimer’s Disease via Peroxisome Proliferator-Activated Receptor α. J. Alzheimers Dis. Rep. 2019, 3, 149–168. [Google Scholar] [CrossRef]
- Luo, R.; Su, L.-Y.; Li, G.; Yang, J.; Liu, Q.; Yang, L.-X.; Zhang, D.-F.; Zhou, H.; Xu, M.; Fan, Y.; et al. Activation of PPARA-Mediated Autophagy Reduces Alzheimer Disease-like Pathology and Cognitive Decline in a Murine Model. Autophagy 2020, 16, 52–69. [Google Scholar] [CrossRef]
- Cassano, T.; Magini, A.; Giovagnoli, S.; Polchi, A.; Calcagnini, S.; Pace, L.; Lavecchia, M.A.; Scuderi, C.; Bronzuoli, M.R.; Ruggeri, L.; et al. Early Intrathecal Infusion of Everolimus Restores Cognitive Function and Mood in a Murine Model of Alzheimer’s Disease. Exp. Neurol. 2019, 311, 88–105. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Shi, R.; She, X.; Gu, C.; Chong, L.; Zhang, L.; Li, R. Mineralocorticoid Receptor Antagonist-Mediated Cognitive Improvement in a Mouse Model of Alzheimer’s Type: Possible Involvement of BDNF-H2 S-Nrf2 Signaling. Fundam. Clin. Pharmacol. 2020, 34, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Jiao, B.; Shen, L. The Epigenetics of Alzheimer’s Disease: Factors and Therapeutic Implications. Front. Genet. 2018, 9, 579. [Google Scholar] [CrossRef] [PubMed]
- Wood, I.C. The Contribution and Therapeutic Potential of Epigenetic Modifications in Alzheimer’s Disease. Front. Neurosci. 2018, 12, 649. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.-F.; Jhao, Y.-T.; Chiu, C.-H.; Sun, L.-H.; Chou, T.-K.; Shiue, C.-Y.; Cheng, C.-Y.; Ma, K.-H. Bezafibrate Exerts Neuroprotective Effects in a Rat Model of Sporadic Alzheimer’s Disease. Pharm. Basel Switz. 2022, 15, 109. [Google Scholar] [CrossRef]
- Stuve, O.; Weideman, R.A.; McMahan, D.M.; Jacob, D.A.; Little, B.B. Diclofenac Reduces the Risk of Alzheimer’s Disease: A Pilot Analysis of NSAIDs in Two US Veteran Populations. Ther. Adv. Neurol. Disord. 2020, 13, 1756286420935676. [Google Scholar] [CrossRef]
- Yulug, B.; Hanoglu, L.; Ozansoy, M.; Isık, D.; Kilic, U.; Kilic, E.; Schabitz, W.R. Therapeutic Role of Rifampicin in Alzheimer’s Disease. Psychiatry Clin. Neurosci. 2018, 72, 152–159. [Google Scholar] [CrossRef]
- Umeda, T.; Tanaka, A.; Sakai, A.; Yamamoto, A.; Sakane, T.; Tomiyama, T. Intranasal Rifampicin for Alzheimer’s Disease Prevention. Alzheimers Dement. N. Y. 2018, 4, 304–313. [Google Scholar] [CrossRef]
- Fu, Q.; Gao, N.; Che, F.; Xu, G.; Han, H.; Su, Q.; Ma, G. Diphenyleneiodonium Chloride Synergizes with Diazoxide to Enhance Protection against Amyloid β Induced Neurotoxicity. J. Integr. Neurosci. 2019, 18, 445–449. [Google Scholar] [CrossRef]
- Liu, D.; Pitta, M.; Lee, J.-H.; Ray, B.; Lahiri, D.K.; Furukawa, K.; Mughal, M.; Jiang, H.; Villarreal, J.; Cutler, R.G.; et al. The KATP Channel Activator Diazoxide Ameliorates Amyloid-β and Tau Pathologies and Improves Memory in the 3xTgAD Mouse Model of Alzheimer’s Disease. J. Alzheimers Dis. JAD 2010, 22, 443–457. [Google Scholar] [CrossRef]
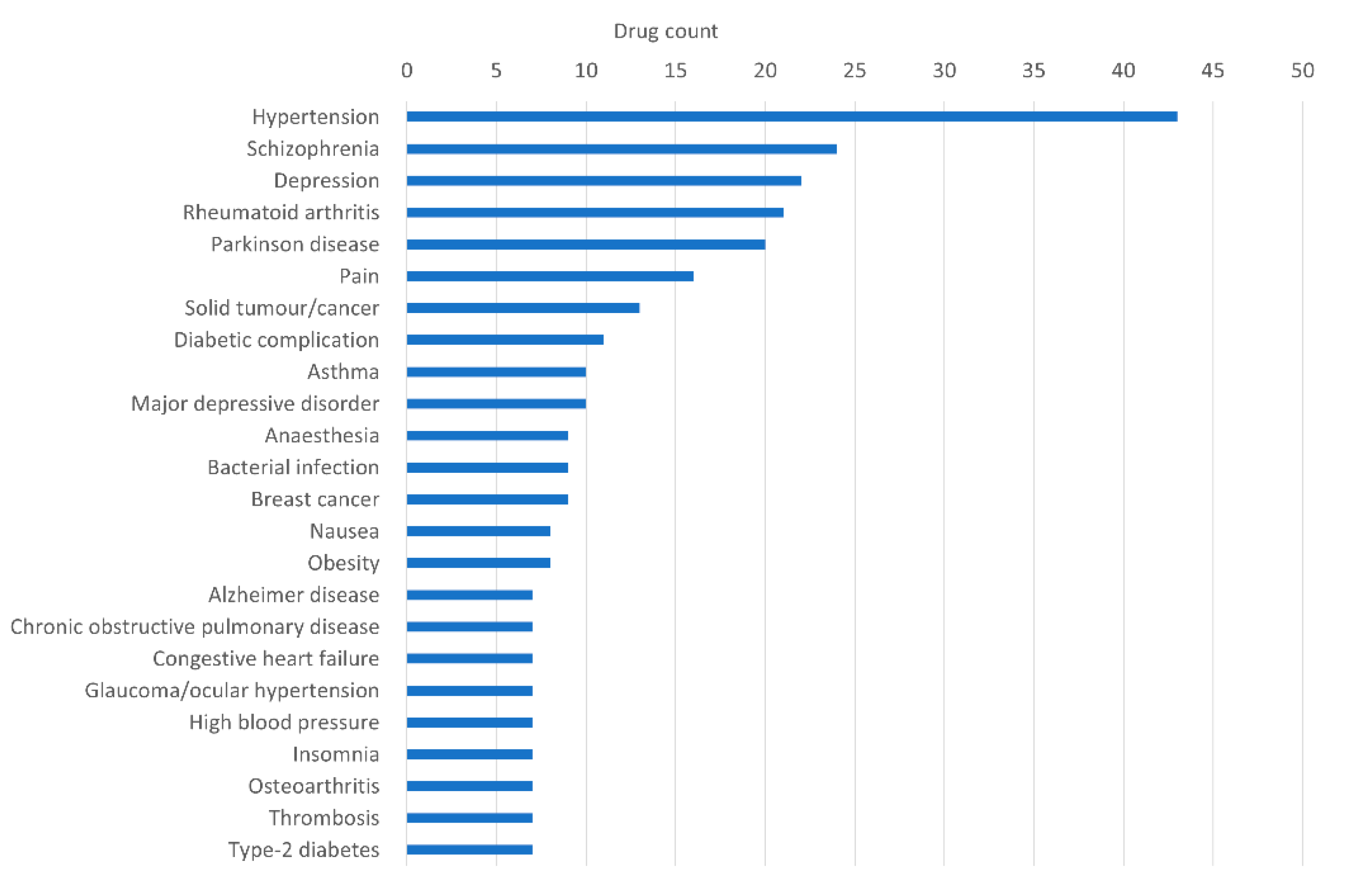
| Disease | Number of Disease Genes | Database |
|---|---|---|
| Alzheimer’s disease | 1749 | Phenopedia |
| Amyloid neuropathies | 8 | Phenopedia |
| Behavior disorders | 77 | DisGeNET |
| Cardiomyopathy, dilated | 148 | Phenopedia |
| Choice behavior | 33 | Phenopedia |
| Executive dysfunction | 33 | DisGeNET |
| Huntington’s disease | 79 | DisGeNET |
| Language disorders | 112 | Phenopedia |
| Lewy Body disease | 54 | DisGeNET |
| Memory disorders | 120 | Phenopedia |
| Mild cognitive disorder | 430 | DisGeNET |
| Parkinson disease | 629 | DisGeNET |
| Pattern recognition, visual | 31 | Phenopedia |
| Supranuclear palsy progressive | 43 | Phenopedia |
| Predicted Drug for AD | Adjusted Similarity | Drug Class | Original Medical Indication | GSEA Score | Shared with |
|---|---|---|---|---|---|
| regorafenib | 0.99 | multi-kinase inhibitor | Metastatic colorectal cancer | 3 | Cardiomyopathy, Dilated, Language Disorders |
| dexamethasone | 0.99 | corticosteroids | Rheumatoid arthritis | 3 | Memory Disorders |
| tamoxifen | 0.98 | estrogen receptor modulator | Breast cancer | 3 | Specifically predicted for AD |
| clopidogrel | 0.99 | platelet inhibitor | Thrombosis | 2 | Specifically predicted for AD |
| sirolimus (rapamycin) | 0.99 | mTOR inhibitor immunosuppressant | Organ transplant Rejection | 2 | Memory Disorders |
| everolimus | 0.99 | mTOR inhibitor | Kidney cancer | 2 | Specifically predicted for AD |
| gemfibrozil | 0.98 | PPAR-alpha agonist | Hyperlipidaemia | 2 | Specifically predicted for AD |
| spironolactone | 0.95 | aldosterone receptor antagonist | Congestive heart failure | 2 | Specifically predicted for AD |
| azacitidine | 0.99 | DNA methyltransferases inhibitor | Myelodysplastic syndrome | 1 | Specifically predicted for AD |
| bezafibrate | 0.99 | PPAR-alpha agonist | Hyperlipidaemia | 1 | Specifically predicted for AD |
| diclofenac | 0.99 | cyclooxygenase-1 and -2 inhibitor | Osteoarthritis | 1 | Specifically predicted for AD |
| rifampicin | 0.99 | DNA-dependent RNA polymerase inhibitor | Tuberculosis and Tuberculosis-related mycobacterial infections | 1 | Specifically predicted for AD |
| diazoxide | 0.98 | Potassium channel activator | Hyperthension | 1 | Memory disorders |
| betaxolol | 0.85 | Beta adrenergic antagonist | Hyperthension | 1 | Behavior disorders, cardiomyopathy, dilated, choice behavior, language disorders, memory disorders |
| bisoprolol | 0.85 | Beta adrenergic antagonist | Hyperthension | 1 | Behavior disorders, cardiomyopathy, dilated, choice behavior, language disorders, memory disorders, amyloid neuropathies |
| metoprolol | 0.85 | Beta adrenergic antagonist | Hyperthension | 1 | Behavior disorders, cardiomyopathy, dilated, choice behavior, language disorders, memory disorders, amyloid neuropathies |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fiscon, G.; Sibilio, P.; Funari, A.; Conte, F.; Paci, P. Identification of Potential Repurposable Drugs in Alzheimer’s Disease Exploiting a Bioinformatics Analysis. J. Pers. Med. 2022, 12, 1731. https://doi.org/10.3390/jpm12101731
Fiscon G, Sibilio P, Funari A, Conte F, Paci P. Identification of Potential Repurposable Drugs in Alzheimer’s Disease Exploiting a Bioinformatics Analysis. Journal of Personalized Medicine. 2022; 12(10):1731. https://doi.org/10.3390/jpm12101731
Chicago/Turabian StyleFiscon, Giulia, Pasquale Sibilio, Alessio Funari, Federica Conte, and Paola Paci. 2022. "Identification of Potential Repurposable Drugs in Alzheimer’s Disease Exploiting a Bioinformatics Analysis" Journal of Personalized Medicine 12, no. 10: 1731. https://doi.org/10.3390/jpm12101731
APA StyleFiscon, G., Sibilio, P., Funari, A., Conte, F., & Paci, P. (2022). Identification of Potential Repurposable Drugs in Alzheimer’s Disease Exploiting a Bioinformatics Analysis. Journal of Personalized Medicine, 12(10), 1731. https://doi.org/10.3390/jpm12101731







