Laboratory Cross-Comparison and Ring Test Trial for Tumor BRCA Testing in a Multicenter Epithelial Ovarian Cancer Series: The BORNEO GEICO 60-0 Study
Abstract
1. Introduction
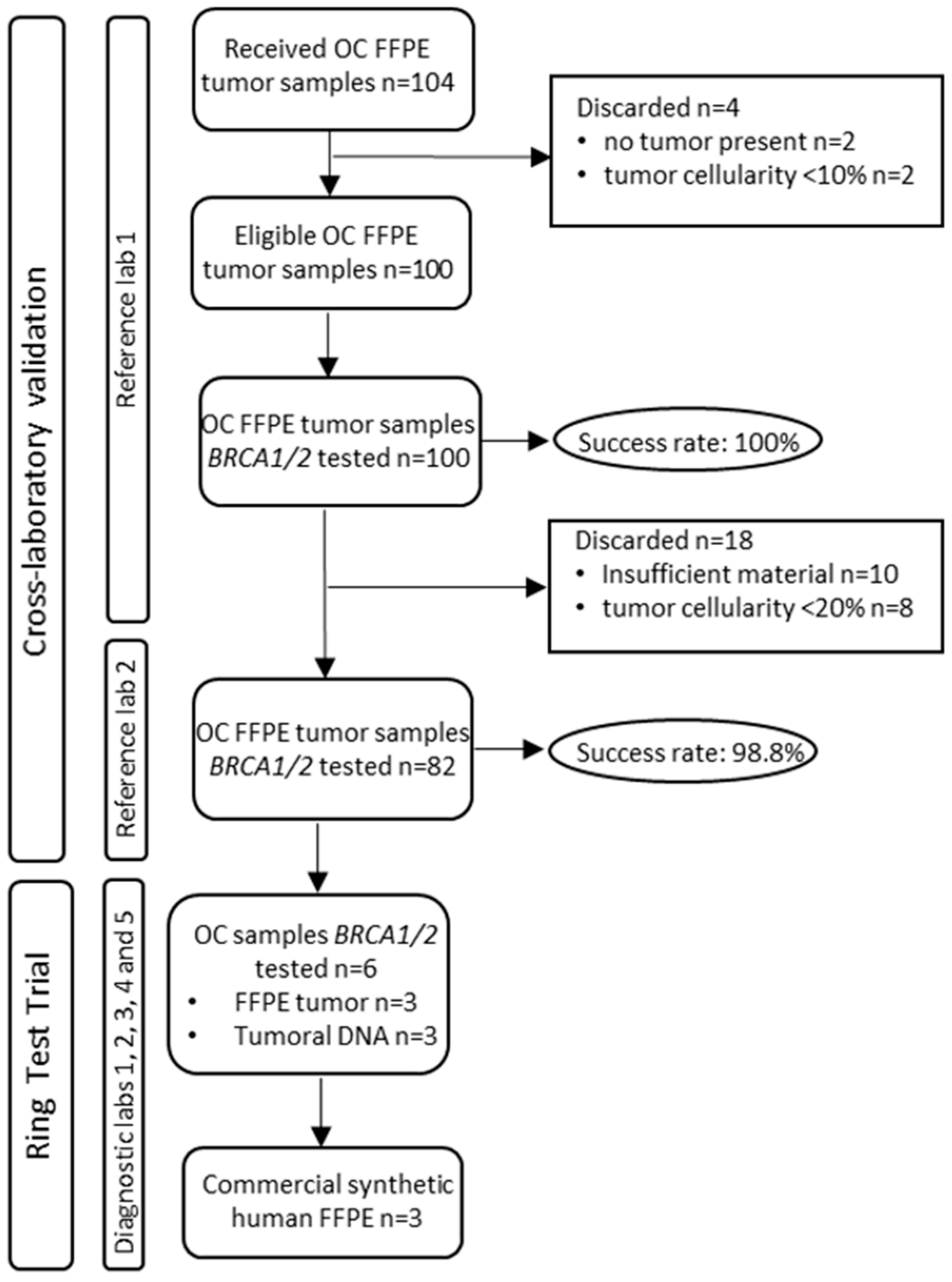
2. Materials and Methods
2.1. Study Design
- Written informed consent signed by all patients participating in the study.
- Histological diagnosis of non-mucinous epithelial ovarian, primary peritoneal, or tubal carcinoma confirmed not more than 2 years prior to the date of ICF signature.
- Adult women (18 years old or older at the time of diagnosis).
- Patients may have had other malignancies and have received or are receiving any anticancer therapy, including investigational drugs.
- Availability of FFPE tumor blocks from the primary tumor for genetic analysis and willingness (100 valid cases).
- Patients with gBRCA testing performed at the site according to current clinical guidelines or willing to be tested centrally if local testing is not available.
- Patients without an available medical record (lost, empty, or unretrievable clinical information).
2.2. Tumor BRCA1/2 Testing Cross-Laboratory Validation
2.2.1. RL1 DNA Extraction
2.2.2. RL1 NGS
2.2.3. RL2 DNA Extraction
2.2.4. RL2 NGS
2.3. Variant Analysis
2.4. Ring Test Trial (RTT)
3. Results
3.1. Tissue BRCA Testing in the Bilateral RL Comparison
3.1.1. Pathogenic and Likely Pathogenic Findings (P and LP)
- PID#81: BRCA1 c.3352C>T p.(Gln1118Ter), variant present at VAF < 5%, hence, below limit of reporting of RL2 and not called. Would not be a discrepancy, since RL1 and RL2 reported correctly according to their own specifications.
- PID#93: BRCA1 c.3770_3771del p.(Glu1257fs), detection failure in RL2 as a consequence of sample handling and insufficient tumor material.
- PID#15: BRCA2 c.8802_8828del p.(Met2935_Gln2943del), the different criterion in the interpretation, initially classified as VUS by the RL1.
3.1.2. Variants of Uncertain Significance (VUS)
- PID#15: BRCA2 c.8802_8828del p.(Met2935_Gln2943del), an in-frame deletion initially considered as VUS by RL1 and after consensus was reported as LP (already mentioned in the previous section).
- PID#13: BRCA2 c.2771A>T p.(Asn924Ile), an interpretation disagreement, classified as VUS and likely benign by RL1 and RL2, respectively.
- PID#57 and PID#79: BRCA1 c.80+6T>A and c.4986+9A>C, both variants located in intronic regions called by RL1 that was removed by the RL2 intronic threshold setting (+/− 3) in the bioinformatic pipeline.
- PID#73: BRCA2 c.353G>A p.(Arg118His), a missense change not reported by RL1 due to a bioinformatic error.
3.2. BRCA Ring Test Trial
- Variant 2: BRCA2 c.8802_8828del; p.(Met2935_Gln2943del), frameshift mutation not called in Lab2 as a consequence of bioinformatic filters.
- Variant 4: BRCA1 c.80+6T>A, intronic VUS reported only by two laboratories; in the remaining laboratories it was missed due to the data analysis methods and filters applied for intronic sequences.
- Variant 6: BRCA2 c.5351dupA; p.(Asn1784Lysfs), pathogenic mutation missed by three labs that used the Ion S5™ System (ThermoFisher Scientific) for sequencing (see Table 2). The alteration is an insertion located within a homopolymeric (polyA) region.
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Cabasag, C.J.; Fagan, P.J.; Ferlay, J.; Vignat, J.; Laversanne, M.; Liu, L.; van der Aa, M.A.; Bray, F.; Soerjomataram, I. Ovarian cancer today and tomorrow: A global assessment by world region and Human Development Index using GLOBOCAN 2020. Int. J. Cancer 2022, 151, 1535–1541. Available online: https://pubmed.ncbi.nlm.nih.gov/35322413/ (accessed on 11 May 2022). [CrossRef] [PubMed]
- Ledermann, J.A.; Harter, P.; Gourley, C.; Friedlander, M.; Vergote, I.; Rustin, G.; Scott, C.; Meier, W.; Shapira-Frommer, R.; Safra, T.; et al. Overall survival in patients with platinum-sensitive recurrent serous ovarian cancer receiving olaparib maintenance monotherapy: An updated analysis from a randomised, placebo-controlled, double-blind, phase 2 trial. Lancet Oncol. 2016, 17, 1579–1589. Available online: https://pubmed.ncbi.nlm.nih.gov/27617661/ (accessed on 25 March 2022). [CrossRef]
- Lheureux, S.; Lai, Z.; Dougherty, B.A.; Runswick, S.; Hodgson, D.R.; Timms, K.M.; Lanchbury, J.S.; Kaye, S.; Gourley, C.; Bowtell, D.; et al. Long-term responders on olaparib maintenance in high-grade serous ovarian cancer: Clinical and molecular characterization. Clin. Cancer Res. 2017, 23, 4086–4094. Available online: https://pubmed.ncbi.nlm.nih.gov/28223274/ (accessed on 11 May 2022). [CrossRef] [PubMed]
- Vergote, I.; González-Martín, A.; Ray-Coquard, I.; Harter, P.; Colombo, N.; Pujol, P.; Lorusso, D.; Mirza, M.; Brasiuniene, B.; Madry, R.; et al. European experts consensus: BRCA/homologous recombination deficiency testing in first-line ovarian cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2022, 33, 276–287. Available online: https://pubmed.ncbi.nlm.nih.gov/34861371/ (accessed on 11 May 2022). [CrossRef] [PubMed]
- Bell, D.; Berchuck, A.; Birrer, M.; Chien, J.; Cramer, D.W.; Dao, F.; Dhir, R.; Disaia, P.; Gabra, H.; Glenn, P.; et al. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609–615. Available online: https://pubmed.ncbi.nlm.nih.gov/21720365/ (accessed on 25 March 2022).
- Pennington, K.P.; Walsh, T.; Harrell, M.I.; Lee, M.K.; Pennil, C.C.; Rendi, M.H.; Thornton, A.; Norquist, B.M.; Casadei, S.; Nord, A.S.; et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin. Cancer Res. 2014, 20, 764–775. Available online: https://pubmed.ncbi.nlm.nih.gov/24240112/ (accessed on 25 March 2022). [CrossRef]
- Alsop, K.; Fereday, S.; Meldrum, C.; DeFazio, A.; Emmanuel, C.; George, J.; Dobrovic, A.; Birrer, M.J.; Webb, P.M.; Stewart, C.; et al. BRCA mutation frequency and patterns of treatment response in BRCA mutation-positive women with ovarian cancer: A report from the Australian Ovarian Cancer Study Group. J. Clin. Oncol. 2012, 30, 2654–2663. Available online: https://pubmed.ncbi.nlm.nih.gov/22711857/ (accessed on 11 May 2022). [CrossRef]
- Dann, R.B.; DeLoia, J.A.; Timms, K.M.; Zorn, K.K.; Potter, J.; Flake, D.D., II; Lanchbury, J.S.; Krivak, T.C. BRCA1/2 mutations and expression: Response to platinum chemotherapy in patients with advanced stage epithelial ovarian cancer. Gynecol. Oncol. 2012, 125, 677–682. Available online: https://pubmed.ncbi.nlm.nih.gov/22406760/ (accessed on 11 May 2022). [CrossRef]
- Sánchez-Lorenzo, L.; Salas-Benito, D.; Villamayor, J.; Patiño-García, A.; González-Martín, A. The BRCA gene in epithelial ovarian cancer. Cancers 2022, 14, 1235. Available online: https://pubmed.ncbi.nlm.nih.gov/35267543/ (accessed on 11 May 2022).
- Capoluongo, E.; Ellison, G.; López-Guerrero, J.A.; Penault-Llorca, F.; Ligtenberg, M.J.; Banerjee, S.; Singer, C.; Friedman, E.; Markiefka, B.; Schirmacher, P.; et al. Guidance statement on BRCA1/2 tumor testing in ovarian cancer patients. Semin. Oncol. 2017, 44, 187–197. Available online: https://pubmed.ncbi.nlm.nih.gov/29248130/ (accessed on 25 March 2022). [CrossRef]
- Konstantinopoulos, P.A.; Norquist, B.; Lacchetti, C.; Armstrong, D.; Grisham, R.N.; Goodfellow, P.J.; Kohn, E.C.; Levine, D.A.; Liu, J.F.; Lu, K.H.; et al. Germline and somatic tumor testing in epithelial ovarian cancer: ASCO guideline. J. Clin. Oncol. 2020, 38, 1222–1245. Available online: https://pubmed.ncbi.nlm.nih.gov/31986064/ (accessed on 11 May 2022). [CrossRef]
- Ellison, G.; Ahdesmäki, M.; Luke, S.; Waring, P.M.; Wallace, A.; Wright, R.; Röthlisberger, B.; Ludin, K.; Merkelbach-Bruse, S.; Heydt, C.; et al. An evaluation of the challenges to developing tumor BRCA1 and BRCA2 testing methodologies for clinical practice. Hum. Mutat. 2018, 39, 394–405. Available online: https://pubmed.ncbi.nlm.nih.gov/29215764/ (accessed on 11 May 2022). [CrossRef]
- Stegel, V.; Blatnik, A.; Škof, E.; Dragoš, V.Š.; Krajc, M.; Gregorič, B.; Škerl, P.; Strojnik, K.; Klančar, G.; Banjac, M.; et al. Real-world data on detection of germline and somatic pathogenic/likely pathogenic variants in BRCA1/2 and other susceptibility genes in ovarian cancer patients using next generation sequencing. Cancers 2022, 14, 1434. Available online: https://pubmed.ncbi.nlm.nih.gov/35326583/ (accessed on 11 May 2022). [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. Available online: https://pubmed.ncbi.nlm.nih.gov/25741868/ (accessed on 25 March 2022). [CrossRef]
- Den Dunnen, J.T.; Dalgleish, R.; Maglott, D.R.; Hart, R.K.; Greenblatt, M.S.; McGowan-Jordan, J.; Roux, A.F.; Smith, T.; Antonarakis, S.E.; Taschner, P.E.; et al. HGVS recommendations for the description of sequence variants: 2016 update. Hum. Mutat. 2016, 37, 564–569. Available online: https://pubmed.ncbi.nlm.nih.gov/26931183/ (accessed on 25 March 2022). [CrossRef]
- Palacios, J.; De La Hoya, M.; Bellosillo, B.; De Juan, I.; Matias-Guiu, X.; Lázaro, C.; Palanca, S.; Osorio, A.; Rojo, F.; Rosa-Rosa, J.M.; et al. Mutational screening of BRCA1/2 genes as a predictive factor for therapeutic response in epithelial ovarian cancer: A consensus guide from the Spanish Society of Pathology (SEAP-IAP) and the Spanish Society of Human Genetics (AEGH). Virchows Arch. 2020, 476, 195–207. Available online: https://pubmed.ncbi.nlm.nih.gov/31797087/ (accessed on 25 March 2022). [CrossRef]
- Stratton, J.F.; Gayther, S.A.; Russell, P.; Dearden, J.; Gore, M.; Blake, P.; Easton, D.; Ponder, B.A. Contribution of BRCA1 mutations to ovarian cancer. N. Engl. J. Med. 1997, 336, 1125–1130. Available online: https://pubmed.ncbi.nlm.nih.gov/9099656/ (accessed on 25 March 2022). [CrossRef]
- Norquist, B.M.; Harrell, M.I.; Brady, M.F.; Walsh, T.; Lee, M.K.; Gulsuner, S.; Bernards, S.S.; Casadei, S.; Yi, Q.; Burger, R.A.; et al. Inherited mutations in women with ovarian carcinoma. JAMA Oncol. 2016, 2, 482–490. Available online: https://pubmed.ncbi.nlm.nih.gov/26720728/ (accessed on 25 March 2022). [CrossRef]
- Petrillo, M.; Marchetti, C.; De Leo, R.; Musella, A.; Capoluongo, E.D.; Paris, I.; Panici, P.B.; Scambia, G.; Fagotti, A. BRCA mutational status, initial disease presentation, and clinical outcome in high-grade serous advanced ovarian cancer: A multicenter study. Am. J. Obstet. Gynecol. 2017, 217, 334.e1–334.e9. Available online: https://pubmed.ncbi.nlm.nih.gov/28549976/ (accessed on 25 March 2022). [CrossRef]
- Zhang, S.; Royer, R.; Li, S.; McLaughlin, J.R.; Rosen, B.; Risch, H.A.; Fan, I.; Bradley, L.; Shaw, P.A.; Narod, S.A. Frequencies of BRCA1 and BRCA2 mutations among 1,342 unselected patients with invasive ovarian cancer. Gynecol. Oncol. 2011, 121, 353–357. Available online: https://pubmed.ncbi.nlm.nih.gov/21324516/ (accessed on 25 March 2022). [CrossRef]
- Weren, R.D.; Mensenkamp, A.R.; Simons, M.; Eijkelenboom, A.; Sie, A.S.; Ouchene, H.; van Asseldonk, M.; Gomez-Garcia, E.B.; Blok, M.J.; de Hullu, J.A.; et al. Novel BRCA1 and BRCA2 tumor test as basis for treatment decisions and referral for genetic counselling of patients with ovarian carcinomas. Hum. Mutat. 2017, 38, 226–235. Available online: https://pubmed.ncbi.nlm.nih.gov/27767231/ (accessed on 25 March 2022). [CrossRef]
- Chandrasekaran, D.; Sobocan, M.; Blyuss, O.; Miller, R.E.; Evans, O.; Crusz, S.M.; Mills-Baldock, T.; Sun, L.; Hammond, R.F.L.; Gaba, F.; et al. Implementation of multigene germline and parallel somatic genetic testing in epithelial ovarian cancer: SIGNPOST study. Cancers 2021, 13, 4344. Available online: https://pubmed.ncbi.nlm.nih.gov/34503154/ (accessed on 25 March 2022). [CrossRef] [PubMed]
- Mafficini, A.; Simbolo, M.; Parisi, A.; Rusev, B.; Luchini, C.; Cataldo, I.; Piazzola, E.; Sperandio, N.; Turri, G.; Franchi, M.; et al. BRCA somatic and germline mutation detection in paraffin embedded ovarian cancers by next-generation sequencing. Oncotarget 2016, 7, 1076–1083. Available online: https://pubmed.ncbi.nlm.nih.gov/26745875/ (accessed on 25 March 2022). [CrossRef] [PubMed]
- Turashvili, G.; Lazaro, C.; Ying, S.; Charames, G.; Wong, A.; Hamilton, K.; Yee, D.; Agro, E.; Chang, M.; Pollett, A.; et al. Tumor BRCA testing in high grade serous carcinoma: Mutation rates and optimal tissue requirements. Cancers 2020, 12, 3468. Available online: https://pubmed.ncbi.nlm.nih.gov/33233347/ (accessed on 25 March 2022). [CrossRef] [PubMed]
- McAlpine, J.N.; Porter, H.; Kobel, M.; Nelson, B.H.; Prentice, L.M.; Kalloger, S.E.; Senz, J.; Milne, K.; Ding, J.; Shah, S.P.; et al. BRCA1 and BRCA2 mutations correlate with TP53 abnormalities and presence of immune cell infiltrates in ovarian high-grade serous carcinoma. Mod. Pathol. 2012, 25, 740–750. Available online: https://pubmed.ncbi.nlm.nih.gov/22282309/ (accessed on 25 March 2022). [CrossRef] [PubMed]
- Yang, D.; Khan, S.; Sun, Y.; Hess, K.; Shmulevich, I.; Sood, A.K.; Zhang, W. Association of BRCA1 and BRCA2 mutations with survival, chemotherapy sensitivity, and gene mutator phenotype in patients with ovarian cancer. JAMA 2011, 306, 1557–1565. Available online: https://pubmed.ncbi.nlm.nih.gov/21990299/ (accessed on 25 March 2022). [CrossRef]
- Hennessy, B.T.; Timms, K.M.; Carey, M.S.; Gutin, A.; Meyer, L.A.; Flake, D.D., 2nd; Abkevich, V.; Potter, J.; Pruss, D.; Glenn, P.; et al. Somatic mutations in BRCA1 and BRCA2 could expand the number of patients that benefit from poly (ADP ribose) polymerase inhibitors in ovarian cancer. J. Clin. Oncol. 2010, 28, 3570–3576. Available online: https://pubmed.ncbi.nlm.nih.gov/20606085/ (accessed on 25 March 2022). [CrossRef]
- Bragg, L.M.; Stone, G.; Butler, M.K.; Hugenholtz, P.; Tyson, G.W. Shining a light on dark sequencing: Characterising errors in Ion Torrent PGM data. PLoS Comput. Biol. 2013, 9, e1003031. Available online: https://pubmed.ncbi.nlm.nih.gov/23592973/ (accessed on 25 March 2022). [CrossRef]
- Yeo, Z.X.; Wong, J.C.L.; Rozen, S.G.; Lee, A.S.G. Evaluation and optimisation of indel detection workflows for ion torrent sequencing of the BRCA1 and BRCA2 genes. BMC Genom. 2014, 15, 516. Available online: https://pubmed.ncbi.nlm.nih.gov/24962530/ (accessed on 25 March 2022). [CrossRef]
- Spurdle, A.B.; Healey, S.; Devereau, A.; Hogervorst, F.B.L.; Monteiro, A.N.A.; Nathanson, K.L.; Radice, P.; Stoppa-Lyonnet, D.; Tavtigian, S.; Wappenschmidt, B.; et al. ENIGMA—Evidence-based network for the interpretation of germline mutant alleles: An international initiative to evaluate risk and clinical significance associated with sequence variation in BRCA1 and BRCA2 genes. Hum. Mutat. 2012, 33, 2–7. Available online: https://pubmed.ncbi.nlm.nih.gov/21990146/ (accessed on 25 March 2022). [CrossRef]
- Li, M.M.; Datto, M.; Duncavage, E.J.; Kulkarni, S.; Lindeman, N.I.; Roy, S.; Tsimberidou, A.M.; Vnencak-Jones, C.L.; Wolff, D.J.; Younes, A.; et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: A joint consensus recommendation of the association for molecular pathology, American Society of Clinical Oncology, and College of American Pathologists. J. Mol. Diagn. 2017, 19, 4–23. Available online: https://pubmed.ncbi.nlm.nih.gov/27993330/ (accessed on 30 March 2022). [CrossRef]
- de Jonge, M.M.; Ruano, D.; van Eijk, R.; van der Stoep, N.; Nielsen, M.; Wijnen, J.T.; Ter Haar, N.T.; Baalbergen, A.; Bos, M.E.; Kagie, M.J.; et al. Validation and implementation of BRCA1/2 variant screening in ovarian tumor tissue. J. Mol. Diagn. 2018, 20, 600–611. Available online: https://pubmed.ncbi.nlm.nih.gov/29936257/ (accessed on 25 March 2022). [CrossRef]
- Fumagalli, C.; Tomao, F.; Betella, I.; Rappa, A.; Calvello, M.; Bonanni, B.; Bernard, L.; Peccatori, F.; Colombo, N.; Viale, G.; et al. Tumor BRCA test for patients with epithelial ovarian cancer: The role of molecular pathology in the era of PARP inhibitor therapy. Cancers 2019, 11, 1641. Available online: https://pubmed.ncbi.nlm.nih.gov/31653094/ (accessed on 25 March 2022). [CrossRef]
- Bekos, C.; Grimm, C.; Kranawetter, M.; Polterauer, S.; Oberndorfer, F.; Tan, Y.; Müllauer, L.; Singer, C. Reliability of tumor testing compared to germline testing for detecting BRCA1 and BRCA2 mutations in patients with epithelial ovarian cancer. J. Pers. Med. 2021, 11, 593. Available online: https://pubmed.ncbi.nlm.nih.gov/34202525/ (accessed on 25 March 2022). [CrossRef]
- Vos, J.R.; Fakkert, I.E.; de Hullu, J.A.; van Altena, A.M.; Sie, A.S.; Ouchene, H.; Willems, R.W.; Nagtegaal, I.D.; Jongmans, M.C.; Mensenkamp, A.R.; et al. Universal tumor DNA BRCA1/2 testing of ovarian cancer: Prescreening PARPi treatment and genetic predisposition. J. Natl. Cancer Inst. 2020, 112, 161–169. Available online: https://pubmed.ncbi.nlm.nih.gov/31076742/ (accessed on 25 March 2022). [CrossRef]
| Clinicopathological Parameters | RL1 | RL2 |
|---|---|---|
| n = 100 | n = 82 | |
| Mean age at diagnosis (range) | 56 (25–84) years | 56 (25–84) years |
| Histology | ||
| High-grade serous | 83 (83%) | 69 (84.1%) |
| Low-grade serous | 4 (4%) | 2 (2.4%) |
| Endometrioid G1 | 3 (3%) | 3 (3.7%) |
| Endometrioid G2 | 4 (4%) | 3 (3.7%) |
| Endometrioid G3 | 0 | 0 |
| Clear cells | 4 (4%) | 3 (3.7%) |
| Seromucinous | 1 (1%) | 1 (1.2%) |
| Carcinosarcoma | 1 (1%) | 1 (1.2%) |
| FIGO Stage | ||
| I | 17 (17%) | 15 (18.3%) |
| II | 9 (9%) | 6 (7.3%) |
| III | 39 (39%) | 32 (39%) |
| IV | 21 (21%) | 17 (20.7%) |
| NA | 14 (14%) | 12 (14.6%) |
| gBRCA | ||
| wt-BRCA | 70 (70%) | 58 (70.7%) |
| VUS-BRCA | 13 (13%) | 10 (12.2%) |
| mt-BRCA | 16 (16%) | 13 (15.9%) |
| NA | 1 (1%) | 1 (1.2%) |
| Laboratory | Chemistry | NGS-Panel | NGS-Instrument |
|---|---|---|---|
| Lab1 | Multiplex-PCR | Oncomine Comprehensive Assay v3 (Thermo Fisher) | Ion S5™ System (Thermo Fisher Scientific) |
| Lab2 | Multiplex-PCR | BRCA MASTR Plus Dx (Multiplicom) | MiSeq (Illumina) |
| Lab3 | Hybrid Capture | Sure Select XT (Agilent) | Ion S5™ System (ThermoFisher Scientific) |
| Lab4 | Hybrid Capture | MiniHRS (Sophia Genetics) | MiSeq (Illumina) |
| Lab5 | Multiplex-PCR | Oncomine BRCA Research Assay (Thermo Fisher) | Ion S5™ System (Thermo Fisher Scientific) |
| Variant Index | Sample | Variant | Clinical Classification | Expected Allelic Frequency (%) |
|---|---|---|---|---|
| 1 | DNA_1 | BRCA1:c.3334G>T p.(Glu1112Ter) | P | 49.1 |
| 2 | DNA_2 | BRCA2:c.8802_8828del p.(Met2935_Gln2943del) | LP | 8.2 |
| 3 | DNA_3 | No pathogenic variant | ||
| 4 | FFPE_1 | BRCA1:c.80+6T>A | VUS | 47.4 |
| 5 | FFPE_2 | No pathogenic variant | ||
| 6 | FFPE_3 | BRCA2:c.5351dupA p.(Asn1784Lysfs) | P | 20.1 |
| 7 | CC_1 | BRCA2:c.5351del p.(Asn1784fs) | P | 40 |
| 8 | CC_1 | BRCA1:c.4327C>T p.(Arg1443Ter) | P | 32.5 |
| 9 | CC_1 | BRCA2:c.5073del p.(Lys1691fs) | P | 32.5 |
| 10 | CC_1 | BRCA2:c.8021dup p.(Ile2675fs) | P | 10 |
| 11 | CC_1 | BRCA1:c.1303G>T p.(Asp435Tyr) | VUS | 7.5 |
| 12 | CC_2 | BRCA2:c.5351del p.(Asn1784fs) | P | 10.2 |
| 13 | CC_2 | BRCA1:c.4327C>T p.(Arg1443Ter) | P | 3.9 |
| 14 | CC_2 | BRCA2:c.5073delA p.(Lys1691AsnfsTer15) | P | 3.1 |
| 15 | CC_2 | BRCA2:c.8021dup p.(Ile2675fs) | P | 4.5 |
| 16 | CC_2 | BRCA1:c.1303G>T p.(Asp435Tyr) | VUS | 8.2 |
| 17 | CC_3 | No pathogenic variant |
| Patient ID | Description | RL1 | RL2 | Germline | ||||
|---|---|---|---|---|---|---|---|---|
| VAF (%) | Reads | Class | VAF (%) | Reads | Class | |||
| 7 | NM_007294.3(BRCA1):c.3627dupA p.(Glu1210Argfs) rs80357729 | 71 | 4287 | P | 46 | 361 | P | Yes |
| 11 | NM_007294.3(BRCA1):c.1674del p.(Gly559fs) rs80357600 | 59 | 13,641 | P | 52 | 362 | P | Yes |
| 13 | NM_007294(BRCA1): c.2843_2849del p.(Gly948Valfs*50) | 26 | 5823 | P | 13 | 313 | P | No |
| 14 | NM_007294.3(BRCA1):c.66_67AG p.(Glu23fs) rs80357914 | 70 | 13,914 | P | 64 | 342 | P | Yes |
| 15 | NM_000059.3(BRCA2):c.8802_8828del p.(Met2935_Gln2943del) | 8.2 | 6380 | VUS | 6 | 870 | P | No |
| 19 | NM_007294.3(BRCA1):c.115T>A p.(Cys39Ser) rs80357164 | 75 | 3148 | P | 78 | 125 | P | Yes |
| 21 | NM_007294.3(BRCA1):c.3331_3334del p.(Gln1111fs) rs80357701 | 68 | 557 | P | 57 | 65 | P | Yes |
| 28 | NM_007294.3(BRCA1):c.3648dupA p.(Ser1217Ilefs) rs80357902 | 80 | 5391 | P | 72 | 556 | P | No |
| 31 | NM_007294.3(BRCA1):c.3752_3755GTCT p.(Ser1253fs) rs80357868 | 55 | 4391 | P | 40 | 292 | P | Yes |
| 35 | NM_000059.3(BRCA2):c.715dup p.(Ser239fs) rs431825350 | 51 | 5313 | P | 52 | 257 | P | Yes |
| 48 | NM_007294.3(BRCA1): c.3334G>T p.(Glu1112Ter) | 49 | 18,219 | P | 53 | 704 | P | No |
| 62 | NM_000059.3(BRCA2):c.1128del p.(Phe376fs) rs80359263 | 73 | 4935 | P | 57 | 393 | P | Yes |
| 70 | NM_000059.3(BRCA2):c.5351dupA p.(Asn1784Lysfs) rs80359507 | 21 | 16,034 | P | 23 | 220 | P | No |
| 71 | NM_007294.3(BRCA1):c.845C>A p.(Ser282Ter) rs786203027 | 42 | 298 | P | 52 | 639 | P | Yes |
| 72 | NM_007294.3(BRCA1): c.5578del p.(His1860Thrfs*?) | 48 | 2399 | P | 36 | 110 | LP | NA |
| 79 | NM_007294.4(BRCA1):c.211A>G p.(Arg71Gly) rs80357382 | 71 | 6629 | P | 72 | 185 | P | Yes |
| 80 | NM_007294.4(BRCA1):c.211A>G p.(Arg71Gly) rs80357382 | 71 | 283 | P | 69 | 131 | P | Yes |
| 81 | NM_007294.3(BRCA1):c.3352C>T p.(Gln1118Ter) rs397507215 | 15 | 3636 | P | UR | No | ||
| 93 | NM_007294.3(BRCA1):c.3770_3771del p.(Glu1257fs) | 60 | 3503 | P | UR | Yes | ||
| 5 | NM_000059.3(BRCA2):c.9026_9030del p.(Tyr3009fs) | 45 | 994 | NT | Yes | |||
| 27 | NM_007294.3(BRCA1):c.3627dupA p.(Glu1210Argfs) | 45 | 3351 | NT | Yes | |||
| 36 | NM_000059.3(BRCA2):c.6275_6276del p.(Leu2092fs) | 53 | 21,450 | NT | Yes | |||
| 63 | NM_007294.3(BRCA1):c.1A>G p.(Met1Val) | 18 | 701 | NT | No | |||
| 110 | NM_000059.3(BRCA2): c.3022del p.(Ser1008Alafs*35) | 94 | 4321 | NT | No | |||
| RTT Laboratory | Data Analysis Tools/Pipelines | VAF | Minimum Coverage | Intron Flanking Region | Variant Annotation Databases |
|---|---|---|---|---|---|
| Lab1 | Ion ReporterTM Software Version 5.10 | 5% | 500× | ±10 bp | ClinVar, Varsome; COSMIC |
| Lab2 | MASTR Reporter 1.3.0 | 5% | 1000× | No | ClinVar; BRCA Exchange |
| Lab3 | novocraft V3.07.01, bamtools-2.4.1, VCFtools (0.1.15), bedtools v2.26.0-40, samtools 1.8, picardtools 2.8.3, ensembl vep release 94, CONTRA.v2.0.8, gatk-3.4.46 | 10% | 20× | NCBI, ClinVar, Ensembl, BRCA Exchange, cBioPortal | |
| Lab4 | Sophia DDM v3-Sophia Genetics | 5% | 500× | ClinVar, COSMIC, dbSNP, EXAC, g1000, ESP, EpiCov, GnomAD, | |
| Lab5 | Ion Reporter Software Version 5.16 | 5% | 100× | ±100 | dbSNP, BIC database, BRCA Exchange, BRCA Mutation Database |
| Variant | Type of Variant | Sample | Lab1 | Lab2 | Lab3 | Lab4 | Lab5 | Detection Concordance to Reference Genotype (%) | Interpretation Concordance to Reference Genotype (%) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | nonsense | DNA_1 | * | 100% | 60% | ||||
| 2 | frameshift | DNA_2 | * | 80% | 75% | ||||
| 3 | wt | DNA_3 | 100% | 100% | |||||
| 4 | splicing | FFPE_1 | 40% | 100% | |||||
| 5 | wt | FFPE_2 | 100% | 100% | |||||
| 6 | frameshift | FFPE_3 | 40% | 100% | |||||
| 7 | frameshift | CC_1 | 20% | 100% | |||||
| 8 | nonsense | CC_1 | 100% | 100% | |||||
| 9 | frameshift | CC_1 | 60% | 100% | |||||
| 10 | frameshift | CC_1 | 20% | 100% | |||||
| 11 | missense | CC_1 | 20% | 100% | |||||
| 12 | frameshift | CC_2 | * | 50% | 50% | ||||
| 13 | nonsense | CC_2 | 50% | 100% | |||||
| 14 | frameshift | CC_2 | 25% | 100% | |||||
| 15 | frameshift | CC_2 | 0% | 100% | |||||
| 16 | missense | CC_2 | 25% | 100% | |||||
| 17 | wt | CC_3 | 100% | 100% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garcia-Casado, Z.; Oaknin, A.; Mendiola, M.; Alkorta-Aranburu, G.; Antunez-Lopez, J.R.; Moreno-Bueno, G.; Palacios, J.; Yubero, A.; Marquez, R.; Gallego, A.; et al. Laboratory Cross-Comparison and Ring Test Trial for Tumor BRCA Testing in a Multicenter Epithelial Ovarian Cancer Series: The BORNEO GEICO 60-0 Study. J. Pers. Med. 2022, 12, 1842. https://doi.org/10.3390/jpm12111842
Garcia-Casado Z, Oaknin A, Mendiola M, Alkorta-Aranburu G, Antunez-Lopez JR, Moreno-Bueno G, Palacios J, Yubero A, Marquez R, Gallego A, et al. Laboratory Cross-Comparison and Ring Test Trial for Tumor BRCA Testing in a Multicenter Epithelial Ovarian Cancer Series: The BORNEO GEICO 60-0 Study. Journal of Personalized Medicine. 2022; 12(11):1842. https://doi.org/10.3390/jpm12111842
Chicago/Turabian StyleGarcia-Casado, Zaida, Ana Oaknin, Marta Mendiola, Gorka Alkorta-Aranburu, Jose Ramon Antunez-Lopez, Gema Moreno-Bueno, Jose Palacios, Alfonso Yubero, Raul Marquez, Alejandro Gallego, and et al. 2022. "Laboratory Cross-Comparison and Ring Test Trial for Tumor BRCA Testing in a Multicenter Epithelial Ovarian Cancer Series: The BORNEO GEICO 60-0 Study" Journal of Personalized Medicine 12, no. 11: 1842. https://doi.org/10.3390/jpm12111842
APA StyleGarcia-Casado, Z., Oaknin, A., Mendiola, M., Alkorta-Aranburu, G., Antunez-Lopez, J. R., Moreno-Bueno, G., Palacios, J., Yubero, A., Marquez, R., Gallego, A., Sanchez-Heras, A. B., Lopez-Guerrero, J. A., Perez-Segura, C., Barretina-Ginesta, P., Alarcon, J., Gaba, L., Marquez, A., Matito, J., Cueva, J., ... Vivancos, A. (2022). Laboratory Cross-Comparison and Ring Test Trial for Tumor BRCA Testing in a Multicenter Epithelial Ovarian Cancer Series: The BORNEO GEICO 60-0 Study. Journal of Personalized Medicine, 12(11), 1842. https://doi.org/10.3390/jpm12111842







