A Comprehensive Analysis of Recent Deep and Federated-Learning-Based Methodologies for Brain Tumor Diagnosis
Abstract
:1. Introduction
2. Research Method
2.1. Objectives of Research
- Focusing on the latest research on brain tumor diagnosis using deep and federated learning.
- Identifying current research trends, open issues, and challenges for brain tumor diagnosis.
- Investigating current brain tumor diagnosis approaches based on similarities and discrepancies.
- Proposing a taxonomy for brain tumor detection subsequent to an analysis of effective methods.
2.2. Research Questions
2.3. Search Strategy
2.4. Study Screening Criteria
- Research articles not based on binary disease classification.
- Research articles diagnosing brain tumors without medical images.
- Research articles not identifying data sources or employing ambiguous methods of data collection.
- Research articles based on non-human samples.
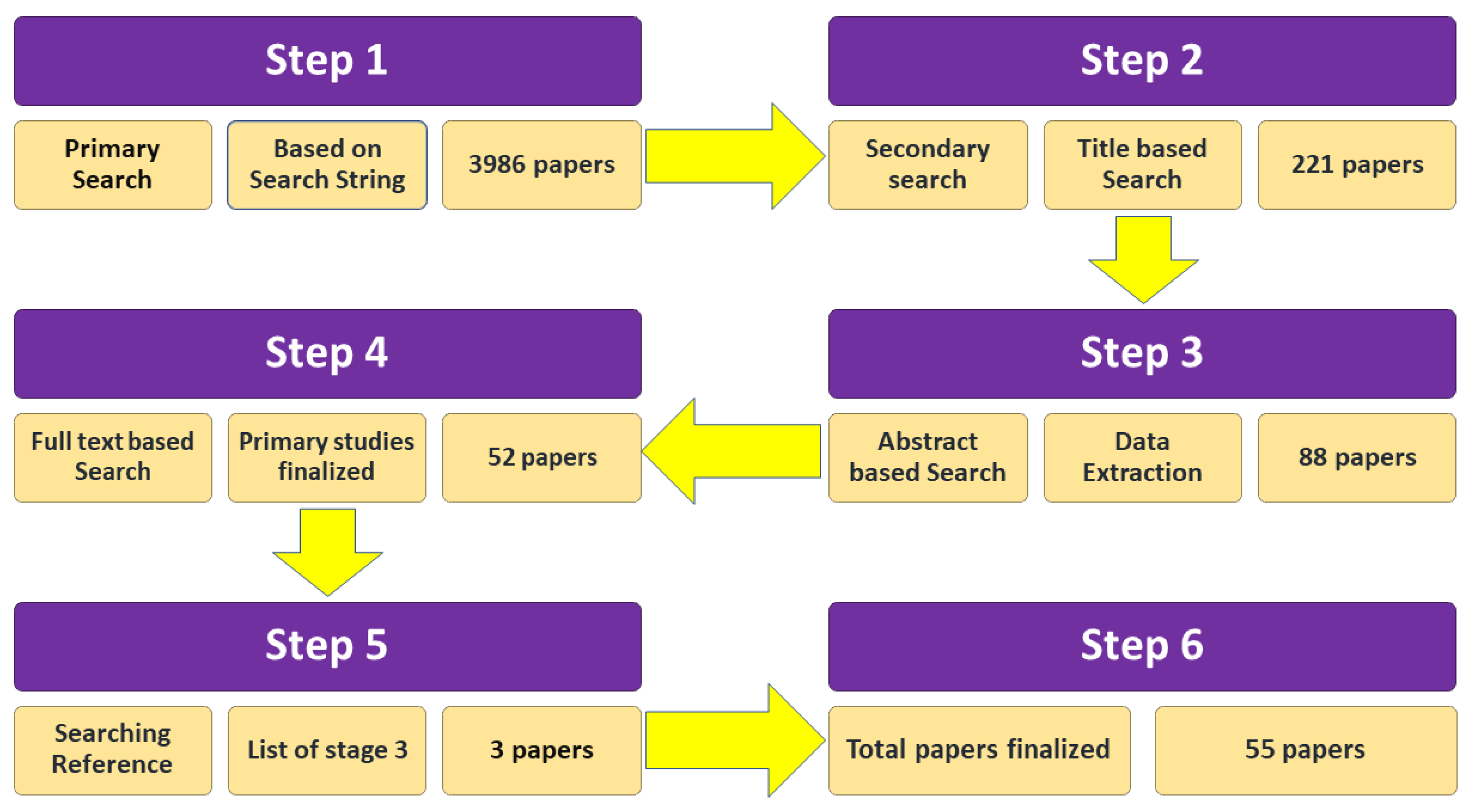
2.5. Study Selection Process
3. Data Analysis and Results
3.1. Search Results
3.2. Discussion and Evaluation of Research Questions
3.3. Analysis of RQ1: What Are the Best Available Methods for the Detection of Brain Tumors?
3.3.1. Pretrained Classifiers
3.3.2. Handcrafted Classifiers
3.3.3. Ensemble Classifiers
3.3.4. Federated Learning
3.4. RQ2 Assessment: What Are the Metrics Used to Determine the Performance of Different Methods Applied to Brain Tumor Diagnosis?
3.4.1. Performance Evaluation of Ensemble Methods
3.4.2. Performance Evaluation on Pretrained Method
3.4.3. Performance Evaluation on Transfer Learning Method
3.4.4. Performance Evaluation on Handcrafted Method
3.5. RQ3 Assessment: What Types of Datasets Are Available to Diagnose Brain Tumors?
3.5.1. Benchmark Datasets
3.5.2. Figshare Dataset
3.5.3. TCGA-GBM Dataset
3.5.4. BraTS 2012 & 2013 Datasets
3.5.5. BraTS 2014 Dataset
3.5.6. BraTS 2015 Dataset
3.5.7. BraTS 2016 and 2017 Dataset
3.5.8. BraTS 2018 Dataset
3.5.9. ISLES 2015 Dataset
3.5.10. ISLES 2017 Dataset
3.5.11. Brain MRI Dataset
3.5.12. BraTS 2019 Dataset
3.6. Non-Public Dataset
3.6.1. Combined Dataset
3.6.2. BRAINIX Dataset
3.7. RQ4 Assessment: What Is the Quality of the Selected Papers?
- (1)
- Has deep learning algorithms been used for diagnosing brain tumors? The response for the potential answer was ‘Yes (1)’ or ‘No (0)’.
- (2)
- Does the research provide a simple approach for the detection of disease with data sets? The response for a potential answer was ‘Yes (1)’ or ‘No (0)’.
- (3)
- A well-known and renowned publication source published the article. Quartile rankings (Q1, Q2, Q3, and Q4) were used to create the Journal Citation Reports, whereas (CORE) (A, B, and C) were used for computer science conference rankings
- 2.0 marks for CORE A-rank conference;
- 1.5 marks for CORE B rank conference;
- 1.0 mark for CORE C rank conference.
- 2.0 marks for Q1 rank journal;
- 1.5 marks for Q2 rank journal;
- 1.0 mark for Q3 rank journal;
- 0.5 marks for Q4 rank journal.
3.8. RQ5 Assessment: What Is the Impact of the Selected Papers on Brain Tumor Detection?
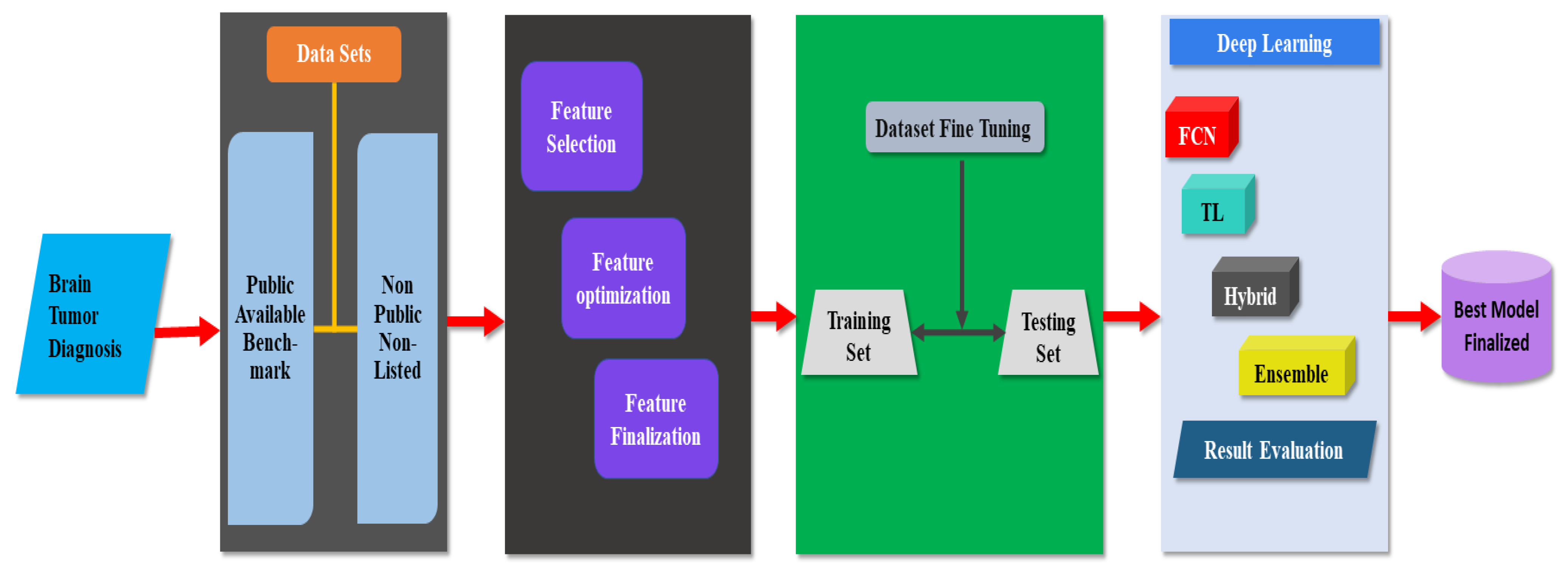
4. Discussion
4.1. Taxonomy for Brain Tumor Diagnosis
4.2. Common Model Used for Brain Tumor Diagnosis
5. Open Issues and Challenges
5.1. Dataset Variations
5.2. Number of Images in Dataset
5.3. Size of Tumor
5.4. Age of Patients
6. Principal Findings
6.1. Best Classifiers for Brain Tumor Detection
6.2. Accuracy Evaluation of Classifiers
6.3. Widely Used Datasets
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Publication Number | Source of Publication | Publication Year | Criteria for Quality | |||
|---|---|---|---|---|---|---|
| a | b | c | Final Score | |||
| [23] | Conference paper | 2019 | 0.5 | 1.0 | 1.5 | 3.0 |
| [24] | Journal paper | 2017 | 1.0 | 1.0 | 1.5 | 3.5 |
| [25] | Journal paper | 2019 | 1.0 | 1.0 | 1.5 | 3.5 |
| [26] | Conference paper | 2019 | 0.5 | 0.5 | 2.0 | 3.0 |
| [27] | Conference paper | 2019 | 0.5 | 0.5 | 2.0 | 3.0 |
| [28] | Conference paper | 2019 | 1.0 | 1.0 | 1.5 | 3.5 |
| [29] | Conference paper | 2019 | 1.0 | 1.0 | 1.5 | 3.5 |
| [30] | Conference paper | 2020 | 1.0 | 1.0 | 1.0 | 3.0 |
| [31] | Conference paper | 2017 | 1.0 | 1.0 | 1.5 | 3.5 |
| [32] | Journal paper | 2020 | 0.5 | 0.5 | 2.0 | 3.0 |
| [33] | Journal paper | 2020 | 1.0 | 1.0 | 2.0 | 4.0 |
| [34] | Journal paper | 2019 | 0.5 | 0.5 | 2.0 | 3.0 |
| [35] | Conference paper | 2019 | 1.0 | 1.0 | 1.5 | 3.5 |
| [36] | Conference paper | 2019 | 1.0 | 1.0 | 2.0 | 4.0 |
| [37] | Journal paper | 2020 | 1.0 | 1.0 | 1.5 | 3.5 |
| [38] | Conference paper | 2018 | 1.0 | 1.0 | 0.5 | 2.5 |
| [39] | Conference paper | 2019 | 0.5 | 0.5 | 1.5 | 2.5 |
| [40] | Conference paper | 2021 | 0.5 | 1.0 | 1.5 | 3.0 |
| [41] | Conference paper | 2019 | 1.0 | 1.0 | 2.0 | 4.0 |
| [42] | Journal paper | 2019 | 0.5 | 1.0 | 1.5 | 3.0 |
| [43] | Journal paper | 2019 | 1.0 | 0.5 | 2.0 | 3.5 |
| [44] | Journal paper | 2020 | 1.0 | 1.0 | 1.5 | 3.5 |
| [45] | Journal paper | 2020 | 0.5 | 1.0 | 1.5 | 3.0 |
| [46] | Journal paper | 2019 | 1.0 | 0.5 | 2.0 | 3.5 |
| [47] | Journal paper | 2020 | 1.0 | 1.0 | 0.0 | 2.0 |
| [48] | Journal paper | 2019 | 1.0 | 1.0 | 2.0 | 4.0 |
| [49] | Journal paper | 2021 | 1.0 | 1.0 | 2.0 | 4.0 |
| [50] | Journal paper | 2019 | 1.0 | 1.0 | 2.0 | 4.0 |
| [51] | Journal paper | 2019 | 1.0 | 1.0 | 2.0 | 4.0 |
| [52] | Journal paper | 2020 | 1.0 | 1.0 | 1.0 | 4.0 |
| [37] | Journal paper | 2020 | 1.0 | 1.0 | 2.0 | 4.0 |
| [53] | Journal paper | 2020 | 1.0 | 1.0 | 1.0 | 3.0 |
| [54] | Journal paper | 2020 | 1.0 | 1.0 | 1.0 | 3.0 |
| [55] | Journal paper | 2021 | 1.0 | 1.0 | 1.5 | 3.5 |
| [56] | Conference paper | 2019 | 1.0 | 1.0 | 1.0 | 3.0 |
| [57] | Journal paper | 2019 | 1.0 | 1.0 | 1.5 | 3.5 |
| [58] | Journal paper | 2018 | 1.0 | 1.0 | 1.5 | 3.5 |
| [59] | Journal paper | 2020 | 1.0 | 1.0 | 1.5 | 3.5 |
| [60] | Journal paper | 2019 | 1.0 | 1.0 | 1.5 | 3.5 |
| [61] | Journal paper | 2020 | 1.0 | 1.0 | 1.5 | 3.5 |
| [62] | Journal paper | 2019 | 1.0 | 1.0 | 2.0 | 4.0 |
| [63] | Journal paper | 2021 | 1.0 | 1.0 | 1.5 | 3.5 |
| [64] | Journal paper | 2019 | 1.0 | 1.0 | 1.5 | 3.5 |
| [65] | Journal paper | 2020 | 1.0 | 1.0 | 2.0 | 4.0 |
| [66] | Journal paper | 2019 | 1.0 | 1.0 | 2.0 | 4.0 |
| [67] | Journal paper | 2021 | 1.0 | 1.0 | 2.0 | 4.0 |
| [68] | Conference paper | 2019 | 1.0 | 1.0 | 2.0 | 4.0 |
| [69] | Journal paper | 2020 | 1.0 | 1.0 | 2.0 | 4.0 |
| [70] | Journal paper | 2019 | 1.0 | 1.0 | 2.0 | 4.0 |
| [71] | Journal paper | 2019 | 1.0 | 1.0 | 2.0 | 4.0 |
| [55] | Journal paper | 2021 | 1.0 | 1.0 | 2.0 | 4.0 |
References
- Cancer Research UK. Brain Tumor. Available online: https://www.cancerresearchuk.org/about-cancer/brain-tumours (accessed on 30 January 2022).
- World Health Organization. Fact Sheet Cancer. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 10 December 2021).
- Australian Institute of Health and Welfare. Cancer in Australia 2017; Cancer Series no. 101; Australian Institute of Health and Welfare: Canberra, Australia, 2017.
- Maile, E.J.; Barnes, I.; Finlayson, A.E.; Sayeed, S.; Ali, R. Nervous system and intracranial tumor incidence by ethnicity in England, 2001–2007: A descriptive epidemiological study. PLoS ONE 2016, 11, e0154347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Logeswari, T.; Karnan, M. An improved implementation of brain tumor detection using segmentation based on hierarchical self-organizing map. Int. J. Comput. Theory Eng. 2010, 2, 591–598. [Google Scholar] [CrossRef] [Green Version]
- Abd-Ellah, M.K.; Awad, A.I.; Khalaf, A.A.M.; Hamed, H.F.A. Classification of brain tumor MRIs using a kernel support vector machine. In Proceedings of the Building Sustainable Health Ecosystems: 6th International Conference on Well-Being in the Information Society, WIS 2016; Communications in Computer and Information Science; Springer: Berlin, Germany, 2016; Volume 636, pp. 151–160. [Google Scholar] [CrossRef]
- Brain Tumor Informatics| Home. Available online: https://braintumor.org/brain-tumor-information (accessed on 10 December 2021).
- Gordillo, N.; Montseny, E.; Sobrevilla, P. State of the art survey on MRI brain tumor segmentation. Magn. Reson. Imaging 2013, 31, 1426–1438. [Google Scholar] [CrossRef] [PubMed]
- Jayadevappa, D.; Kumar, S.S.; Murty, D.S. Medical image segmentation algorithms using deformable models: A review. IETE Tech. Rev. 2011, 28, 248–255. [Google Scholar] [CrossRef]
- Yazdani, S.; Yusof, R.; Karimian, A.; Pashna, M.; Hematian, A. Image segmentation methods and applications in MRI brain images. IETE Tech. Rev. 2015, 32, 413–427. [Google Scholar] [CrossRef]
- Nalepa, J.; Marcinkiewicz, M.; Kawulok, M. Data Augmentation for Brain-Tumor Segmentation: A Review. Front. Comput. Neurosci. 2019, 13, 83. [Google Scholar] [CrossRef] [Green Version]
- Ritchie, H. How Many People in the World Die from Cancer| Our World in Data| Institute for Health Metrics and Evaluation (IHME). Available online: https://ourworldindata.org/how-many-people-in-the-world-die-from-cancer (accessed on 10 December 2021).
- Xu, J.; Glicksberg, B.S. Federated Learning for Healthcare Informatics. J. Health Inform. Res. 2021, 5, 1–19. [Google Scholar] [CrossRef]
- Abd-Ellah, M.K.; Awad, A.I.; Khalaf, A.A.; Hamed, H.F. A review on brain tumor diagnosis from MRI images: Practical implications, key achievements, and lessons learned. Magn. Reson. Imaging 2019, 61, 300–318. [Google Scholar] [CrossRef]
- Ng, D.; Lan, X.; Yao, M.M.-S.; Chan, W.P.; Feng, M. Federated learning: A collaborative effort to achieve better medical imaging models for individual sites that have small labelled datasets. Quant. Imaging Med. Surg. 2021, 11, 852–857. [Google Scholar] [CrossRef]
- Kitchenham, B. Procedures for Undertaking Systematic Reviews: Joint Technical Report; Computer Science Department, Keele University (TR/SE-0401): Keele, UK; National ICT Australia Ltd.: Sydney, Australia, 2004. [Google Scholar]
- Aryani, A.; Peake, I.D.; Hamilton, M. Domain-based change propagation analysis: An enterprise system case study. In Proceedings of the 2010 IEEE International Conference on Software Maintenance 2010, Timișoara, Romania, 12–18 September 2010; IEEE: Piscataway, NJ, USA, 2010; pp. 1–9. [Google Scholar]
- Naeem, A.; Farooq, M.S.; Khelifi, A.; Abid, A. Malignant Melanoma Classification Using Deep Learning: Datasets, Performance Measurements, Challenges and Opportunities. IEEE Access 2020, 8, 110575–110597. [Google Scholar] [CrossRef]
- Dybå, T.; Dingsøyr, T. Empirical studies of agile software development: A systematic review. Inf. Softw. Technol. 2008, 50, 833–859. [Google Scholar] [CrossRef]
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; Mulrow, C.; Gøtzsche, P.C.; Ioannidis, J.P.; Clarke, M.; Devereaux, P.J.; Kleijnen, J.; Moher, D. The PRISMA Statement for Reporting Systematic Reviews and Meta-Analyses of Studies That Evaluate Health Care Interventions: Explanation and Elaboration. Ann. Intern. Med. 2009, 151, W65–W94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malik, H.; Farooq, M.S.; Khelifi, A.; Abid, A.; Qureshi, J.N.; Hussain, M. A Comparison of Transfer Learning Performance Versus Health Experts in Disease Diagnosis from Medical Imaging. IEEE Access 2020, 8, 139367–139386. [Google Scholar] [CrossRef]
- Centre for Reviews and Dissemination | Home. Available online: http://www.york.ac.uk/inst/crd/faq4.htm (accessed on 10 December 2021).
- Afshar, P.; Plataniotis, K.N.; Mohammadi, A. Capsule networks for brain tumor classification based on mri images and coarse tumor boundaries. In Proceedings of the ICASSP 2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1368–1372. [Google Scholar]
- Dong, H.; Yang, G.; Liu, F.; Mo, Y.; Guo, Y. Automatic brain tumor detection and segmentation using u-net based fully convolutional networks. In Proceedings of the Annual Conference on Medical Image Understanding and Analysis, Liverpool, UK, 24–26 July 2019; Springer: Cham, Switzerland, 2017; pp. 506–517. [Google Scholar]
- Laukamp, K.R.; Thiele, F.; Shakirin, G.; Zopfs, D.; Faymonville, A.; Timmer, M.; Maintz, D.; Perkuhn, M.; Borggrefe, J. Fully automated detection and segmentation of meningiomas using deep learning on routine multiparametric MRI. Eur. Radiol. 2019, 29, 124–132. [Google Scholar] [CrossRef] [Green Version]
- Kotowski, K.; Nalepa, J.; Dudzik, W. Detection and Segmentation of Brain Tumors from MRI Using U-Nets. In Proceedings of the International MICCAI Brainlesion Workshop 2019 May 19; Springer: Berlin, Germany, 2019; pp. 179–190. [Google Scholar]
- Cui, B.; Xie, M.; Wang, C. A Deep Convolutional Neural Network Learning Transfer to SVM-Based Segmentation Method for Brain Tumor. In Proceedings of the 2019 IEEE 11th International Conference on Advanced Infocomm Technology (ICAIT), Jinan, China, 18–20 October 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1–5. [Google Scholar]
- Pathak, M.K.; Pavthawala, M.; Patel, M.N.; Malek, D.; Shah, V.; Vaidya, B. Classification of Brain Tumor Using Convolutional Neural Network. In Proceedings of the 2019 3rd International Conference on Electronics, Communication and Aerospace Technology (ICECA), Coimbatore, India, 12–14 June 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 128–132. [Google Scholar]
- Zhao, H.; Guo, Y.; Zheng, Y. A Compound Neural Network for Brain Tumor Segmentation. In Proceedings of the 2019 IEEE International Conference on Image Processing (ICIP), Taipei, Taiwan, 22–25 September 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1435–1439. [Google Scholar]
- Krishnammal, P.M.; Raja, S.S. Convolutional Neural Network based Image Classification and Detection of Abnormalities in MRI Brain Images. In Proceedings of the 2019 International Conference on Communication and Signal Processing (ICCSP), Melmaruvathur, India, 4–6 April 2019; IEEE: Piscataway, NJ, USA; pp. 548–553. [Google Scholar]
- Soleymanifard, M.; Hamghalam, M. Segmentation of Whole Tumor Using Localized Active Contour and Trained Neural Network in Boundaries. In Proceedings of the 2019 5th Conference on Knowledge Based Engineering and Innovation (KBEI), Tehran, Iran, 28 February–1 March 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 739–744. [Google Scholar]
- Özyurt, F.; Sert, E.; Avcı, D. An expert system for brain tumor detection: Fuzzy C-means with super resolution and convolutional neural network with extreme learning machine. Med. Hypotheses 2020, 134, 109433. [Google Scholar] [CrossRef]
- Sharif, M.; Amin, J.; Raza, M.; Anjum, M.A.; Afzal, H.; Shad, S.A. Brain tumor detection based on extreme learning. Neural Comput. Appl. 2020, 32, 15975–15987. [Google Scholar] [CrossRef]
- Thillaikkarasi, R.; Saravanan, S. An enhancement of deep learning algorithm for brain tumor segmentation using kernel-based CNN with M-SVM. J. Med. Syst. 2019, 43, 84. [Google Scholar] [CrossRef]
- Myronenko, A.; Hatamizadeh, A. Robust semantic segmentation of brain tumor regions from 3d mris. In Proceedings of the International MICCAI Brainlesion Workshop 2019; Springer: Cham, Switzerland, 2019; pp. 82–89. [Google Scholar]
- Madhupriya, G.; Guru, N.M.; Praveen, S.; Nivetha, B. Brain Tumor Segmentation with Deep Learning Technique. In Proceedings of the 2019 3rd International Conference on Trends in Electronics and Informatics (ICOEI), Tirunelveli, India, 23–25 April 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 758–763. [Google Scholar]
- Vijh, S.; Sharma, S.; Gaurav, P. Brain tumor segmentation using OTSU embedded adaptive particle swarm optimization method and convolutional neural network. In Data Visualization and Knowledge Engineering; Springer: Cham, Switzerland, 2020; pp. 171–194. [Google Scholar]
- Sheller, M.J.; Reina, G.A.; Edwards, B.; Martin, J.; Bakas, S. Multi-institutional deep learning modeling without sharing patient data: A feasibility study on brain tumor segmentation. In Proceedings of the International MICCAI Brain lesion Workshop 2018; Springer: Cham, Switzerland, 2018; pp. 92–104. [Google Scholar]
- Li, W.; Milletarì, F.; Xu, D.; Rieke, N.; Hancox, J.; Zhu, W.; Baust, M.; Cheng, J.; Ourselim, S.; Jorge Cardoso, M.; et al. Privacy-preserving federated brain tumour segmentation. In Proceedings of the International Workshop on Machine Learning in Medical Imaging 2019; Springer: Cham, Switzerland, 2019; pp. 133–141. [Google Scholar]
- Guo, P.; Wang, P.; Zhou, J.; Jiang, S.; Patel, V.M. Multi-institutional collaborations for improving deep learning-based magnetic resonance image reconstruction using federated learning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition 2021, Nashville, TN, USA, 20–25 June 2021; pp. 2423–2432. [Google Scholar]
- Siar, M.; Teshnehlab, M. Brain Tumor Detection Using Deep Neural Network and Machine Learning Algorithm. In Proceedings of the 2019 9th International Conference on Computer and Knowledge Engineering (ICCKE), Mashhad, Iran, 24–25 October 2019; IEEE: Piscataway, NJ, USA; pp. 363–368. [Google Scholar]
- Sultan, H.H.; Salem, N.M.; Al-Atabany, W. Multi-Classification of Brain Tumor Images Using Deep Neural Network. IEEE Access 2019, 7, 69215–69225. [Google Scholar] [CrossRef]
- Li, M.; Kuang, L.; Xu, S.; Sha, Z. Brain Tumor Detection Based on Multimodal Information Fusion and Convolutional Neural Network. IEEE Access 2019, 7, 180134–180146. [Google Scholar] [CrossRef]
- Noreen, N.; Palaniappan, S.; Qayyum, A.; Ahmad, I.; Imran, M.; Shoaib, M. A Deep Learning Model Based on Concatenation Approach for the Diagnosis of Brain Tumor. IEEE Access 2020, 8, 55135–55144. [Google Scholar] [CrossRef]
- Liu, P.; Dou, Q.; Wang, Q.; Heng, P.-A. An Encoder-Decoder Neural Network With 3D Squeeze-and-Excitation and Deep Supervision for Brain Tumor Segmentation. IEEE Access 2020, 8, 34029–34037. [Google Scholar] [CrossRef]
- Shakeel, P.M.; Tobely, T.E.E.; Al-Feel, H.; Manogaran, G.; Baskar, S. Neural Network Based Brain Tumor Detection Using Wireless Infrared Imaging Sensor. IEEE Access 2019, 7, 5577–5588. [Google Scholar] [CrossRef]
- Deng, W.; Shi, Q.; Wang, M.; Zheng, B.; Ning, N. Deep Learning-Based HCNN and CRF-RRNN Model for Brain Tumor Segmentation. IEEE Access 2020, 8, 26665–26675. [Google Scholar] [CrossRef]
- Ozyurt, F.; Sert, E.; Avci, E.; Dogantekin, E. Brain tumor detection based on Convolutional Neural Network with neutrosophic expert maximum fuzzy sure entropy. Measurement 2019, 147, 106830. [Google Scholar] [CrossRef]
- Majib, M.S.; Rahman, M.; Sazzad, T.M.S.; Khan, N.I.; Dey, S.K. VGG-SCNet: A VGG Net-Based Deep Learning Framework for Brain Tumor Detection on MRI Images. IEEE Access 2021, 9, 116942–116952. [Google Scholar] [CrossRef]
- Amin, J.; Sharif, M.; Raza, M.; Saba, T.; Anjum, M.A. Brain tumor detection using statistical and machine learning method. Comput. Methods Programs Biomed. 2019, 177, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Mittal, M.; Goyal, L.M.; Kaur, S.; Kaur, I.; Verma, A.; Hemanth, D.J. Deep learning based enhanced tumor segmentation approach for MR brain images. Appl. Soft Comput. 2019, 78, 346–354. [Google Scholar] [CrossRef]
- Saba, T.; Mohamed, A.S.; El-Affendi, M.; Amin, J.; Sharif, M. Brain tumor detection using fusion of hand crafted and deep learning features. Cogn. Syst. Res. 2020, 59, 221–230. [Google Scholar] [CrossRef]
- Amin, J.; Sharif, M.; Gul, N.; Raza, M.; Anjum, M.A.; Nisar, M.W.; Bukhari, S.A.C. Brain Tumor Detection by Using Stacked Autoencoders in Deep Learning. J. Med. Syst. 2020, 44, 32. [Google Scholar] [CrossRef]
- Rehman, A.; Naz, S.; Razzak, M.I.; Akram, F.; Imran, M. A deep learning-based framework for automatic brain tumors classification using transfer learning. Circuits Syst. Signal Process. 2020, 39, 757–775. [Google Scholar] [CrossRef]
- Sharif, M.I.; Khan, M.A.; Alhussein, M.; Aurangzeb, K.; Raza, M. A decision support system for multimodal brain tumor classification using deep learning. Complex Intell. Syst. 2021. [Google Scholar] [CrossRef]
- Das, S.; Aranya, O.R.R.; Labiba, N.N. Brain Tumor Classification Using Convolutional Neural Network. In Proceedings of the 2019 1st International Conference on Advances in Science, Engineering and Robotics Technology (ICASERT), Dhaka, Bangladesh, 3–5 May 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1–5. [Google Scholar]
- Gumaei, A.; Hassan, M.M.; Hassan, R.; Alelaiwi, A.; Fortino, G. A Hybrid Feature Extraction Method with Regularized Extreme Learning Machine for Brain Tumor Classification. IEEE Access 2019, 7, 36266–36273. [Google Scholar] [CrossRef]
- Amin, J.; Sharif, M.; Yasmin, M.; Fernandes, S.L. Big data analysis for brain tumor detection: Deep convolutional neural networks. Future Gener. Comput. Syst. 2018, 87, 290–297. [Google Scholar] [CrossRef]
- Toğaçar, M.; Ergen, B.; Cömert, Z. BrainMRNet: Brain tumor detection using magnetic resonance images with a novel convolutional neural network model. Med. Hypotheses 2020, 134, 109531. [Google Scholar] [CrossRef] [PubMed]
- Sajjad, M.; Khan, S.; Muhammad, K.; Wu, W.; Ullah, A.; Baik, S.W. Multi-grade brain tumor classification using deep CNN with extensive data augmentation. J. Comput. Sci. 2019, 30, 174–182. [Google Scholar] [CrossRef]
- Nema, S.; Dudhane, A.; Murala, S.; Naidu, S. RescueNet: An unpaired GAN for brain tumor segmentation. Biomed. Signal Process. Control. 2020, 55, 101641. [Google Scholar] [CrossRef]
- Sajid, S.; Hussain, S.; Sarwar, A. Brain tumor detection and segmentation in MR images using deep learning. Arab. J. Sci. Eng. 2019, 44, 9249–9261. [Google Scholar] [CrossRef]
- Díaz-Pernas, F.J.; Martínez-Zarzuela, M.; Antón-Rodríguez, M.; González-Ortega, D. A Deep Learning Approach for Brain Tumor Classification and Segmentation Using a Multiscale Convolutional Neural Network. Healthcare 2021, 9, 153. [Google Scholar] [CrossRef]
- Swati, Z.N.K.; Zhao, Q.; Kabir, M.; Ali, F.; Ali, Z.; Ahmed, S.; Lu, J. Content-based brain tumor retrieval for MR images using transfer learning. IEEE Access 2019, 7, 17809–17822. [Google Scholar] [CrossRef]
- Sharif, M.; Amin, J.; Raza, M.; Yasmin, M.; Satapathy, S.C. An integrated design of particle swarm optimization (PSO) with fusion of features for detection of brain tumor. Pattern Recognit. Lett. 2020, 129, 150–157. [Google Scholar] [CrossRef]
- Amin, J.; Sharif, M.; Yasmin, M.; Saba, T.; Anjum, M.A.; Fernandes, S.L. A new approach for brain tumor segmentation and classification based on score level fusion using transfer learning. J. Med. Syst. 2019, 43, 326. [Google Scholar] [CrossRef] [PubMed]
- Sadad, T.; Rehman, A.; Munir, A.; Saba, T.; Tariq, U.; Ayesha, N.; Abbasi, R. Brain tumor detection and multi-classification using advanced deep learning techniques. Microsc. Res. Tech. 2021, 84, 1296–1308. [Google Scholar] [CrossRef] [PubMed]
- Siar, H.; Teshnehlab, M. Diagnosing and Classification Tumors and MS Simultaneous of Magnetic Resonance Images Using Convolution Neural Network. In Proceedings of the 2019 7th Iranian Joint Congress on Fuzzy and Intelligent Systems (CFIS); IEEE: Piscataway, NJ, USA, 2019; pp. 1–4. [Google Scholar]
- Sharif, M.I.; Li, J.P.; Khan, M.A.; Saleem, M.A. Active deep neural network features selection for segmentation and recognition of brain tumors using MRI images. Pattern Recognit. Lett. 2020, 129, 181–189. [Google Scholar] [CrossRef]
- Deng, W.; Shi, Q.; Luo, K.; Yang, Y.; Ning, N. Brain tumor segmentation based on improved convolutional neural network in combination with non-quantifiable local texture feature. J. Med. Syst. 2019, 43, 152. [Google Scholar] [CrossRef] [PubMed]
- Nie, D.; Lu, J.; Zhang, H.; Adeli, E.; Wang, J.; Yu, Z.; Liu, L.; Wang, Q.; Wu, J.; Shen, D. Multi-Channel 3D Deep Feature Learning for Survival Time Prediction of Brain Tumor Patients Using Multi-Modal Neuroimages. Sci. Rep. 2019, 9, 1103. [Google Scholar] [CrossRef] [Green Version]
- Cheng, J.; Huang, W.; Cao, S.; Yang, R.; Yang, W.; Yun, Z.; Wang, Z.; Feng, Q. Enhanced performance of brain tumor classification via tumor region augmentation and partition. PLoS ONE 2015, 10, e0140381. [Google Scholar] [CrossRef]
- Scarpace, L.; Mikkelsen, L.; Cha, T.; Rao, S.; Tekchandani, S.; Gutman, S.; Pierce, D. Radiology data from the cancer genome atlas glioblastoma multiforme [TCGA-GBM] collection. Cancer Imaging Arch. 2016, 11, 1. [Google Scholar]
- Cham.BraTS Challenge | Start 2012. Available online: https://www.smir.ch/BRATS/Start2012 (accessed on 10 December 2021).
- BraTS Challenge | Start 2013. Available online: https://www.smir.ch/BRATS/Start2013 (accessed on 10 December 2021).
- BraTS Challenge | Start 2014. Available online: https://www.virtualskeleton.ch/BRATS/Start2014 (accessed on 10 December 2021).
- BraTS Challenge | Start 2015. Available online: https://www.smir.ch/BraTS/Start2015 (accessed on 10 December 2021).
- BraTS Challenge | Start 2017. Available online: https://www.med.upenn.edu/sbia/brats2017/data.html (accessed on 10 December 2021).
- BraTS Challenge | Start 2016. Available online: https://www.smir.ch/BraTS/Start2016 (accessed on 10 December 2021).
- Zhao, X.; Wu, Y.; Song, G.; Li, Z.; Zhang, Y.; Fan, Y. A deep learning model integrating FCNNs and CRFs for brain tumor segmentation. Med. Image Anal. 2018, 43, 98–111. [Google Scholar] [CrossRef]
- Ischemic Stroke Lesion Segmentation | ISLES 2015. Available online: http://www.isles-challenge.org/ISLES2015/ (accessed on 10 December 2021).
- Ischemic Stroke Lesion Segmentation | ISLES 2017. Available online: http://www.isles-challenge.org/ISLES2017/ (accessed on 10 December 2021).
- Kaggle. Navoneel | Brain Tumor. Available online: https://www.kaggle.com/navoneel/brain-mri-images-for-brain-tumor-detection (accessed on 10 December 2021).
- BraTS Challenge | Start 2019. Available online: https://www.med.upenn.edu/cbica/brats-2019 (accessed on 10 December 2021).
- DICOM Image Library | Home. Available online: https://www.osirix-viewer.com/resources/dicom-image-library/brainix (accessed on 10 December 2021).
- Guadagno, E.; Presta, I.; Maisano, D.; Donato, A.; Pirrone, C.K.; Cardillo, G.; Corrado, S.D.; Mignogna, C.; Mancuso, T.; Donato, G.; et al. Role of Macrophages in Brain Tumor Growth and Progression. Int. J. Mol. Sci. 2018, 19, 1005. [Google Scholar] [CrossRef] [Green Version]
- Ostrom, Q.T.; Fahmideh, M.A.; Cote, D.J.; Muskens, I.S.; Schraw, J.; Scheurer, M.; Bondy, M.L. Risk factors for childhood and adult primary brain tumors. Neuro-Oncology 2019, 21, 1357–1375. [Google Scholar] [CrossRef]




| Statement of Research Question | Motivation | |
|---|---|---|
| Q1 | What are the best available methods for the detection of a brain tumor? | This question investigates a deep and federated-learning-based method for the diagnosis of brain tumors. |
| Q2 | What are the metrics used to determine the performance of different methods used for brain tumor diagnosis? | This question determines the research efficacy of deep learning and federated-learning-based methods for brain tumor diagnosis. |
| Q3 | What datasets are used in recent research for the diagnosis of brain tumors? | This question identifies the available benchmark, public, and non-public datasets for brain tumor diagnosis. |
| Q4 | What is the quality of the selected papers? | This question investigates the quality of the selected studies. |
| Q5 | What is the impact of the selected papers on brain tumor detection? | This question investigates the impact of selected papers on the detection of brain tumors with a minimum intervention of radiologists |
| Repository Name | Search Strings |
|---|---|
| ACM | ((“deep learning” OR “machine learning” OR “artificial intelligence” OR “convolutional neural network” OR “federated learning”) AND (“glioblastoma,” OR “astrocytoma,” OR “brain cancer,” OR “brain tumor”) AND (“detection” OR “classification”)) Publication Year: 2017–2021 |
| IEEE Xplore | ((“document title”: “deep learning” OR “machine learning” OR “artificial intelligence” OR “convolutional neural network” OR “federated learning” OR “supervised learning” OR “Bayesian”) AND (“abstract”: “glioblastoma,” OR “astrocytoma,” OR “brain cancer,” OR “brain tumor”) AND (“detection” OR “classification”)) Publication Year: 2017–2021 |
| Medline | (“deep learning”(All Fields) OR “machine learning”(All Fields) OR “artificial intelligence”(All Fields) OR “convolutional neural network”(All Fields) OR “federated learning”(All Fields) AND (“glioblastoma,”(All Fields) OR (“astrocytoma “(MeSH Terms) OR (“brain”(All Fields) AND “tumor”(All Fields)) OR “brain tumor”[All Fields] OR (“brain”(All Fields) AND “cancer”(All Fields)) OR “brain cancer”(All Fields)) OR “brain tumor”(All Fields)) AND (“detection”(All Fields) OR “diagnosis”(All Fields) OR “classification”(All Fields)) Publication Year: 2017–2021 |
| Elsevier | (“deep learning” OR “machine learning” OR “artificial intelligence” OR “convolutional neural network” OR “federated learning”) AND (“glioblastoma,” OR “astrocytoma,” OR “brain cancer,” OR “brain tumor”) AND (“detection” OR “classification”) Publication Year: 2017–2021 |
| Springer | ((“deep learning” OR “machine learning” OR “artificial intelligence” OR “convolutional neural network” OR “federated learning”) AND (“glioblastoma,” OR “astrocytoma,” OR “brain cancer,” OR “brain tumor”) AND (“detection” OR “classification”)) Publication Year: 2017–2021 |
| Scopus | TITLE-ABS-KEY (“deep learning” OR “machine learning” OR “artificial intelligence” OR “convolutional neural network” OR “federated learning”) AND (“glioblastoma,” OR “astrocytoma,” OR “brain cancer,” OR “brain tumor”) AND (“detection” OR “diagnosis” OR “classification”)) Year: 2017–2021 |
| Wiley | deep-learning OR machine learning OR artificial intelligence OR convolutional neural network OR federated learning AND glioblastoma, OR astrocytoma, OR brain cancer OR brain tumor AND detection OR diagnosis OR classification Year: 2017–2021 |
| Publication Contribution | Architecture | Training Algorithm | Dataset | Source |
|---|---|---|---|---|
| A revamped CapsNet architecture for the detection of brain tumors that carry the coarse tumor borders into the extra pipeline to improve the emphasis of CapsNet. | CNN | CapsNets | CE-MRI | [23] |
| Automatic and efficient brain tumor segmentation and detection is achieved by using U-Net. | CNN | U-Net + Resnet50 | BraTS 2015 | [24] |
| The performance of the deep learning model was investigated on MRI data from various institutions. | CNN | Deep learning | BraTS benchmark | [25] |
| Two-stage cascaded U-Net for brain tumor detection and segmentation. | CNN | U-Net | [26] | |
| Brain tumor classification by transferring CNN-based learning to SVM based classifier. | CNN | CNN + SVM | BraTS challenge 2019 | [27] |
| Brain tumor classified using CNN, and for the segmentation of tumor, the watershed technique was implemented. | CNN | CNN + Watershed | Non-published brain MRI dataset | [28] |
| Fine and coarse features were extracted using hybrid two-path convolution with a modified down-sampling structure. | CNN | Hybrid two-path CNN | Non-published brain MRI dataset | [29] |
| CNN was used with the curvelet domain, which extracts features of reasonable resolution and direction. | CNN | CNN + Curvelet domain | BraTS 2017 | [30] |
| Active contours used with CNN to automatically segment the tumor faster, independent of image type | DCNN | CNN + Active contour | Non-published brain MRI dataset | [31] |
| The fuzzy c-means efficiently segment the tumor, whereas pretrained SqueezNet effectively detects the tumor. | CNN | Fuzzy c-mean + SqueezNet | BraTS 2015 | [32] |
| Accurate segmentation was achieved using triangular fuzzy median filters, whereas classification was done using ELM. | CNN | ST + ELM | BraTS 2012. 2013, 2014, 2015 | [33] |
| Pre-processing, feature extraction, imaging classification and brain tumor segmentation were achieved using CNN with SVM. | CNN | CNN + SVM | 40 MRI image dataset | [34] |
| Brain tumor detection using 3D semantic segmentation with conventional encoder–decoder architecture. | CNN | 3D Semantic with encoder decoder | BraTS 2019 | [35] |
| CNN and PNN have been utilized to make an intelligent system that can detect tumors of any shape and size efficiently. | CNN | PNN + CNN | BraTS 2013 | [36] |
| Optimal threshold value with OTSU for the optimization of the adaptive swarm. This system uses an anisotropic diffusion filter to remove noise, while classification is done by the CNN. | CNN | CNN + OTSU | IBSR dataset + MS free brain dataset | [37] |
| Federated learning to improve the training process; for this purpose, multiple organizations collaborated with the privacy of patient data retained. The performance of federated semantic segmentation is demonstrated using a deep learning model. | FL | DNN | Different institutions; collaborated dataset | [38] |
| Federated learning based on deep neural network (DNN) for the segmentation of brain tumor using BraTS dataset. | FL | DNN | BraTS 2018 | [39] |
| A cross-site modeling platform using FL for the reconstruction of MR images collected from multiple institutions using different scanners and acquisition protocols. The concealed features extracted from different sub-sites are aligned with the concealed features of the main site | FL | DNN | Multiple datasets | [40] |
| Classifier | Sensitivity /Recall | Specifity | Precision | Accuracy | Dice | Dataset | Source |
|---|---|---|---|---|---|---|---|
| CNN + SoftMax | 100% | 96.42% | 98.83% | 99.12% | ----- | CE-MRI | [41] |
| CNN + GA | 95.5% | 98.7% | 95.8% | 97.54% | ----- | Combined dataset | [42] |
| Information Fusion + CNN | 99.81% | ----- | 92.7% | ----- | 92.7% | BraTS 2018 | [43] |
| Inception V3 + SoftMax | ----- | ----- | 99.0% | ----- | 99.34% | CE-MRI | [44] |
| Encoder-decoder neural network | ----- | ----- | ----- | ----- | 89.28% | BraTS 2017 | [45] |
| MLBPNN | 95.10% | 99.8% | ----- | 93.33% | ----- | Infrared imaging technology | [46] |
| CRF—HCNN | 97.8% | ----- | 96.5% | ----- | ----- | BraTS 2013 & 2015 | [47] |
| NS—CNN | 96.25% | 95% | ----- | 95.62% | ----- | TCGA-GBM dataset | [48] |
| VGG + Stack classifier | 99.1% | ----- | 99.2% | ----- | ----- | Private collected | [49] |
| Statistical learning | 92% | 100% | ----- | 96% | 96% | BraTS 2013 | [50] |
| Statistical learning | 91% | 90% | ----- | 90% | 95% | BraTS 2015 | [50] |
| SWT + GCNN | 98.23% | ----- | 98.81% | ----- | ----- | BRAINIX dataset | [51] |
| Handcrafted + Deep learning | 99% | ----- | ----- | 98.78% | 96.36% | BraTS 2015 | [52] |
| Handcrafted + Deep learning | 100% | 100% | 100% | 99..63% | 99.62% | BraTS 2016 | [52] |
| Handcrafted + Deep learning | ----- | ----- | ----- | 99.69% | 95.06% | BraTS 2017 | [52] |
| OTSU +CNN | ----- | ----- | ----- | 98% | ----- | IBSR | [37] |
| Stack autoencoder in DL | 88% | 100% | ----- | 90% | 94% | BraTS 2012 | [53] |
| Stack autoencoder in DL | 100% | 100% | 100% | BraTS 2012 | [53] | ||
| Stack autoencoder in DL | 100% | 90% | ----- | 95% | 100% | BraTS 2013 | [53] |
| Stack autoencoder in DL | 98% | 96% | ----- | 97% | 96% | BraTS 2014 | [53] |
| Stack autoencoder in DL | 93% | 100% | ----- | 95% | 98% | BraTS 2015 | [53] |
| Ensemble | ----- | ----- | ----- | 98.69% | ----- | CE-MRI | [54] |
| Densenet201 with EKbHFV & MGA | 99.9% | ----- | 99.9% | 99.9% | ----- | BraTS 2019 | [55] |
| CNN | 94.56% | 89% | 93.33% | 94.39% | ----- | CE-MRI | [56] |
| Extreme learning | 91.6% | ----- | ----- | ----- | 94.93% | CE-MRI | [57] |
| DNN | 98.4% | 98.4% | 99.9% | 98.6% | 98.4% | BraTS 2012 | [58] |
| DNN | 99.8% | 98.9% | 98.9% | 99.8% | 99.8% | BraTS 2013 | [58] |
| DNN | 92.01% | 95.5% | 95.5% | 93.1% | 92.9% | BraTS 2014 | [58] |
| DNN | 95% | 97.2% | ----- | 95.1% | 96% | BraTS 2015 | [58] |
| DNN | 99.05% | 98.20% | ----- | 100% | ISLES 2015 | [58] | |
| DNN | 99.44% | 100% | ----- | 98.8.7% | 94.63% | ISLES 2017 | [58] |
| Brain MRNet | 96.0% | 96.08% | 92.31% | 96.05% | 84.2% | BrainMRI dataset | [59] |
| Pretrained CNN | 88.41% | 96.12% | ----- | 94.58% | ----- | CE-MRI | [60] |
| RescueNet | 94.89% | ----- | ----- | ----- | 94.29% | BraTS 2015 | [61] |
| RescueNet | 99% | ----- | ----- | ----- | ----- | BraTS 2017 | [61] |
| Deep learning | 90% | 94% | ----- | ----- | 88% | BraTS 2013 | [62] |
| MultiScale CNN | 94% | 97.3% | 82.8% | CE-MRI | [63] | ||
| CBIR—TL | ----- | ----- | 96.13% | ----- | ----- | CE-MRI | [64] |
| Transfer learning | 80% | 98.1% | ----- | 97% | ----- | BraTS 2015 | [65] |
| Score Level Fusion using TL | 95.31% | 96.30% | ----- | ----- | 96.44% | BraTS 2014 | [66] |
| Score Level Fusion using TL | 97.62% | 95.05% | ----- | ----- | 97.74% | BraTS 2013 | [66] |
| Score Level Fusion using TL | ----- | ----- | ----- | ----- | ----- | BraTS 2015 | [66] |
| Score Level Fusion using TL | 99.9% | ----- | ----- | ----- | 100% | BraTS 2016 | [66] |
| Score Level Fusion using TL | 91.27% | ----- | ----- | ----- | 99.80% | BraTS 2017 | [66] |
| Resnet50 + Unet | ----- | ----- | ----- | 99.61% | ----- | CE-MRI | [67] |
| Fine-tuned CNN) | 94.64% | 100% | ----- | 96.88% | ----- | CE-MRI | [68] |
| Active DNN | ----- | ----- | ----- | 98.3% | ----- | BraTS 2013 | [69] |
| Active DNN | ----- | ----- | 97.2% | 97.8% | 95.0% | BraTS 2015 | [69] |
| Active DNN | 98.39% | 96.06% | ----- | 96.9% | 99.59% | BraTS 2017 | [69] |
| Active DNN | 98.7% | 99.0% | 99% | 92.5% | 99.94% | BraTS 2018 | [69] |
| CNN with non-quantifiable local texture | 90.12% | ----- | ----- | ----- | 85.25% | BraTS 2015 | [70] |
| Dtf + Fc7 | 88.9% | 87.5% | ----- | 88% | ----- | 68% patient data collected from 2010–2015 Haushan hospital | [71] |
| Densenet with MGA +EKbHFV | 99.7% | ----- | 99.7% | 99.7% | ----- | BraTS 2018 | [55] |
| Densenet with MGA + EKbHFV | ----- | ----- | ----- | 99.8% | 98.7% | BraTS 2019 | [55] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naeem, A.; Anees, T.; Naqvi, R.A.; Loh, W.-K. A Comprehensive Analysis of Recent Deep and Federated-Learning-Based Methodologies for Brain Tumor Diagnosis. J. Pers. Med. 2022, 12, 275. https://doi.org/10.3390/jpm12020275
Naeem A, Anees T, Naqvi RA, Loh W-K. A Comprehensive Analysis of Recent Deep and Federated-Learning-Based Methodologies for Brain Tumor Diagnosis. Journal of Personalized Medicine. 2022; 12(2):275. https://doi.org/10.3390/jpm12020275
Chicago/Turabian StyleNaeem, Ahmad, Tayyaba Anees, Rizwan Ali Naqvi, and Woong-Kee Loh. 2022. "A Comprehensive Analysis of Recent Deep and Federated-Learning-Based Methodologies for Brain Tumor Diagnosis" Journal of Personalized Medicine 12, no. 2: 275. https://doi.org/10.3390/jpm12020275
APA StyleNaeem, A., Anees, T., Naqvi, R. A., & Loh, W.-K. (2022). A Comprehensive Analysis of Recent Deep and Federated-Learning-Based Methodologies for Brain Tumor Diagnosis. Journal of Personalized Medicine, 12(2), 275. https://doi.org/10.3390/jpm12020275








