Education and Training of Non-Genetics Providers on the Return of Genome Sequencing Results in a NICU Setting
Abstract
:1. Introduction
2. Materials and Methods
2.1. SouthSeq Study Protocol and Purposes
2.2. Study Population
2.3. Training Protocol and Objectives
- Explain the benefits and limitations of genome sequencing and how it compares to other types of genetic tests;
- State the purpose of the SouthSeq study and the hypothesis being tested through result disclosure;
- Identify the role of the non-genetics NICU provider in the SouthSeq study;
- Demonstrate familiarity and proficiency completing provider tasks in the online Genome Gateway platform;
- Interpret a SouthSeq genome sequencing result letter and report;
- Develop a plan for disclosing various types of genome sequencing results (positive, negative, and uncertain) including key points and next steps;
- Describe common questions among patients receiving genome sequencing results;
- Attend to psychosocial needs of families surrounding genome sequencing result disclosure;
- Identify and critique patient support resources relevant to genome sequencing results.
2.4. Survey Instrumentation
3. Results
3.1. SouthSeq Non-Genetics Provider Participants
3.1.1. Demographics
3.1.2. Prior Experience with Genetic and Genomic Testing
3.2. Impact of the SouthSeq Training Intervention
3.2.1. Provider Understanding and Skills
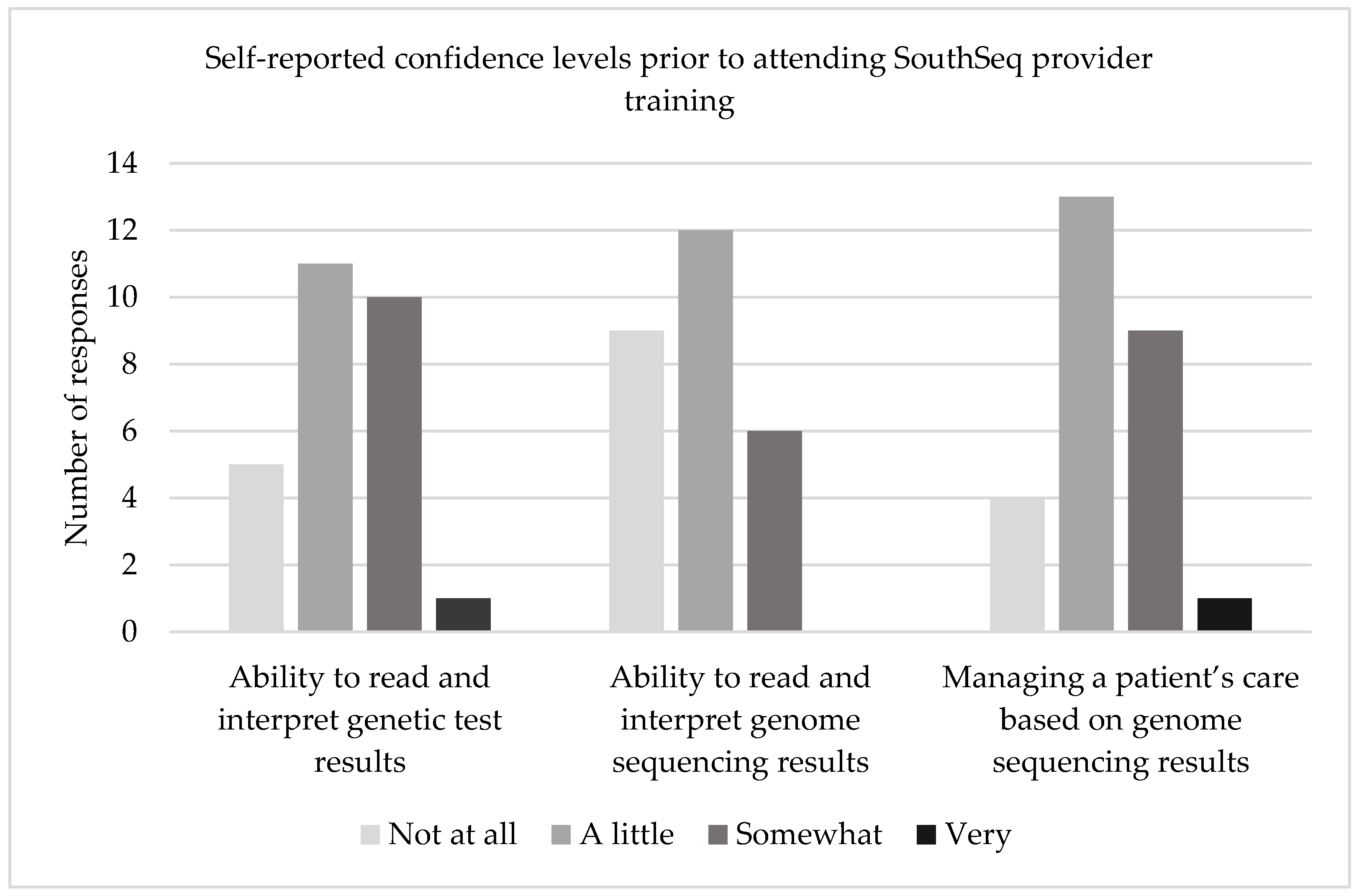
3.2.2. Provider Confidence
3.3. Qualitative Feedback
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Soneda, A.; Teruya, H.; Furuya, N.; Yoshihashi, H.; Enomoto, K.; Ishikawa, A.; Matsui, K.; Kurosawa, K. Proportion of malformations and genetic disorders among cases encountered at a high-care unit in a children’s hospital. Eur. J. Pediatr. 2011, 171, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, D.A.; Carey, J.C. Contribution of malformations and genetic disorders to mortality in a children’s hospital. Am. J. Med Genet. Part A 2004, 126, 393–397. [Google Scholar] [CrossRef] [PubMed]
- Willig, L.; Petrikin, J.E.; Smith, L.D.; Saunders, C.J.; Thiffault, I.; Miller, N.A.; Soden, S.E.; Cakici, J.; Herd, S.M.; Twist, G.; et al. Whole-genome sequencing for identification of Mendelian disorders in critically ill infants: A retrospective analysis of diagnostic and clinical findings. Lancet Respir. Med. 2015, 3, 377–387. [Google Scholar] [CrossRef] [Green Version]
- Dimmock, D.P.; Clark, M.M.; Gaughran, M.; Cakici, J.A.; Caylor, S.A.; Clarke, C.; Feddock, M.; Chowdhury, S.; Salz, L.; Cheung, C.; et al. An RCT of Rapid Genomic Sequencing among Seriously Ill Infants Results in High Clinical Utility, Changes in Management, and Low Perceived Harm. Am. J. Hum. Genet. 2020, 107, 942–952. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, N.M.; Nahas, S.A.; Chowdhury, S.; Batalov, S.; Clark, M.; Caylor, S.; Cakici, J.; Nigro, J.J.; Ding, Y.; Veeraraghavan, N.; et al. Rapid whole genome sequencing impacts care and resource utilization in infants with congenital heart disease. NPJ Genom. Med. 2021, 6, 29. [Google Scholar] [CrossRef]
- Bowling, K.M.; Thompson, M.L.; Finnila, C.R.; Hiatt, S.M.; Latner, D.R.; Amaral, M.D.; Lawlor, J.M.; East, K.M.; Cochran, M.E.; Greve, V.; et al. Genome sequencing as a first-line diagnostic test for hospitalized infants. Genet. Med. 2021, S1098360021054009. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.D.; Willig, L.K.; Kingsmore, S.F. Whole-Exome Sequencing and Whole-Genome Sequencing in Critically Ill Neonates Suspected to Have Single-Gene Disorders. Cold Spring Harb. Perspect. Med. 2016, 6, a023168. [Google Scholar] [CrossRef] [Green Version]
- Vassy, J.L.; Lautenbach, D.M.; McLaughlin, H.M.; Kong, S.W.; Christensen, K.D.; Krier, J.; Kohane, I.S.; Feuerman, L.Z.; Blumenthal-Barby, J.; Roberts, J.S.; et al. The MedSeq Project: A randomized trial of integrating whole genome sequencing into clinical medicine. Trials 2014, 15, 85. [Google Scholar] [CrossRef] [Green Version]
- Maiese, D.R.; Keehn, A.; Lyon, M.; Flannery, D.; Watson, M. Current conditions in medical genetics practice. Genet. Med. 2019, 21, 1874–1877. [Google Scholar] [CrossRef] [Green Version]
- Hoskovec, J.M.; Bennett, R.L.; Carey, M.E.; DaVanzo, J.E.; Dougherty, M.; Hahn, S.E.; LeRoy, B.S.; O’Neal, S.; Richardson, J.G.; Wicklund, C.A. Projecting the Supply and Demand for Certified Genetic Counselors: A Workforce Study. J. Genet. Couns. 2018, 27, 16–20. [Google Scholar] [CrossRef]
- Villegas, C.; Haga, S.B. Access to Genetic Counselors in the Southern United States. J. Pers. Med. 2019, 9, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shields, A.E.; Burke, W.; Levy, D.E. Differential use of available genetic tests among primary care physicians in the United States: Results of a national survey. Genet. Med. 2008, 10, 404–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greendale, K.; Pyeritz, R.E. Empowering primary care health professionals in medical genetics: How soon? How fast? How far? Am. J. Med Genet. 2001, 106, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Plunkett-Rondeau, J.; Hyland, K.; Dasgupta, S. Training future physicians in the era of genomic medicine: Trends in undergraduate medical genetics education. Genet. Med. 2015, 17, 927–934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dasgupta, S.; Feldman, G.L.; Powell, C.M.; Toriello, H.V.; Westman, J.; Wilson, W.G.; Waggoner, D.J. Training the next generation of genomic medicine providers: Trends in medical education and national assessment. Genet. Med. 2020, 22, 1718–1722. [Google Scholar] [CrossRef] [PubMed]
- Bensend, T.A.; Veach, P.M.; Niendorf, K.B. What’s the Harm? Genetic Counselor Perceptions of Adverse Effects of Genetics Service Provision by Non-Genetics Professionals. J. Genet. Couns. 2013, 23, 48–63. [Google Scholar] [CrossRef]
- Brierley, K.L.; Blouch, E.; Cogswell, W.; Homer, J.P.; Pencarinha, D.; Stanislaw, C.L.; Matloff, E.T. Adverse Events in Cancer Genetic Testing: Medical, Ethical, Legal, and Financial Implications. Cancer J. 2012, 18, 303–309. [Google Scholar] [CrossRef]
- Farmer, M.B.; Bonadies, D.C.; Mahon, S.M.; Baker, M.J.; Ghate, S.M.; Munro, C.; Nagaraj, C.B.; Besser, A.G.; Bui, K.; Csuy, C.M.; et al. Errors in Genetic Testing: The Fourth Case Series. Cancer J. 2019, 25, 231–236. [Google Scholar] [CrossRef]
- Knapp, B.; Decker, C.; Lantos, J.D. Neonatologists’ Attitudes about Diagnostic Whole-Genome Sequencing in the NICU. Pediatrics 2019, 143, S54–S57. [Google Scholar] [CrossRef] [Green Version]
- Stoll, K.; Kubendran, S.; Cohen, S.A. The past, present and future of service delivery in genetic counseling: Keeping up in the era of precision medicine. Am. J. Med Genet. Part C Semin. Med Genet. 2018, 178, 24–37. [Google Scholar] [CrossRef]
- Boothe, E.; West, B.; Hendon, L.G.; Kaplan, J.D.; Kirmse, B. Asynchronous telemedicine for clinical genetics consultations in the NICU: A single center’s solution. J. Perinatol. 2021, 42, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Suther, S.; Goodson, P. Barriers to the provision of genetic services by primary care physicians: A systematic review of the literature. Genet. Med. 2003, 5, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, J.G.; Abdiwahab, E.; Edwards, H.M.; Fang, M.-L.; Jdayani, A.; Breslau, E.S. Primary care providers’ cancer genetic testing-related knowledge, attitudes, and communication behaviors: A systematic review and research agenda. J. Gen. Intern. Med. 2017, 32, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Dekanek, E.W.; Thull, D.L.; Massart, M.; Grubs, R.; Rajkovic, A.; Mai, P.L. Knowledge and opinions regarding BRCA1 and BRCA2 genetic testing among primary care physicians. J. Genet. Couns. 2019, 29, 122–130. [Google Scholar] [CrossRef]
- Wilkes, M.S.; Day, F.C.; Fancher, T.L.; McDermott, H.; Lehman, E.; Bell, R.A.; Green, M.J. Increasing confidence and changing behaviors in primary care providers engaged in genetic counselling. BMC Med. Educ. 2017, 17, 163. [Google Scholar] [CrossRef] [Green Version]
- Cornel, M.C. Evidence-Based Genetic Education of Non-Genetic-Expert Physicians: Experiences Over Three Decades in Amsterdam. Front. Genet. 2019, 10, 712. [Google Scholar] [CrossRef]
- Jackson, L.; O’Connor, A.; Paneque, M.; Curtisova, V.; Lunt, P.W.; Pourova, R.K.; Macek, M.; Stefansdottir, V.; Turchetti, D.; Campos, M.; et al. The Gen-Equip Project: Evaluation and impact of genetics e-learning resources for primary care in six European languages. Genet. Med. 2018, 21, 718–726. [Google Scholar] [CrossRef] [Green Version]
- Campion, M.; Goldgar, C.; Hopkin, R.J.; Prows, C.A.; Dasgupta, S. Genomic education for the next generation of health-care providers. Genet. Med. 2019, 21, 2422–2430. [Google Scholar] [CrossRef]
- Miller, D.T.; Lee, K.; Chung, W.K.; Gordon, A.S.; Herman, G.E.; Klein, T.E.; Stewart, D.R.; Amendola, L.M.; Adelman, K.; Bale, S.J.; et al. ACMG SF v3.0 list for reporting of secondary findings in clinical exome and genome sequencing: A policy statement of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 2021, 23, 1381–1390. [Google Scholar] [CrossRef]
- Cohen, J. Statistical Power Analysis for the Behavioral Sciences, 2nd ed.; Lawrence Erlbaum Associates: Hillsdale, NJ, USA, 1988. [Google Scholar]
- Metcalfe, S.; Hurworth, R.; Newstead, J.; Robins, R. Needs assessment study of genetics education for general practitioners in Australia. Genet. Med. 2002, 4, 71–77. [Google Scholar] [CrossRef] [Green Version]
- Barnabé, C.; Kirk, P. A Needs Assessment for Southern Manitoba Physicians for Palliative Care Education. J. Palliat. Care 2002, 18, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Botes, J.; Bezuidenhout, J.; Steinberg, W.J.; Joubert, G. The needs and preferences of general practitioners regarding their CPD learning: A Free State perspective. S. Afr. Fam. Pract. 2016, 58, 114–118. [Google Scholar] [CrossRef] [Green Version]
- Stark, Z.; Nisselle, A.; McClaren, B.; Lynch, F.; Best, S.; Long, J.C.; Martyn, M.; Patel, C.; Schlapbach, L.J.; Barnett, C.; et al. Attitudes of Australian health professionals towards rapid genomic testing in neonatal and paediatric intensive care. Eur. J. Hum. Genet. 2019, 27, 1493–1501. [Google Scholar] [CrossRef] [PubMed]
- Franck, L.S.; Kriz, R.M.; Rego, S.; Garman, K.; Hobbs, C.; Dimmock, D. Implementing Rapid Whole-Genome Sequencing in Critical Care: A Qualitative Study of Facilitators and Barriers to New Technology Adoption. J. Pediatr. 2021, 237, 237–243. [Google Scholar] [CrossRef]
- Accreditation Council for Genetic Counseling Practice-based Competencies for Genetic Counselors. 2019. Retrieved 15 December 2021. Available online: https://www.gceducation.org/practice-based-competencies/ (accessed on 1 January 2022).
- National Society of Genetic Counselors Professional Status Survey 2021: Work Environment. 2021. Retrieved 15 December 2021. Available online: https://www.nsgc.org/Policy-Research-and-Publications/Professional-Status-Survey (accessed on 1 January 2022).


| Clinical Site | Frequency (%) |
|---|---|
| University of Louisville, Louisville, KY, USA | 10 (31%) |
| Woman’s Hospital, Baton Rouge, LA, USA | 8 (24%) |
| University of Alabama at Birmingham, Birmingham, AL, USA | 7 (21%) |
| University of Mississippi Medical Center, Jackson, MS, USA | 5 (15%) |
| Children’s Hospital, New Orleans, LA, USA | 3 (9%) |
| Race | Frequency (%) |
| White | 21 (78%) |
| Black | 4 (15%) |
| Asian | 2 (7%) |
| Years of Experience | Frequency (%) |
| 0–5 years | 10 (37%) |
| 6–10 years | 4 (15%) |
| 11–15 years | 3 (11%) |
| 16–20 years | 4 (15%) |
| 21–25 years | 2 (7%) |
| 25+ years | 4 (15%) |
| Barrier | n (%) |
|---|---|
| Test cost | 22 (81%) |
| Lack of insurance coverage | 22 (81%) |
| Turnaround time | 22 (81%) |
| Lack of healthcare provider knowledge/training | 15 (56%) |
| Possibility of uncertain results | 13 (48%) |
| Possibility of unexpected results | 9 (33%) |
| Limited healthcare provider time | 3 (11%) |
| Limited diagnostic value | 2 (7%) |
| Other | 0 (0%) |
| There are no barriers | 0 (0%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
East, K.M.; Cochran, M.E.; Kelley, W.V.; Greve, V.; Finnila, C.R.; Coleman, T.; Jennings, M.; Alexander, L.; Rahn, E.J.; Danila, M.I.; et al. Education and Training of Non-Genetics Providers on the Return of Genome Sequencing Results in a NICU Setting. J. Pers. Med. 2022, 12, 405. https://doi.org/10.3390/jpm12030405
East KM, Cochran ME, Kelley WV, Greve V, Finnila CR, Coleman T, Jennings M, Alexander L, Rahn EJ, Danila MI, et al. Education and Training of Non-Genetics Providers on the Return of Genome Sequencing Results in a NICU Setting. Journal of Personalized Medicine. 2022; 12(3):405. https://doi.org/10.3390/jpm12030405
Chicago/Turabian StyleEast, Kelly M., Meagan E. Cochran, Whitley V. Kelley, Veronica Greve, Candice R. Finnila, Tanner Coleman, Mikayla Jennings, Latonya Alexander, Elizabeth J. Rahn, Maria I. Danila, and et al. 2022. "Education and Training of Non-Genetics Providers on the Return of Genome Sequencing Results in a NICU Setting" Journal of Personalized Medicine 12, no. 3: 405. https://doi.org/10.3390/jpm12030405
APA StyleEast, K. M., Cochran, M. E., Kelley, W. V., Greve, V., Finnila, C. R., Coleman, T., Jennings, M., Alexander, L., Rahn, E. J., Danila, M. I., Barsh, G., Korf, B., & Cooper, G. (2022). Education and Training of Non-Genetics Providers on the Return of Genome Sequencing Results in a NICU Setting. Journal of Personalized Medicine, 12(3), 405. https://doi.org/10.3390/jpm12030405






