Deep Multi-Objective Learning from Low-Dose CT for Automatic Lung-RADS Report Generation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Acquisition
2.2. Radiomics Feature Extraction
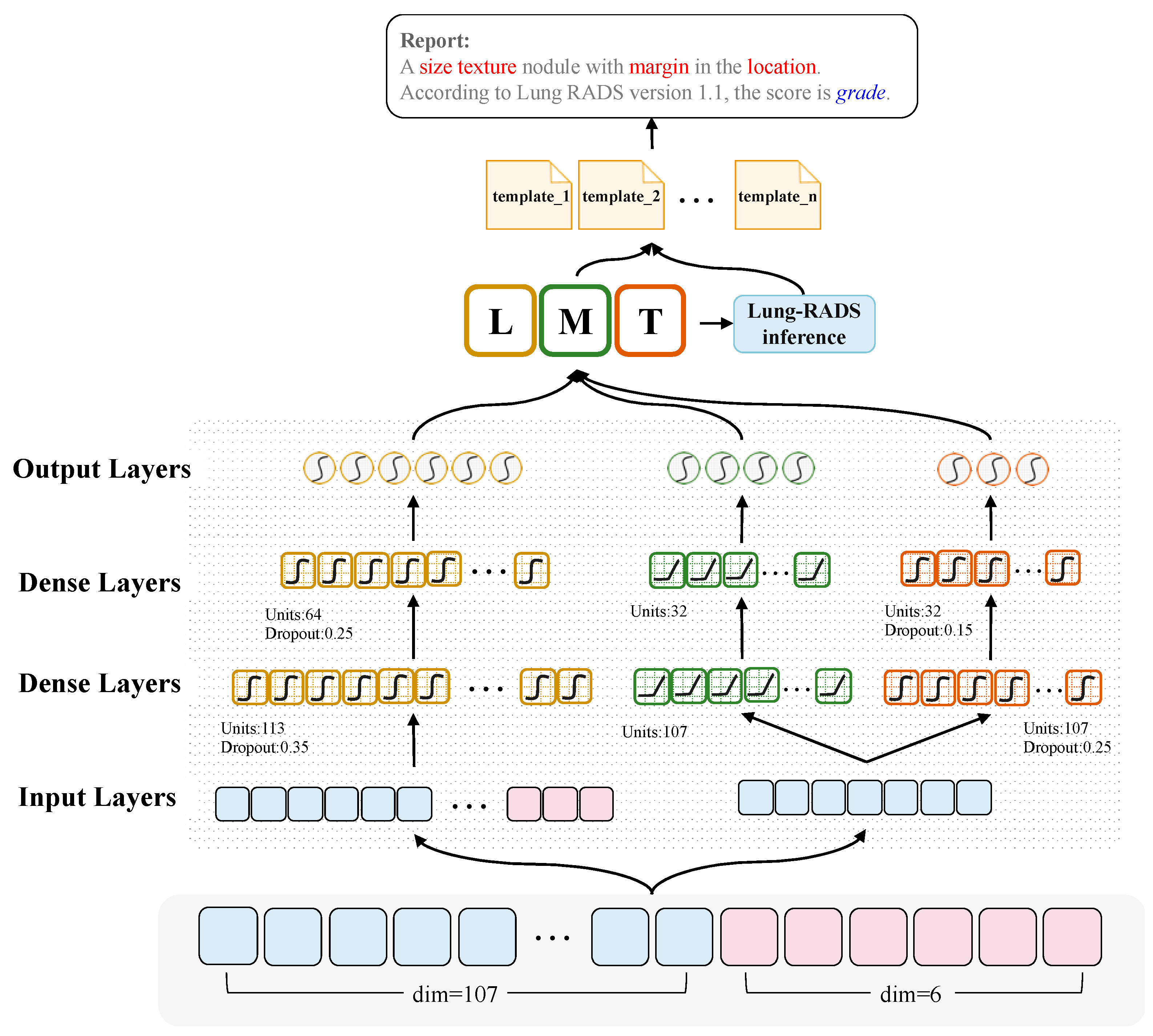
2.3. CT2Rep: An Effective Multi-Objective Learning Method for Radiology Report Generation
3. Results
3.1. Semantic Feature Prediction
3.2. Satisfaction Analysis for Radiology Report Generation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Allemani, C.; Matsuda, T.; Di Carlo, V.; Harewood, R.; Matz, M.; Nikšić, M.; Bonaventure, A.; Valkov, M.; Johnson, C.J.; Estève, J. Global surveillance of trends in cancer survival 2000–2014 (CONCORD-3): Analysis of individual records for 37,513,025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet 2018, 391, 1023–1075. [Google Scholar] [CrossRef] [Green Version]
- De Koning, H.J.; van der Aalst, C.M.; de Jong, P.A.; Scholten, E.T.; Nackaerts, K.; Heuvelmans, M.A.; Lammers, J.-W.J.; Weenink, C.; Yousaf-Khan, U.; Horeweg, N. Reduced lung-cancer mortality with volume CT screening in a randomized trial. N. Engl. J. Med. 2020, 382, 503–513. [Google Scholar] [CrossRef] [PubMed]
- National Lung Screening Trial Research Team. Lung cancer incidence and mortality with extended follow-up in the National Lung Screening Trial. J. Thorac. Oncol. 2019, 14, 1732–1742. [Google Scholar] [CrossRef] [PubMed]
- McKee, B.J.; Regis, S.M.; McKee, A.B.; Flacke, S.; Wald, C. Performance of ACR Lung-RADS in a clinical CT lung screening program. J. Am. Coll. Radiol. 2016, 13, R25–R29. [Google Scholar] [CrossRef] [PubMed]
- Sinjab, F.; Kong, K.; Gibson, G.; Varma, S.; Williams, H.; Padgett, M.; Notingher, I. Tissue diagnosis using power-sharing multifocal Raman micro-spectroscopy and auto-fluorescence imaging. Biomed. Opt. Express 2016, 7, 2993–3006. [Google Scholar] [CrossRef] [PubMed]
- Qian, C.; Wang, L.; Gao, Y.; Yousuf, A.; Yang, X.; Oto, A.; Shen, D. In vivo MRI based prostate cancer localization with random forests and auto-context model. Comput. Med. Imaging Graph. 2016, 52, 44–57. [Google Scholar] [CrossRef] [Green Version]
- Huda, S.; Yearwood, J.; Jelinek, H.F.; Hassan, M.M.; Fortino, G.; Buckland, M. A hybrid feature selection with ensemble classification for imbalanced healthcare data: A case study for brain tumor diagnosis. IEEE Access 2016, 4, 9145–9154. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Yao, C.; Yu, H.; Wu, T. Deep reinforcement learning with its application for lung cancer detection in medical Internet of Things. Future Gener. Comput. Syst. 2019, 97, 1–9. [Google Scholar] [CrossRef]
- Yasaka, K.; Abe, O. Deep learning and artificial intelligence in radiology: Current applications and future directions. PLoS Med. 2018, 15, e1002707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, Z.; Huang, R.; Ding, Y.; Lan, T.; Dong, R.; Qin, Z.; Zhang, X.; Wang, W. A deep learning-based segmentation method for brain tumor in MR images. In Proceedings of the 2016 IEEE 6th International Conference on Computational Advances in Bio and Medical Sciences (ICCABS), Atlanta, GA, USA, 13–16 October 2016; pp. 1–6. [Google Scholar]
- Mohsen, H.; El-Dahshan, E.-S.A.; El-Horbaty, E.-S.M.; Salem, A.-B.M. Classification using deep learning neural networks for brain tumors. Future Comput. Inform. J. 2018, 3, 68–71. [Google Scholar] [CrossRef]
- Bhandary, A.; Prabhu, G.A.; Rajinikanth, V.; Thanaraj, K.P.; Satapathy, S.C.; Robbins, D.E.; Shasky, C.; Zhang, Y.-D.; Tavares, J.M.R.; Raja, N.S.M. Deep-learning framework to detect lung abnormality—A study with chest X-ray and lung CT scan images. Pattern Recognit. Lett. 2020, 129, 271–278. [Google Scholar] [CrossRef]
- Xie, H.; Yang, D.; Sun, N.; Chen, Z.; Zhang, Y. Automated pulmonary nodule detection in CT images using deep convolutional neural networks. Pattern Recognit. 2019, 85, 109–119. [Google Scholar] [CrossRef]
- Zhu, W.; Liu, C.; Fan, W.; Xie, X. Deeplung: Deep 3d dual path nets for automated pulmonary nodule detection and classification. In Proceedings of the 2018 IEEE Winter Conference on Applications of Computer Vision (WACV), Lake Tahoe, NV, USA, 12–15 March 2018; pp. 673–681. [Google Scholar]
- Hunter, J.; Freer, Y.; Gatt, A.; Reiter, E.; Sripada, S.; Sykes, C. Automatic generation of natural language nursing shift summaries in neonatal intensive care: BT-Nurse. Artif. Intell. Med. 2012, 56, 157–172. [Google Scholar] [CrossRef] [Green Version]
- Monshi, M.M.A.; Poon, J.; Chung, V. Deep learning in generating radiology reports: A survey. Artif. Intell. Med. 2020, 106, 101878. [Google Scholar] [CrossRef]
- Xue, Y.; Xu, T.; Long, L.R.; Xue, Z.; Antani, S.; Thoma, G.R.; Huang, X. Multimodal recurrent model with attention for automated radiology report generation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Granada, Spain, 16–20 September 2018; pp. 457–466. [Google Scholar]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Summers, R.M. TieNet: Text-image embedding network for common thorax disease classification and reporting in chest x-rays. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 9049–9058. [Google Scholar]
- Chen, C.-H.; Chang, C.-K.; Tu, C.-Y.; Liao, W.-C.; Wu, B.-R.; Chou, K.-T.; Chiou, Y.-R.; Yang, S.-N.; Zhang, G.; Huang, T.-C. Radiomic features analysis in computed tomography images of lung nodule classification. PLoS ONE 2018, 13, e0192002. [Google Scholar] [CrossRef] [Green Version]
- Lambin, P.; Leijenaar, R.T.; Deist, T.M.; Peerlings, J.; De Jong, E.E.; Van Timmeren, J.; Sanduleanu, S.; Larue, R.T.; Even, A.J.; Jochems, A. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- Van Griethuysen, J.J.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [Green Version]
- Rizzo, S.; Botta, F.; Raimondi, S.; Origgi, D.; Fanciullo, C.; Morganti, A.G.; Bellomi, M. Radiomics: The facts and the challenges of image analysis. Eur. Radiol. Exp. 2018, 2, 1–8. [Google Scholar] [CrossRef]
- Li, Q.; Cai, W.; Wang, X.; Zhou, Y.; Feng, D.D.; Chen, M. Medical image classification with convolutional neural network. In Proceedings of the 2014 13th International Conference on Control Automation Robotics & Vision (ICARCV), Singapore, 10–12 December 2014; pp. 844–848. [Google Scholar]
- Donner, R.V.; Zou, Y.; Donges, J.F.; Marwan, N.; Kurths, J. Recurrence networks—A novel paradigm for nonlinear time series analysis. New J. Phys. 2010, 12, 033025. [Google Scholar] [CrossRef] [Green Version]
- Tang, J.; Deng, C.; Huang, G.-B. Extreme learning machine for multilayer perceptron. IEEE Trans. Neural Netw. Learn. Syst. 2015, 27, 809–821. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.K.; Mitra, S. Multilayer perceptron, fuzzy sets, classifiaction. IEEE Trans. Neural Netw. Learn. Syst. 1992, 3, 683–697. [Google Scholar] [CrossRef] [PubMed]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning representations by back-propagating errors. Nature 1986, 323, 533–536. [Google Scholar] [CrossRef]
- Gardner, M.W.; Dorling, S. Artificial neural networks (the multilayer perceptron)—A review of applications in the atmospheric sciences. Atmos. Environ. 1998, 32, 2627–2636. [Google Scholar] [CrossRef]
- Glorot, X.; Bordes, A.; Bengio, Y. Deep sparse rectifier neural networks. In Proceedings of the 14th International Conference on Artificial Intelligence and Statistics, Lauderdale, FL, USA, 11–13 April 2011; pp. 315–323. [Google Scholar]
- Kalman, B.L.; Kwasny, S.C. Why tanh: Choosing a sigmoidal function. In Proceedings of the IJCNN International Joint Conference on Neural Networks, Baltimore, MD, USA, 7–11 June 1992; pp. 578–581. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Dahl, G.E.; Sainath, T.N.; Hinton, G.E. Improving deep neural networks for LVCSR using rectified linear units and dropout. In Proceedings of the 2013 IEEE International Conference on Acoustics, Speech and Signal Processing, Vancouver, BC, Canada, 26–31 May 2013; pp. 8609–8613. [Google Scholar]
- Taqi, A.M.; Awad, A.; Al-Azzo, F.; Milanova, M. The impact of multi-optimizers and data augmentation on TensorFlow convolutional neural network performance. In Proceedings of the 2018 IEEE Conference on Multimedia Information Processing and Retrieval (MIPR), Miami, FL, USA, 10–12 April 2018; pp. 140–145. [Google Scholar]
- Rady, H. Classification of multilayer neural networks using cross entropy and mean square errors. J. Am. Chem. Soc. 2008, 2, 29–48. [Google Scholar]
- Pinsky, P.F.; Gierada, D.S.; Black, W.; Munden, R.; Nath, H.; Aberle, D.; Kazerooni, E. Performance of Lung-RADS in the National Lung Screening Trial: A retrospective assessment. Ann. Intern. Med. 2015, 162, 485–491. [Google Scholar] [CrossRef] [Green Version]
- Han, J.; Kamber, M.; Pei, J. Data Mining: Concepts and Techniques, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2011. [Google Scholar]
- Parmar, C.; Grossmann, P.; Bussink, J.; Lambin, P.; Aerts, H.J. Machine learning methods for quantitative radiomic biomarkers. Sci. Rep. 2015, 5, 13087. [Google Scholar] [CrossRef]
- Liao, S.; Wang, J.; Yu, R.; Sato, K.; Cheng, Z. CNN for situations understanding based on sentiment analysis of twitter data. Procedia Comput. Sci. 2017, 111, 376–381. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Lai, S.; Xu, L.; Liu, K.; Zhao, J. Recurrent convolutional neural networks for text classification. In Proceedings of the Twenty-Ninth AAAI Conference on Artificial Intelligence, Austin, TX, USA, 25–30 January 2015. [Google Scholar]
- LeCun, Y.; Kavukcuoglu, K.; Farabet, C. Convolutional networks and applications in vision. In Proceedings of the 2010 IEEE International Symposium on Circuits and Systems, Paris, France, 30 May–2 June 2010; pp. 253–256. [Google Scholar]
- Zeiler, M.D.; Fergus, R. Visualizing and understanding convolutional networks. In Proceedings of the European Conference on Computer Vision, Zurich, Switzerland, 6–12 September 2014; pp. 818–833. [Google Scholar]
- Bar, Y.; Diamant, I.; Wolf, L.; Lieberman, S.; Konen, E.; Greenspan, H. Chest pathology detection using deep learning with non-medical training. In Proceedings of the 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), New York, NY, USA, 16–19 April 2015; pp. 294–297. [Google Scholar]
- Cicero, M.; Bilbily, A.; Colak, E.; Dowdell, T.; Gray, B.; Perampaladas, K.; Barfett, J. Training and validating a deep convolutional neural network for computer-aided detection and classification of abnormalities on frontal chest radiographs. Investig. Radiol. 2017, 52, 281–287. [Google Scholar] [CrossRef] [PubMed]
- DeLong, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Gold, K.A.; Lin, H.Y.; Swisher, S.G.; Xing, Y.; Lee, J.J.; Kim, E.S.; William, W.N., Jr. Relationship between tumor size and survival in non-small-cell lung cancer (NSCLC): An analysis of the surveillance, epidemiology, and end results (SEER) registry. J. Thorac. Oncol. 2015, 10, 682–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohri, M.; Rostamizadeh, A.; Talwalkar, A. Foundations of Machine Learning; MIT Press: Cambridge, MA, USA, 2018. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Fitzgerald, R. Error in radiology. Clin. Radiol. 2001, 56, 938–946. [Google Scholar] [CrossRef]
- Maselli, G.; Bertamino, E.; Capalbo, C.; Mancini, R.; Orsi, G.; Napoli, C.; Napoli, C. Hierarchical convolutional models for automatic pneu-monia diagnosis based on X-ray images: New strategies in public health. Ann. Ig. 2021, 33, 644–655. [Google Scholar]
- Donovan, T.; Litchfield, D. Looking for cancer: Expertise related differences in searching and decision making. Appl. Cogn. Psychol. 2013, 27, 43–49. [Google Scholar] [CrossRef] [Green Version]
- Oberije, C.; Nalbantov, G.; Dekker, A.; Boersma, L.; Borger, J.; Reymen, B.; van Baardwijk, A.; Wanders, R.; De Ruysscher, D.; Steyerberg, E. A prospective study comparing the predictions of doctors versus models for treatment outcome of lung cancer patients: A step toward individualized care and shared decision making. Radiother. Oncol. 2014, 112, 37–43. [Google Scholar] [CrossRef] [Green Version]
- Doi, K. Computer-aided diagnosis in medical imaging: Historical review, current status and future potential. Comput. Med. Imaging Graph. 2007, 31, 198–211. [Google Scholar] [CrossRef] [Green Version]





| Semantic Feature | Label |
|---|---|
| Location | RUL |
| RML | |
| RLL | |
| LUL | |
| LLL | |
| Lingular Lobe | |
| Texture | Solid |
| Subsolid | |
| Pure GGO | |
| Margin | Sharp Circumscribed |
| Lobulated | |
| Indistinct | |
| Spiculated |
| Group | Feature | Amount | Total | |
|---|---|---|---|---|
| Radiomics | First-order | Intensity | 18 | 107 |
| shape-based | 14 | |||
| Second-order | GLCM | 24 | ||
| GLRLM | 16 | |||
| GLSZM | 16 | |||
| NGTDM | 5 | |||
| GLDM | 14 | |||
| Segmentation | SISN | 6 | 6 | |
| Method | Location | Margin | Texture | Macro avg. |
|---|---|---|---|---|
| Precision, Recall, F1-Score (%) | ||||
| NB | 49.40/49.78/49.05 | 34.20/33.62/27.10 | 82.37/76.20/76.65 | 55.20/53.20/50.93 |
| KNN | 47.82/50.44/48.99 | 40.64/40.61/39.97 | 84.56/86.90/84.85 | 57.67/86.90/57.94 |
| XGB | 69.23/73.58/70.89 | 40.49/40.83/40.13 | 87.32/88.65/87.45 | 65.68/67.69/66.32 |
| RNN | 64.46/65.07/57.05 | 83.19/78.82/77.22 | 93.36/92.79/91.76 | 80.34/78.89/75.34 |
| MLP | 85.69/85.81/83.32 | 84.12/81.22/79.50 | 92.81/82.14/91.05 | 87.54/86.39/84.62 |
| CNN | 86.39/85.81/82.99 | 86.25/82.75/81.34 | 93.18/92.58/91.62 | 88.61/87.05/85.32 |
| CT2Rep | 85.68/87.12/84.66 | 83.36/83.41/83.10 | 94.11/94.10/94.09 | 87.72/82.74/87.29 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, Y.-C.; Hsing, Y.-C.; Chiu, Y.-W.; Shih, C.-C.; Lin, J.-H.; Hsiao, S.-H.; Sakai, K.; Ko, K.-H.; Chen, C.-Y. Deep Multi-Objective Learning from Low-Dose CT for Automatic Lung-RADS Report Generation. J. Pers. Med. 2022, 12, 417. https://doi.org/10.3390/jpm12030417
Chang Y-C, Hsing Y-C, Chiu Y-W, Shih C-C, Lin J-H, Hsiao S-H, Sakai K, Ko K-H, Chen C-Y. Deep Multi-Objective Learning from Low-Dose CT for Automatic Lung-RADS Report Generation. Journal of Personalized Medicine. 2022; 12(3):417. https://doi.org/10.3390/jpm12030417
Chicago/Turabian StyleChang, Yung-Chun, Yan-Chun Hsing, Yu-Wen Chiu, Cho-Chiang Shih, Jun-Hong Lin, Shih-Hsin Hsiao, Koji Sakai, Kai-Hsiung Ko, and Cheng-Yu Chen. 2022. "Deep Multi-Objective Learning from Low-Dose CT for Automatic Lung-RADS Report Generation" Journal of Personalized Medicine 12, no. 3: 417. https://doi.org/10.3390/jpm12030417
APA StyleChang, Y.-C., Hsing, Y.-C., Chiu, Y.-W., Shih, C.-C., Lin, J.-H., Hsiao, S.-H., Sakai, K., Ko, K.-H., & Chen, C.-Y. (2022). Deep Multi-Objective Learning from Low-Dose CT for Automatic Lung-RADS Report Generation. Journal of Personalized Medicine, 12(3), 417. https://doi.org/10.3390/jpm12030417







