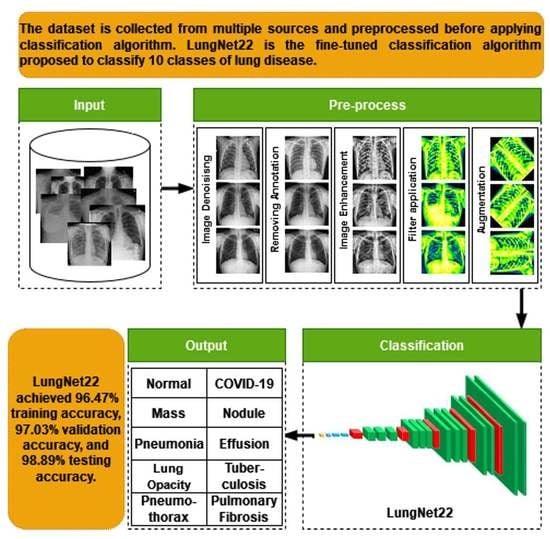
LungNet22: A Fine-Tuned Model for Multiclass Classification and Prediction of Lung Disease Using X-ray Images
Abstract
:1. Introduction
- The raw chest X-ray data for the study is collected from multiple data sources, merged into a collective dataset. To ensure all the data are in the same scale, preprocessing is performed.
- Autoencoder is employed to denoise the noisy images. For removing annotation in the data, the EnsNet method is used.
- CLAHE enhancement methods are used to increase the quality of the X-ray images, and the Green Fire Blue filtering technique is applied to improve the image properties and make them more identical to the model.
- Augmentation is performed to balance each class of the dataset by upsampling the classes with fewer data.
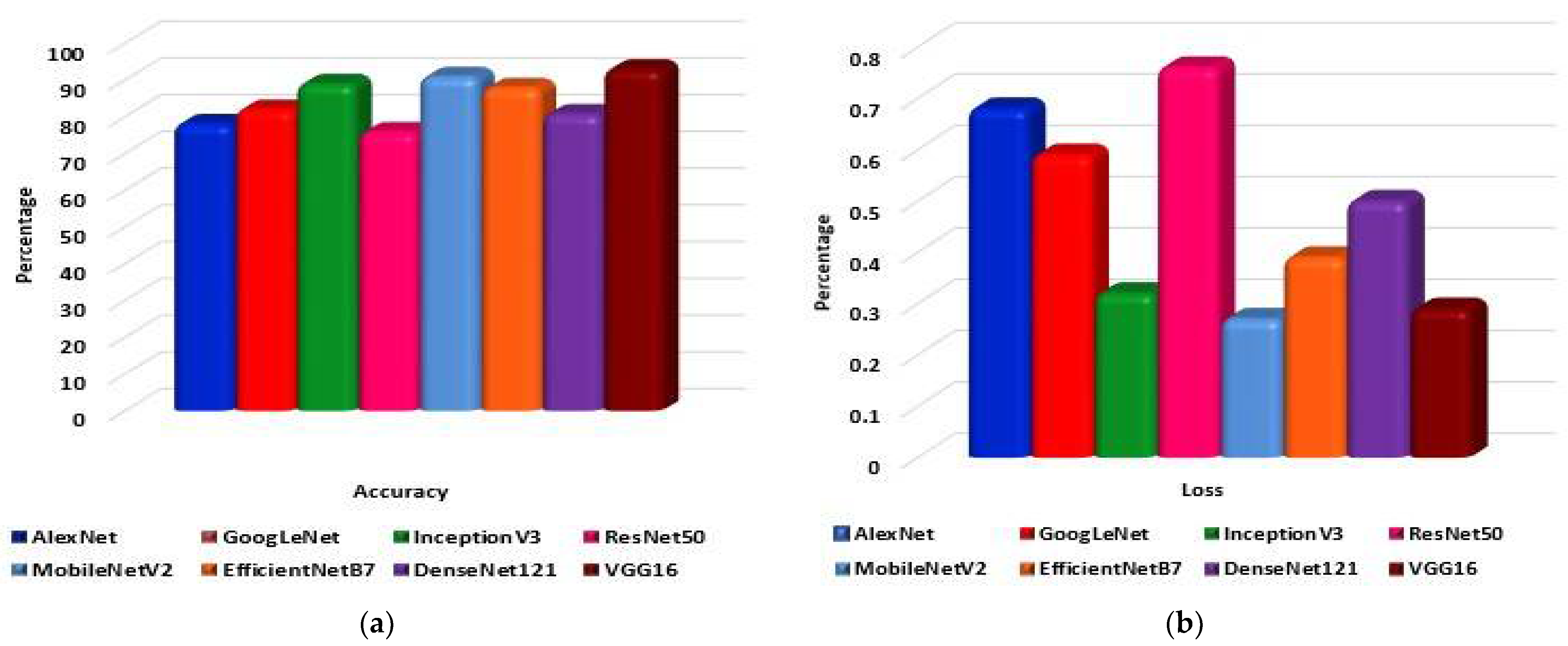
- Eight pre-trained models, namely AlexNet, GoogLeNet, InceptionV3, MobileNetV2, VGG16, ResNet 50, DenseNet121, and EfficientNetB7 are evaluated to determine the highest-performing model.
- A fine-tuned LungNet22 is proposed based on VGG16 architecture.
- To justify the model’s accuracy, the precision, recall, specificity, and f1-score are calculated from the confusion matrix. The loss function is also used to evaluate the performance of the models.
- For the visual description of the model’s outcome, Grad-CAM is produced.
- The proposed model is evaluated by computing the ROC curve and the AUC value.
2. Literature Review
3. Datasets
3.1. Dataset Preprocessing
3.1.1. Image Denoising
3.1.2. Image Annotation Remove
3.1.3. Image Enhancement (CLAHE)
3.1.4. Filter Apply
Ablation Study of the Applied Filters
3.2. Image Augmentation
4. Proposed Model
4.1. Transfer Learning Models
4.1.1. AlexNet
4.1.2. GoogLeNet
4.1.3. InceptionV3
4.1.4. ResNet50
4.1.5. MobileNetV2
4.1.6. EfficientNetB7
4.1.7. DenseNet121
4.1.8. VGG16
4.1.9. LungNet22
4.2. Ablation Study of the Proposed Model (LungNet22)
4.2.1. Ablation Study 1: Changing GlobalAveragePooling2D Layer
4.2.2. Ablation Study 2: Changing the Flatten layer
4.2.3. Ablation Study 3: Changing Loss Functions
4.2.4. Ablation Study 4: Changing Optimizer and Learning Rate
5. Results
6. Discussion
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bhandary, A.; Prabhu, G.A.; Rajinikanth, V.; Thanaraj, K.P.; Satapathy, S.C.; Robbins, D.E.; Shasky, C.; Zhang, Y.D.; Tavares, J.M.; Raja, N.S. Deep-learning framework to detect lung abnormality–A study with chest X-ray and lung CT scan images. Pattern Recognit. Lett. 2020, 129, 271–278. [Google Scholar] [CrossRef]
- Han, T.; Nunes, V.X.; Souza, L.F.D.F.; Marques, A.G.; Silva, I.C.L.; Junior, M.A.A.F.; Sun, J.; Filho, P.P.R. Internet of Medical Things—Based on Deep Learning Techniques for Segmentation of Lung and Stroke Regions in CT Scans. IEEE Access 2020, 8, 71117–71135. [Google Scholar] [CrossRef]
- De Sousa, P.M.; Carneiro, P.C.; Oliveira, M.M.; Pereira, G.M.; Junior, C.A.D.C.; de Moura, L.V.; Mattjie, C.; da Silva, A.M.M.; Patrocinio, A.C. COVID-19 classification in X-ray chest images using a new convolutional neural network: CNN-COVID. Res. Biomed. Eng. 2021, 38, 87–97. [Google Scholar] [CrossRef]
- Tobias, R.R.; De Jesus, L.C.; Mital, M.E.; Lauguico, S.C.; Guillermo, M.A.; Sybingco, E.; Bandala, A.A.; Dadios, E.P. CNN-based deep learning model for chest X-ray health classification using tensorflow. In Proceedings of the 2020 RIVF International Conference on Computing and Communication Technologies (RIVF), Ho Chi Minh City, Vietnam, 14–15 October 2020; pp. 1–6. [Google Scholar]
- Zhou, L.; Yin, X.; Zhang, T.; Feng, Y.; Zhao, Y.; Jin, M.; Peng, M.; Xing, C.; Li, F.; Wang, Z.; et al. Detection and Semiquantitative Analysis of Cardiomegaly, Pneumothorax, and Pleural Effusion on Chest Radiographs. Radiol. Artif. Intell. 2021, 3, e200172. [Google Scholar] [CrossRef] [PubMed]
- Rajaraman, S.; Antani, S. Training deep learning algorithms with weakly labeled pneumonia chest X-ray data for COVID-19 detection. medRxiv 2020. preprint. [Google Scholar]
- Akter, S.; Shamrat, F.M.; Chakraborty, S.; Karim, A.; Azam, S. COVID-19 detection using deep learning algorithm on chest X-ray images. Biology 2021, 10, 1174. [Google Scholar] [CrossRef] [PubMed]
- Hashmi, M.F.; Katiyar, S.; Keskar, A.G.; Bokde, N.D.; Geem, Z.W. Efficient Pneumonia Detection in Chest Xray Images Using Deep Transfer Learning. Diagnostics 2020, 10, 417. [Google Scholar] [CrossRef]
- Chatterjee, S.; Dzitac, S.; Sen, S.; Rohatinovici, N.C.; Dey, N.; Ashour, A.S.; Balas, V.E. Hybrid modified Cuckoo Search-Neural Network in chronic kidney disease classifi cation. In Proceedings of the 2017 14th International Conference on Engineering of Modern Electric Systems (EMES), Oradea, Romania, 1–2 June 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 164–167. [Google Scholar]
- Islam, K.; Wijewickrema, S.; Collins, A.; O’Leary, S. A deep transfer learning frame work for pneumonia detection from chest X-ray images. In Proceedings of the 15th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications, Valletta, Malta, 27–29 February 2020; SCITEPRESS—Science and Technology Publications: Setúbal, Portugal, 2020. [Google Scholar] [CrossRef]
- Salehinejad, H.; Colak, E.; Dowdell, T.; Barfett, J.; Valaee, S. Synthesizing chest X-ray pathology for training deep convolutional neural networks. IEEE Trans. Med. Imaging 2018, 38, 1197–1206. [Google Scholar] [CrossRef]
- Demir, F.; Sengur, A.; Bajaj, V. Convolutional neural networks based efficient approach for classification of lung diseases. Health Inf. Sci. Syst. 2019, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- Jin, B.; Cruz, L.; Goncalves, N. Deep Facial Diagnosis: Deep Transfer Learning from Face Recognition to Facial Diagnosis. IEEE Access 2020, 8, 123649–123661. [Google Scholar] [CrossRef]
- Rauf, H.T.; Saleem, B.A.; Lali, M.I.U.; Khan, M.A.; Sharif, M.; Bukhari, S.A.C. A citrus fruits and leaves dataset for detection and classification of citrus diseases through machine learning. Data Brief 2019, 26, 104340. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wang, H.; Shen, H. Task Failure Prediction in Cloud Data Centers Using Deep Learning. IEEE Trans. Serv. Comput. 2020. [Google Scholar] [CrossRef]
- Gao, J.; Wang, H.; Shen, H. Smartly handling renewable energy instability in supporting a cloud datacenter. In Proceedings of the 2020 IEEE International Parallel and Distributed Processing Symposium (IPDPS), New Orleans, LA, USA, 18–22 May 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 769–778. [Google Scholar]
- Jain, R.; Nagrath, P.; Kataria, G.; Kaushik, V.S.; Hemanth, D.J. Pneumonia detection in chest X-ray images using convolutional neural networks and transfer learning. Measurement 2020, 165, 108046. [Google Scholar] [CrossRef]
- Mongan, J.; Moy, L.; Kahn, C.E. Checklist for Artificial Intelligence in Medical Imaging (CLAIM): A Guide for Authors and Reviewers. Radiol. Artif. Intell. 2020, 2, e200029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, A.-R.; Li, J.; Yang, G.; O’Shea, S.J. A machine learning approach to automatic detection of irregularity in skin lesion border using dermoscopic images. PeerJ Comput. Sci. 2020, 6, e268. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xie, Y.; Li, Y.; Shen, C.; Xia, Y. Covid-19 screening on chest x-ray images using deep learning based anomaly detection. arXiv 2020, arXiv:2003.12338. [Google Scholar]
- Kaggle. Available online: https://www.kaggle.com/prashant268/chest-xray-covid19-pneumonia (accessed on 5 January 2022).
- Kaggle. Available online: https://www.kaggle.com/sid321axn/covid-cxr-image-dataset-research (accessed on 5 January 2022).
- Kaggle. Available online: https://www.kaggle.com/jtiptj/chest-xray-pneumoniacovid19tuberculosis (accessed on 5 January 2022).
- Github. Available online: https://github.com/ieee8023/covid-chestxray-dataset (accessed on 3 January 2022).
- Kaggle. Available online: https://www.kaggle.com/andyczhao/covidx-cxr2?select=train (accessed on 4 January 2022).
- Kaggle. Available online: https://www.kaggle.com/tawsifurrahman/tuberculosis-tb-chest-xray-dataset (accessed on 4 January 2022).
- Kaggle. Available online: https://www.kaggle.com/raddar/tuberculosis-chest-xrays-shenzhen?select=images (accessed on 5 January 2022).
- Kaggle. Available online: https://www.kaggle.com/donjon00/covid19-detection (accessed on 5 January 2022).
- Kaggle. Available online: https://www.kaggle.com/volodymyrgavrysh/pneumothorax-binary-classification-task (accessed on 5 January 2022).
- Kaggle. Available online: https://www.kaggle.com/nih-chest-xrays/sample (accessed on 6 January 2022).
- Kaggle. Available online: https://www.kaggle.com/kamildinleyici/covid-normal-viral-opacity-v2 (accessed on 3 January 2022).
- Kaggle. Available online: https://www.kaggle.com/gauravduttakiit/x-ray-report (accessed on 4 January 2022).
- NIH Dataset. Available online: https://nihcc.app.box.com/v/ChestXray-NIHCC/folder/36938765345 (accessed on 3 January 2022).
- Kaggle. Available online: https://www.kaggle.com/homayoonkhadivi/chest-xray-worldwide-datasets (accessed on 3 January 2022).
- Kaggle. Available online: https://www.kaggle.com/tawsifurrahman/covid19-radiography-database (accessed on 6 January 2022).
- Kaggle. Available online: https://www.kaggle.com/raddar/tuberculosis-chest-xrays-shenzhen (accessed on 6 January 2022).
- Ibrahim, D.M.; Elshennawy, N.M.; Sarhan, A.M. Deep-chest: Multi-classification deep learning model for diagnosing COVID-19, pneumonia, and lung cancer chest diseases. Comput. Biol. Med. 2021, 132, 104348. [Google Scholar] [CrossRef]
- Bharati, S.; Podder, P.; Mondal, M.R.H. Hybrid deep learning for detecting lung diseases from X-ray images. Inform. Med. Unlocked 2020, 20, 100391. [Google Scholar] [CrossRef]
- Chen, J.I.Z. Design of accurate classification of COVID-19 disease in X-ray images using Deep Learning Approach. J. ISMAC 2021, 3, 132–148. [Google Scholar] [CrossRef]
- Sivasamy, J.; Subashini, T. Classification and predictions of lung diseases from chest X-rays using MobileNet. Int. J. Anal. Exp. Modal Anal. 2020, 12, 665–672. [Google Scholar]
- Fauzan, A.R.; Wahyuddin, M.I.; Ningsih, S. Pleural Effusion Classification Based on Chest X-ray Images using Convolutional Neural Network. J. Ilmu Komput. Inf. 2021, 14, 9–16. [Google Scholar] [CrossRef]
- Abbas, A.; Abdelsamea, M.M.; Gaber, M.M. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Appl. Intell. 2021, 51, 854–864. [Google Scholar] [CrossRef] [PubMed]
- Albahli, S.; Rauf, H.T.; Algosaibi, A.; Balas, V.E. AI-driven deep CNN approach for multi-label pathology classification using chest X-rays. PeerJ Comput. Sci. 2021, 7, e495. [Google Scholar] [CrossRef] [PubMed]
- Podder, S.; Bhattacharjee, S.; Roy, A. An efficient method of detection of COVID-19 using Mask R-CNN on chest X-ray images. AIMS Biophys. 2021, 8, 281–290. [Google Scholar] [CrossRef]
- Singh, S.; Sapra, P.; Garg, A.; Vishwakarma, D.K. CNN based COVID-aid: COVID 19 Detection using Chest X-ray. In Proceedings of the 2021 5th International Conference on Computing Methodologies and Communication (ICCMC), Erode, India, 8–10 April 2021; pp. 1791–1797. [Google Scholar]
- Moujahid, H.; Cherradi, B.; El Gannour, O.; Bahatti, L.; Terrada, O.; Hamida, S. Convolutional neural network based classification of patients with pneumonia using X-ray lung images. Adv. Sci. Technol. Eng. Syst. J. 2020, 5, 167–175. [Google Scholar] [CrossRef]
- Das, A.K.; Ghosh, S.; Thunder, S.; Dutta, R.; Agarwal, S.; Chakrabarti, A. Automatic COVID-19 detection from X-ray images using ensemble learning with convolutional neural network. Pattern Anal. Appl. 2021, 24, 1111–1124. [Google Scholar] [CrossRef]
- Irmak, E. COVID-19 disease severity assessment using CNN model. IET Image Proc. 2021, 15, 1814. [Google Scholar] [CrossRef]
- Hassantabar, S.; Ahmadi, M.; Sharifi, A. Diagnosis and detection of infected tissue of COVID-19 patients based on lung X-ray image using convolutional neural network approaches. Chaos Solitons Fractals 2020, 140, 110170. [Google Scholar] [CrossRef]
- Ismael, A.M.; Şengür, A. Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Syst. Appl. 2021, 164, 114054. [Google Scholar] [CrossRef]
- Wang, B.; Zhang, W. MARnet: Multi-scale adaptive residual neural network for chest X-ray images recognition of lung diseases. Math. Biosci. Eng. 2022, 19, 331–350. [Google Scholar] [CrossRef]
- Denoising Autoencoders. Available online: https://omdena.com/blog/denoising-autoencoders/ (accessed on 15 January 2022).
- Denoising Autoencoders. Available online: https://pyimagesearch.com/2020/02/24/denoising-autoencoders-with-keras-tensorflow-and-deep-learning/ (accessed on 16 January 2022).
- Liang, J.; Doermann, D.; Li, H. Camera-based analysis of text and documents: A survey. Int. J. Doc. Anal. Recognit. 2005, 7, 84–104. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, Y.; Jin, L.; Huang, Y.; Lai, S. Ensnet: Ensconce text in the wild. In Proceedings of the AAAI 2019 Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; Volume 33, pp. 801–808. [Google Scholar]
- Mirza, M.; Osindero, S. Conditional generative adversarial nets. arXiv 2014, arXiv:1411.1784. [Google Scholar]
- Shen, X.; Chen, Y.C.; Tao, X.; Jia, J. Convolutional neural pyramid for image processing. arXiv 2017, arXiv:1704.02071. [Google Scholar]
- Dong, C.; Loy, C.C.; Tang, X. Accelerating the super-resolution convolutional neural network. In Proceedings of the 14th European Conference on Computer Vision, Amsterdam, The Netherlands, 8–16 October 2016; Springer: Berlin/Heidelberg, Germany; pp. 391–407. [Google Scholar]
- Yang, C.; Lu, X.; Lin, Z.; Shechtman, E.; Wang, O.; Li, H. High-resolution image inpainting using multi-scale neural patch synthesis. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2017, Honolulu, HI, USA, 21–26 July 2017; Volume 1, pp. 6721–6729. [Google Scholar]
- Johnson, J.; Alahi, A.; Li, F. Perceptual losses for real-time style transfer and super-resolution. In Proceedings of the European Conference on Computer Vision, Amsterdam, The Netherland, 8–16 October 2016; Springer: Berlin/Heidelberg, Germany, 2016; pp. 694–711. [Google Scholar]
- Gatys, L.A.; Ecker, A.S.; Bethge, M. A neural algorithm of artistic style. arXiv 2015, arXiv:1508.06576. [Google Scholar] [CrossRef]
- Gatys, L.A.; Ecker, A.S.; Bethge, M. Image style transfer using convolutional neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2016, Las Vegas, NV, USA, 27–30 June 2016; pp. 2414–2423. [Google Scholar]
- Alshehri, A.; Taileb, M.; Alotaibi, R. DeepAIA: An Automatic Image Annotation Model based on Generative Adversarial Networks and Transfer Learning. IEEE Access 2022, 10, 38437–38445. [Google Scholar] [CrossRef]
- Khvostikov, A.; Aderghal, K.; Benois-Pineau, J.; Krylov, A.; Catheline, G. 3D CNN-based classification using sMRI and MD-DTI images for Alzheimer disease studies. arXiv 2018, arXiv:1801.05968. [Google Scholar]
- Mahmood, T.; Li, J.; Pei, Y.; Akhtar, F. An Automated In-Depth Feature Learning Algorithm for Breast Abnormality Prognosis and Robust Characterization from Mammography Images Using Deep Transfer Learning. Biology 2021, 10, 859. [Google Scholar] [CrossRef]
- Wang, P.; Wang, J.; Li, Y.; Li, P.; Li, L.; Jiang, M. Automatic classification of breast cancer histopathological images based on deep feature fusion and enhanced routing. Biomed. Signal. Process. Control. 2021, 65, 102341. [Google Scholar] [CrossRef]
- Zuiderveld, K. Contrast Limited Adaptive Histogram Equalization; ACM Digital Library: New York, NY, USA, 1994. [Google Scholar]
- Hassan, N.; Ullah, S.; Bhatti, N.; Mahmood, H.; Zia, M. The Retinex based improved underwater image enhancement. Multimed. Tools Appl. 2021, 80, 1839–1857. [Google Scholar] [CrossRef]
- Wang, G.; Li, W.; Aertsen, M.; Deprest, J.; Ourselin, S.; Vercauteren, T. Aleatoric uncertainty estimation with test-time augmentation for medical image segmentation with convolutional neural networks. Neurocomputing 2019, 338, 34–45. [Google Scholar] [CrossRef]
- Nalepa, J.; Marcinkiewicz, M.; Kawulok, M. Data Augmentation for Brain-Tumor Segmentation: A Review. Front. Comput. Neurosci. 2019, 13, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moshkov, N.; Mathe, B.; Kertesz-Farkas, A.; Hollandi, R.; Horvath, P. Test-time augmentation for deep learning-based cell segmentation on microscopy images. Sci. Rep. 2020, 10, 5068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amiri, M.; Brooks, R.; Behboodi, B.; Rivaz, H. Two-stage ultrasound image segmentation using U-Net and test time augmentation. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 981–988. [Google Scholar] [CrossRef]
- Shanmugam, D.; Blalock, D.; Balakrishnan, G.; Guttag, J. Better aggregation in test-time augmentation. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 1214–1223. [Google Scholar]
- Jha, D.; Smedsrud, P.H.; Johansen, D.; de Lange, T.; Johansen, H.D.; Halvorsen, P.; Riegler, M.A. A Comprehensive Study on Colorectal Polyp Segmentation with ResUNet++, Conditional Random Field and Test-Time Augmentation. IEEE J. Biomed. Health Inform. 2021, 25, 2029–2040. [Google Scholar] [CrossRef] [PubMed]
- Kandel, I.; Castelli, M. Improving convolutional neural networks performance for image classification using test time augmentation: A case study using MURA dataset. Health Inf. Sci. Syst. 2021, 9, 33. [Google Scholar] [CrossRef]
- Hoar, D.; Lee, P.Q.; Guida, A.; Patterson, S.; Bowen, C.V.; Merrimen, J.; Wang, C.; Rendon, R.; Beyea, S.D.; Clarke, S.E. Combined Transfer Learning and Test-Time Augmentation Improves Convolutional Neural Network-Based Semantic Segmentation of Prostate Cancer from Multi-Parametric MR Images. Comput. Methods Programs Biomed. 2021, 210, 106375. [Google Scholar] [CrossRef]
- Gonzalo-Martín, C.; García-Pedrero, A.; Lillo-Saavedra, M. Improving deep learning sorghum head detection through test time augmentation. Comput. Electron. Agric. 2021, 186, 106179. [Google Scholar] [CrossRef]
- Cohen, S.; Dagan, N.; Cohen-Inger, N.; Ofer, D.; Rokach, L. ICU Survival Prediction Incorporating Test-Time Augmentation to Improve the Accuracy of Ensemble-Based Models. IEEE Access 2021, 9, 91584–91592. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Menon, L.T.; Laurensi, I.A.; Penna, M.C.; Oliveira, L.E.S.; Britto, A.S. Data Augmentation and Transfer Learning Applied to Charcoal Image Classification. In Proceedings of the 2019 International Conference on Systems, Signals and Image Processing (IWSSIP), Osijek, Croatia, 5–7 June 2019; pp. 69–74. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the 2017 IEEE international conference on computer vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]













| Reference | Year | Classes | Disease | Data Type | Model | Performance Accuracy |
|---|---|---|---|---|---|---|
| [37] | 2021 | 4 | Normal, COVID-19, Pneumonia, Lung cancer | X-ray, CT | VGG19-CNN | 98.05% |
| ResNet152V2 | 95.31% | |||||
| ResNet152V2 + GRU | 96.09% | |||||
| ResNet152V2 + Bi-GRU | 93.36% | |||||
| [38] | 2020 | 15 | No finding, Infiltration, Mass, Effusion, Atelectasis, Nodule, Pneumothorax, Consolidation, Plural thickening, Hernia, Cardiomegaly, Emphysema, Edema, Fibrosis, Pneumonia | X-ray | Vanilla Gray | 67.8% |
| Vanilla RGB | 69% | |||||
| Hybrid CNN VGG | 69.5% | |||||
| VDSNet | 73% | |||||
| Modified CapsNet | 63.8% | |||||
| Basic CapsNet | 60.5% | |||||
| [39] | 2021 | 3 | Normal, Pneumonia, COVID-19 | X-ray | CNN + HOG | 92.95% |
| [40] | 2020 | 10 | Infiltration, Mass, Effusion, Atelectasis, Nodule, Pneumothorax, Consolidation, Emphysema, Edema, Fibrosis | X-ray | UCMobN | 85.62% |
| [41] | 2021 | 2 | Pleural effusion, Normal | X-ray | CNN | 95% |
| [42] | 2021 | 3 | Normal, COVID-19, SARS | X-ray | DeTrac + AlexNet | 95.66% |
| DeTrac + VGG19 | 97.53% | |||||
| DeTrac + ResNet | 95.66% | |||||
| DeTrac + GoogleNet | 94.71% | |||||
| DeTrac + SqueezNet | 94.90% | |||||
| [43] | 2021 | 14 | Atelectasis, Cardiomegaly, Consolidation, Edema, Mass, Effusion, Emphysema, Nodule, Pneumothorax, Fibrosis, Infiltration, Plural thickening, Pneumonia, Normal | X-ray | DenseNet121 | 40.43% |
| InceptionResNetV2 | 41.02% | |||||
| ResNet152V2 | 37.60% | |||||
| [44] | 2021 | 2 | COVID-19, Not-COVID-19 | X-ray | Mask R-CNN | 96.98% |
| [45] | 2021 | 3 | No finding, COVID-19, Pneumonia | X-ray | CNN | 87% |
| [46] | 2020 | 2 | Normal, Pneumonia | X-ray | VGG16 | 96.81% |
| VGG19 | 96.58% | |||||
| NasNetMobile | 83.37% | |||||
| ResNet152V2 | 96.35% | |||||
| InceptionResNetV2 | 94.87% | |||||
| [47] | 2021 | 2 | Normal, COVID-19 | X-ray | INceptionV3 | 90.43% |
| ResNet50V2 | 91.11% | |||||
| DenseNet201 | 91.11% | |||||
| Combined Proposed Model | 91.62% | |||||
| [48] | 2021 | 4 | COVID-19 (Mild), COVID-19 (Moderate), COVID-19 (Severe), COVID-19 (Critical) | X-ray | Novel CNN | 95.52% |
| [49] | 2020 | 2 | Normal, COVID-19 | X-ray | DNN | 83.4% |
| CNN | 93.2% | |||||
| [50] | 2021 | 2 | Normal, COVID-19 | X-ray | ResNet50 + SVM | 94.7% |
| Fine-Tune ResNet50 | 92.6% | |||||
| End-to-End training of CNN | 91.6% | |||||
| [51] | 2022 | 4 | Nodules, Atelectasis, Infection, Normal | X-ray | MARnet | 83.3% |
| No. | Disease | Data | Reference |
|---|---|---|---|
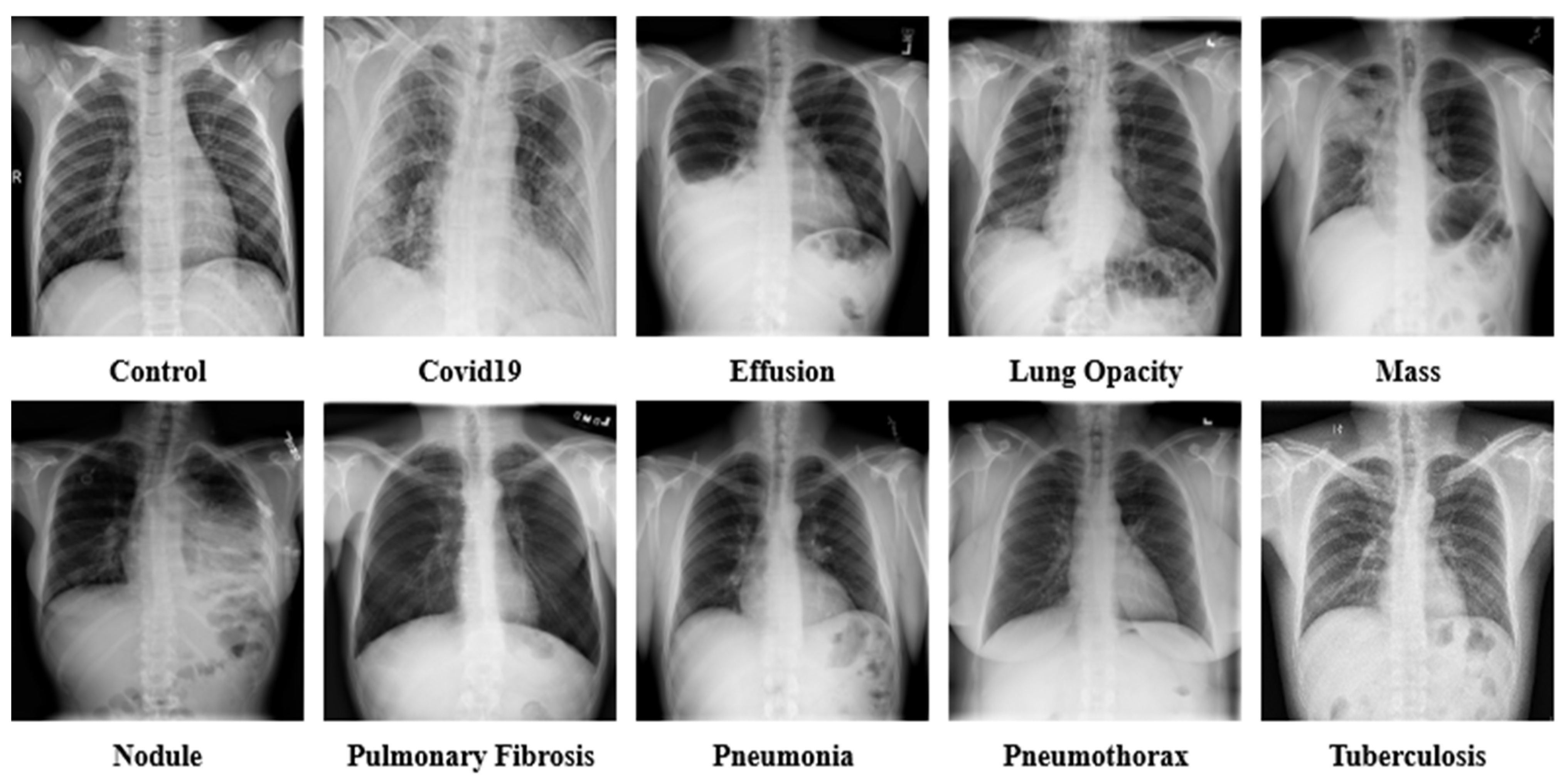
| 1. | Control | 13,672 | [21,22,23,26,27,28,31] |
| 2. | COVID-19 | 15,660 | [21,22,24,25,28,31] |
| 3. | Effusion | 13,501 | [30,32,33,34] |
| 4. | Lung Opacity | 7179 | [31,35] |
| 5. | Mass | 5603 | [30,33] |
| 6. | Nodule | 6201 | [30,33] |
| 7. | Pulmonary Fibrosis | 3357 | [28,30,33] |
| 8. | Pneumonia | 9878 | [21,22,23,28,31] |
| 9. | Pneumothorax | 6870 | [29,30,33] |
| 10. | Tuberculosis | 3184 | [24,26,27,28,36] |
| Filters | Val_Acc (%) | Val_Loss (%) | T_Acc (%) | T_Loss (%) | Finding |
|---|---|---|---|---|---|
| 5 Ramps | 63.88 | 0.89 | 64.32 | 0.87 | Accuracy Dropped |
| 6 Shades | 62.71 | 0.94 | 63.45 | 0.90 | Accuracy Dropped |
| 3-3-2 RGB | 65.33 | 0.86 | 66.01 | 0.83 | Accuracy Dropped |
| Blue Orange icb | 72.59 | 0.72 | 73.77 | 0.68 | Accuracy Dropped |
| 16 colors | 82.63 | 0.61 | 83.09 | 0.59 | Accuracy Dropped |
| Spectrum | 81.19 | 0.65 | 81.80 | 0.61 | Accuracy Dropped |
| Sepia | 85.40 | 0.49 | 86.90 | 0.39 | Accuracy Dropped |
| Cyan Hot | 85.67 | 0.45 | 86.11 | 0.41 | Accuracy Dropped |
| Edges | 83.55 | 0.57 | 83.99 | 0.53 | Accuracy Dropped |
| Fire | 87.92 | 0.37 | 87.47 | 0.40 | Accuracy Dropped |
| Green Fire Blue | 97.03 | 0.11 | 98.89 | 0.09 | Identical Accuracy |
| Phase | 88.76 | 0.25 | 88.23 | 0.29 | Accuracy Dropped |
| Jet | 83.52 | 0.58 | 83.85 | 0.54 | Accuracy Dropped |
| Thallium | 88.19 | 0.28 | 88.99 | 0.22 | Accuracy Dropped |
| Viridis | 95.10 | 0.19 | 95.50 | 0.16 | Accuracy Dropped |
| Augmentation Parameters | Value |
|---|---|
| Rotate left | 90° |
| Rotate right | 90° |
| Horizontal flip | True |
| Rotate | 45° |
| Rotate Horizontal | 45° |
| Rotate Vertical | 45° |
| Vertical flip | True |
| Translate | x, y (29.0, 14.0) |
| Features | Properties |
|---|---|
| Number of Images | 80,000 |
| Number of Classes | 10 |
| Enhancement and Color Grading | CLAHE, Green Fire Blue |
| Number of Augmentation Techniques | 8 |
| Control | 8000 |
| COVID-19 | 8000 |
| Effusion | 8000 |
| Lung Opacity | 8000 |
| Mass | 8000 |
| Nodule | 8000 |
| Pulmonary Fibrosis | 8000 |
| Pneumonia | 8000 |
| Pneumothorax | 8000 |
| Tuberculosis | 8000 |
| Case Study | Layer Name | Val_Acc (%) | Val_Loss (%) | T_Acc (%) | T_Loss (%) | Finding |
|---|---|---|---|---|---|---|
| 1 | GlobalAveragePooling2D | 97.03 | 0.11 | 98.89 | 0.09 | Identical Performance |
| GlobalMaxPooling2D | 95.73 | 0.27 | 96.12 | 0.23 | Accuracy Dropped | |
| AveragePooling2D | 94.92 | 0.35 | 95.30 | 0.27 | Accuracy Dropped | |
| MaxPooling2D | 95.77 | 0.21 | 96.19 | 0.25 | Accuracy Dropped |
| Case Study | Layer Name | Val_Acc (%) | Val_Loss (%) | T_Acc (%) | T_Loss (%) | Finding |
|---|---|---|---|---|---|---|
| 2 | Flatten | 97.03 | 0.11 | 98.89 | 0.09 | Identical Performance |
| GlobalAveragePooling2D | 96.12 | 0.25 | 96.94 | 0.22 | Accuracy Dropped | |
| GlobalMaxPooling2D | 90.73 | 0.67 | 91.12 | 0.58 | Accuracy Dropped | |
| AveragePooling2D | 92.95 | 0.53 | 93.21 | 0.47 | Accuracy Dropped | |
| MaxPooling2D | 94.78 | 0.40 | 95.16 | 0.36 | Accuracy Dropped |
| Case Study | Loss Function | Val_Acc (%) | Val_Loss (%) | T_Acc (%) | T_Loss (%) | Finding |
|---|---|---|---|---|---|---|
| 3 | Categorical Crossentropy | 97.03 | 0.11 | 98.89 | 0.09 | Identical Performance |
| Mean Squared Error | 93.77 | 0.67 | 94.20 | 0.58 | Accuracy Dropped | |
| Cosine Similarity | 95.82 | 0.41 | 96.19 | 0.37 | Accuracy Dropped |
| Case Study | Optimizer | Learning Rate | Val_Acc (%) | Val_Loss (%) | T_Acc (%) | T_Loss (%) | Finding |
|---|---|---|---|---|---|---|---|
| 4 | Adam | 0.000001 | 97.03 | 0.11 | 98.89 | 0.09 | Identical Performance |
| 0.0001 | 94.50 | 0.64 | 95.07 | 0.47 | Accuracy dropped | ||
| 0.00001 | 95.61 | 0.46 | 96.22 | 0.36 | Accuracy dropped | ||
| 0.001 | 93.35 | 0.69 | 93.98 | 0.65 | Accuracy dropped | ||
| 0.01 | 91.96 | 0.77 | 92.35 | 0.71 | Accuracy dropped | ||
| SGD | 0.000001 | 95.12 | 0.27 | 95.88 | 0.21 | Accuracy dropped | |
| 0.0001 | 94.83 | 0.65 | 95.15 | 0.60 | Accuracy dropped | ||
| 0.00001 | 94.97 | 0.63 | 95.70 | 0.58 | Accuracy dropped | ||
| 0.001 | 90.65 | 0.80 | 92.08 | 0.75 | Accuracy dropped | ||
| 0.01 | 92.57 | 0.72 | 93.75 | 0.64 | Accuracy dropped | ||
| RMSprop | 0.000001 | 94.57 | 0.64 | 94.90 | 0.49 | Accuracy dropped | |
| 0.0001 | 92.27 | 0.69 | 93.20 | 0.57 | Accuracy dropped | ||
| 0.00001 | 93.06 | 0.58 | 94.51 | 0.68 | Accuracy dropped | ||
| 0.001 | 92.13 | 0.72 | 93.90 | 0.51 | Accuracy dropped | ||
| 0.01 | 91.74 | 0.80 | 92.56 | 0.62 | Accuracy dropped | ||
| Nadam | 0.000001 | 95.06 | 0.25 | 95.89 | 0.23 | Accuracy dropped | |
| 0.0001 | 94.26 | 0.75 | 95.11 | 0.68 | Accuracy dropped | ||
| 0.00001 | 94.76 | 0.70 | 95.32 | 0.65 | Accuracy dropped | ||
| 0.001 | 93.44 | 0.72 | 94.81 | 0.70 | Accuracy dropped | ||
| 0.01 | 92.35 | 0.65 | 94.20 | 0.67 | Accuracy dropped |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shamrat, F.M.J.M.; Azam, S.; Karim, A.; Islam, R.; Tasnim, Z.; Ghosh, P.; De Boer, F. LungNet22: A Fine-Tuned Model for Multiclass Classification and Prediction of Lung Disease Using X-ray Images. J. Pers. Med. 2022, 12, 680. https://doi.org/10.3390/jpm12050680
Shamrat FMJM, Azam S, Karim A, Islam R, Tasnim Z, Ghosh P, De Boer F. LungNet22: A Fine-Tuned Model for Multiclass Classification and Prediction of Lung Disease Using X-ray Images. Journal of Personalized Medicine. 2022; 12(5):680. https://doi.org/10.3390/jpm12050680
Chicago/Turabian StyleShamrat, F. M. Javed Mehedi, Sami Azam, Asif Karim, Rakibul Islam, Zarrin Tasnim, Pronab Ghosh, and Friso De Boer. 2022. "LungNet22: A Fine-Tuned Model for Multiclass Classification and Prediction of Lung Disease Using X-ray Images" Journal of Personalized Medicine 12, no. 5: 680. https://doi.org/10.3390/jpm12050680
APA StyleShamrat, F. M. J. M., Azam, S., Karim, A., Islam, R., Tasnim, Z., Ghosh, P., & De Boer, F. (2022). LungNet22: A Fine-Tuned Model for Multiclass Classification and Prediction of Lung Disease Using X-ray Images. Journal of Personalized Medicine, 12(5), 680. https://doi.org/10.3390/jpm12050680