Predicting the Treatment Outcomes of Antidepressants Using a Deep Neural Network of Deep Learning in Drug-Naïve Major Depressive Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Measurements of Peripheral Biochemistry and Genotyping
2.2.1. Blood Lipid and Sugar Profile
2.2.2. Leptin
2.2.3. C-Reactive Protein
2.2.4. Oxytocin
2.2.5. SNP Determination and Genotyping
2.3. Questionnaires
2.3.1. World Health Organization Quality of Life (WHOQoL)
2.3.2. Social Support Scale
2.3.3. Life Event Scale
2.4. Cognitive Function
2.4.1. Finger-Tapping Test (FTT)
2.4.2. Continuous Performance Test (CPT)
2.4.3. Wisconsin Card Sorting Test (WCST)
2.5. Statistical Analysis
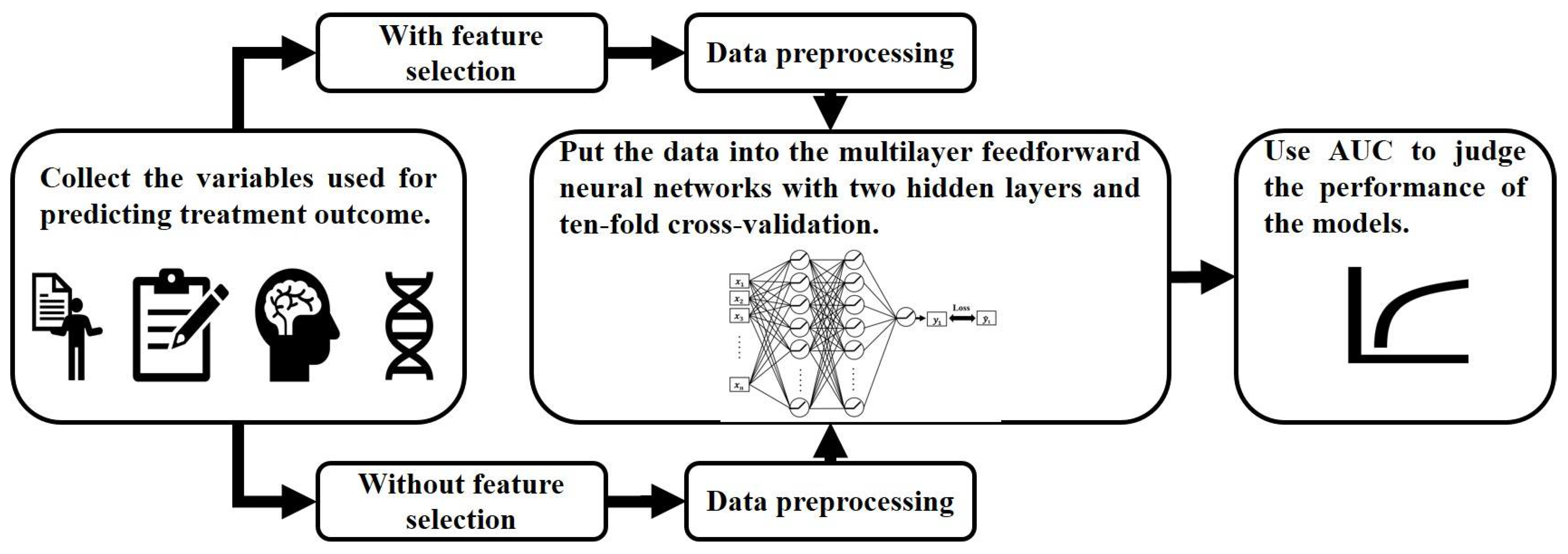
2.6. Machine Learning
2.6.1. Data Preprocessing and Feature Selection
2.6.2. Feedforward Neural Network Model
3. Results
3.1. Demographic Characteristics and Peripheral Biochemistry
3.2. Questionnaire Score
3.3. Cognitive Function
3.4. Genotype Frequencies of SNPs
3.5. The Performance of the Feedforward Neural Network Model in Predicting the Remission of Patients
3.5.1. Training Model without Feature Selection
3.5.2. Training Model after Feature Selection
4. Discussion
4.1. Oxytocin and Cortisol
4.2. Social Support Scale and Quality of Life
4.3. OXTR and Treatment Response
4.4. Antidepressant Treatment Response Prediction Model
4.5. Limitation of the Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Depression. 2021. Available online: https://www.who.int/news-room/fact-sheets/detail/depression (accessed on 1 December 2021).
- Maes, M.; Smith, R.; Christophe, A.; Vandoolaeghe, E.; Van Gastel, A.; Neels, H.; Demedts, P.; Wauters, A.; Meltzer, H.Y. Lower serum high-density lipoprotein cholesterol (HDL-C) in major depression and in depressed men with serious suicidal attempts: Relationship with immune-inflammatory markers. Acta Psychiatr. Scand. 1997, 95, 212–221. [Google Scholar] [CrossRef]
- Andrade, C. Relative efficacy and acceptability of antidepressant drugs in adults with major depressive disorder: Commentary on a network meta-analysis. J. Clin. Psychiatry 2018, 79, 3331. [Google Scholar] [CrossRef] [PubMed]
- Paul, R.; Andlauer, T.F.M.; Czamara, D.; Hoehn, D.; Lucae, S.; Pütz, B.; Lewis, C.M.; Uher, R.; Müller-Myhsok, B.; Ising, M.; et al. Treatment response classes in major depressive disorder identified by model-based clustering and validated by clinical prediction models. Transl. Psychiatry 2019, 9, 187. [Google Scholar] [CrossRef] [Green Version]
- Lin, E.; Kuo, P.H.; Liu, Y.L.; Yu, Y.W.; Yang, A.C.; Tsai, S.J. A deep learning approach for predicting antidepressant response in major depression using clinical and genetic biomarkers. Front. Psychiatry 2018, 9, 290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, E.; Kuo, P.H.; Liu, Y.L.; Yu, Y.W.; Yang, A.C.; Tsai, S.J. Prediction of antidepressant treatment response and remission using an ensemble machine learning framework. Pharmaceuticals 2020, 13, 305. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.; Stewart, R.; Kang, H.J.; Kim, J.W.; Kim, H.K.; Kang, H.C.; Lee, J.Y.; Kim, S.W.; Kim, J.M. Interactive effects of systemic inflammation and life stressors on treatment response of depressive disorders. Brain Behav. Immun. 2021, 95, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Grigolon, R.B.; Brietzke, E.; Mansur, R.B.; Idzikowski, M.A.; Gerchman, F.; De Felice, F.G.; McIntyre, R.S. Association between diabetes and mood disorders and the potential use of anti-hyperglycemic agents as antidepressants. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2019, 95, 109720. [Google Scholar] [CrossRef]
- Yuan, B.; Sun, X.; Xu, Z.; Pu, M.; Yuan, Y.; Zhang, Z. Influence of genetic polymorphisms in homocysteine and lipid metabolism systems on antidepressant drug response. BMC Psychiatry 2020, 20, 408. [Google Scholar] [CrossRef]
- Moradi, Y.; Albatineh, A.N.; Mahmoodi, H.; Gheshlagh, R.G. The relationship between depression and risk of metabolic syndrome: A meta-analysis of observational studies. Clin. Diabetes Endocrinol. 2021, 7, 4. [Google Scholar] [CrossRef]
- Uher, R.; Perroud, N.; Ng, M.Y.; Hauser, J.; Henigsberg, N.; Maier, W.; Mors, O.; Placentino, A.; Rietschel, M.; Souery, D.; et al. Genome-wide pharmacogenetics of antidepressant response in the GENDEP project. Am. J. Psychiatry 2010, 167, 555–564. [Google Scholar] [CrossRef]
- Garriock, H.A.; Kraft, J.B.; Shyn, S.I.; Peters, E.J.; Yokoyama, J.S.; Jenkins, G.D.; Reinalda, M.S.; Slager, S.L.; McGrath, P.J.; Hamilton, S.P. A genomewide association study of citalopram response in major depressive disorder. Biol. Psychiatry 2010, 67, 133–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ising, M.; Lucae, S.; Binder, E.B.; Bettecken, T.; Uhr, M.; Ripke, S.; Kohli, M.A.; Hennings, J.M.; Horstmann, S.; Kloiber, S.; et al. A genomewide association study points to multiple loci that predict antidepressant drug treatment outcome in depression. Arch. Gen. Psychiatry 2009, 66, 966–975. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, C.; Di Girolamo, G.; Serretti, A. Pharmacogenetics of antidepressant drugs: An update after almost 20 years of research. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2013, 162, 487–520. [Google Scholar] [CrossRef] [PubMed]
- Taliaz, D.; Spinrad, A.; Barzilay, R.; Barnett-Itzhaki, Z.; Averbuch, D.; Teltsh, O.; Schurr, R.; Darki-Morag, S.; Lerer, B. Optimizing prediction of response to antidepressant medications using machine learning and integrated genetic, clinical, and demographic data. Transl. Psychiatry 2021, 11, 381. [Google Scholar] [CrossRef] [PubMed]
- So, H.C.; Chau, C.K.; Lau, A.; Wong, S.Y.; Zhao, K. Translating GWAS findings into therapies for depression and anxiety disorders: Gene-set analyses reveal enrichment of psychiatric drug classes and implications for drug repositioning. Psychol. Med. 2019, 49, 2692–2708. [Google Scholar] [CrossRef] [PubMed]
- Iniesta, R.; Hodgson, K.; Stahl, D.; Malki, K.; Maier, W.; Rietschel, M.; Mors, O.; Hauser, J.; Henigsberg, N.; Dernovsek, M.Z.; et al. Antidepressant drug-specific prediction of depression treatment outcomes from genetic and clinical variables. Sci. Rep. 2018, 8, 5530. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Ren, D.; Guo, Z.; Ma, G.; Xu, F.; Chen, Z.; An, L.; Zhang, N.; Ji, L.; Yuan, F.; et al. Influence and interaction of genetic, cognitive, neuroendocrine and personalistic markers to antidepressant response in Chinese patients with major depression. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2021, 104, 110036. [Google Scholar] [CrossRef]
- Kautzky, A.; Möller, H.J.; Dold, M.; Bartova, L.; Seemüller, F.; Laux, G.; Riedel, M.; Gaebel, W.; Kasper, S. Combining machine learning algorithms for prediction of antidepressant treatment response. Acta Psychiatr. Scand. 2021, 143, 36–49. [Google Scholar] [CrossRef]
- Su, C.; Xu, Z.; Pathak, J.; Wang, F. Deep learning in mental health outcome research: A scoping review. Transl. Psychiatry 2020, 10, 116. [Google Scholar] [CrossRef]
- Joyce, J.B.; Grant, C.W.; Liu, D.; MahmoudianDehkordi, S.; Kaddurah-Daouk, R.; Skime, M.; Biernacka, J.; Frye, M.A.; Mayes, T.; Carmody, T.; et al. Multi-omics driven predictions of response to acute phase combination antidepressant therapy: A machine learning approach with cross-trial replication. Transl. Psychiatry 2021, 11, 513. [Google Scholar] [CrossRef]
- Chang, H.H.; Lee, I.H.; Gean, P.W.; Lee, S.-Y.; Chi, M.H.; Yang, Y.K.; Lu, R.-B.; Chen, P.S. Treatment response and cognitive impairment in major depression: Association with C-reactive protein. Brain Behav. Immun. 2012, 26, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Matthews, D.R.; Hosker, J.P.; Rudenski, A.S.; Naylor, B.A.; Treacher, D.F.; Turner, R.C. Homeostasis model assessment: Insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985, 28, 412–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chi, M.-H.; Chang, H.H.; Lee, S.-Y.; Lee, I.H.; Gean, P.-W.; Yang, Y.K.; Chen, P.S.; Lu, R.-B. Brain derived neurotrophic factor gene polymorphism (Val66Met) and short-term treatment response to fluoxetine or venlafaxine in major depressive disorder. In Proceedings of the International College of Neuropsychopharmacology 27th Biennial Meeting, Hong Kong, 2010; Available online: https://www.cinp.org/Past-Events (accessed on 6 June 2010).
- Wang, H.-C.; Yeh, T.L.; Chang, H.H.; Gean, P.W.; Chi, M.H.; Yang, Y.K.; Lu, R.-B.; Chen, P.S. TPH1 is associated with major depressive disorder but not with SSRI/SNRI response in taiwanese patients. Psychopharmacology 2011, 213, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Lin, E.; Chen, P.S.; Chang, H.H.; Gean, P.-W.; Tsai, H.C.; Yang, Y.K.; Lu, R.-B. Interaction of serotonin-related genes affects short-term antidepressant response in major depressive disorder. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2009, 33, 1167–1172. [Google Scholar] [CrossRef]
- Chang, H.H.; Chang, W.-H.; Chen, P.S. Association between the polymorphisms of OXTR and HbA1c in major depressive patients treated with antidepressant. In Proceedings of the 17th World Congress of Basic and Clinical Pharmacology, Cape Town, South Africa, 13 July 2014. [Google Scholar]
- Yao, G.; Chung, C.W.; Yu, C.F.; Wang, J.D. Development and verification of validity and reliability of the WHOQOL-BREF Taiwan version. J. Formos Med. Assoc. 2002, 101, 342–351. [Google Scholar]
- Lee, T.C.; Yang, Y.K.; Chen, P.S.; Hung, N.C.; Lin, S.H.; Chang, F.L.; Cheng, S.H. Different dimensions of social support for the caregivers of patients with schizophrenia: Main effect and stress-buffering models. Psychiatry Clin. Neurosci. 2006, 60, 546–550. [Google Scholar] [CrossRef]
- Miller, M.A.; Rahe, R.H. Life changes scaling for the 1990s. J. Psychosom. Res. 1997, 43, 279–292. [Google Scholar] [CrossRef]
- Pai, L.; Wen, H.; Lu, R.; Kuo, M. The questionnaire of life stress cognition of adults-revised (full text in Chinese). Chin. J. Ment. Health 1985, 3, 195–205. [Google Scholar]
- Taylor, B.P.; Bruder, G.E.; Stewart, J.W.; McGrath, P.J.; Halperin, J.; Ehrlichman, H.; Quitkin, F.M. Psychomotor slowing as a predictor of fluoxetine nonresponse in depressed outpatients. Am. J. Psychiatry 2006, 163, 73–78. [Google Scholar] [CrossRef] [Green Version]
- Etkin, A.; Patenaude, B.; Song, Y.J.C.; Usherwood, T.; Rekshan, W.; Schatzberg, A.F.; Rush, A.J.; Williams, L.M. A cognitive-emotional biomarker for predicting remission with antidepressant medications: A report from the iSPOT-D trial. Neuropsychopharmacology 2015, 40, 1332–1342. [Google Scholar] [CrossRef] [Green Version]
- Dunkin, J.J.; Leuchter, A.F.; Cook, I.A.; Kasl-Godley, J.E.; Abrams, M.; Rosenberg-Thompson, S. Executive dysfunction predicts nonresponse to fluoxetine in major depression. J. Affect. Disord. 2000, 60, 13–23. [Google Scholar] [CrossRef]
- Gudayol-Ferré, E.; Herrera-Guzmán, I.; Camarena, B.; Cortés-Penagos, C.; Herrera-Abarca, J.E.; Martínez-Medina, P.; Asbun-Bojalil, J.; Lira-Islas, Y.; Reyes-Ponce, C.; Guàrdia-Olmos, J. Prediction of remission of depression with clinical variables, neuropsychological performance, and serotonergic/dopaminergic gene polymorphisms. Hum. Psychopharmacol. 2012, 27, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Shimoyama, I.; Ninchoji, T.; Uemura, K. The finger-tapping test: A quantitative analysis. Arch. Neurol. 1990, 47, 681–684. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.J.; Hsiao, C.K.; Hsiao, L.L.; Hwu, H.G. Performance of the continuous performance test among community samples. Schizophr. Bull. 1998, 24, 163–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsieh, P.C.; Chu, C.L.; Yang, Y.K.; Yang, Y.C.; Yeh, T.L.; Lee, I.H.; Chen, P.S. Norms of performance of sustained attention among a community sample: Continuous performance test study. Psychiatry Clin. Neurosci. 2005, 59, 170–176. [Google Scholar] [CrossRef]
- Smid, H.G.O.M.; de Witte, M.R.; Homminga, I.; Van Den Bosch, R.J. Sustained and transient attention in the continuous performance task. J. Clin. Exp. Neuropsychol. 2006, 28, 859–883. [Google Scholar] [CrossRef]
- Heaton, R.K.; Staff, P. Wisconsin card sorting test: Computer version 2. Odessa Psychol. Assess. Resour. 1993, 4, 1–4. [Google Scholar]
- Stratta, P.; Daneluzzo, E.; Prosperini, P.; Bustini, M.; Mattei, P.; Rossi, A. Is wisconsin card sorting test performance related to ‘working memory’ capacity? Schizophr. Res. 1997, 27, 11–19. [Google Scholar] [CrossRef]
- Volkow, N.D.; Gur, R.C.; Wang, G.J.; Fowler, J.S.; Moberg, P.J.; Ding, Y.S.; Hitzemann, R.; Smith, G.; Logan, J. Association between decline in brain dopamine activity with age and cognitive and motor impairment in healthy individuals. Am. J. Psychiatry 1998, 155, 344–349. [Google Scholar]
- Mandrekar, J.N. Receiver operating characteristic curve in diagnostic test assessment. J. Thorac. Oncol. 2010, 5, 1315–1316. [Google Scholar] [CrossRef] [Green Version]
- Gold, P.W.; Chrousos, G.P. Organization of the stress system and its dysregulation in melancholic and atypical depression: High vs low CRH/NE states. Mol. Psychiatry 2002, 7, 254–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keating, C.; Dawood, T.; Barton, D.A.; Lambert, G.W.; Tilbrook, A.J. Effects of selective serotonin reuptake inhibitor treatment on plasma oxytocin and cortisol in major depressive disorder. BMC Psychiatry 2013, 13, 124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gimpl, G.; Fahrenholz, F. The oxytocin receptor system: Structure, function, and regulation. Physiol. Rev. 2001, 81, 629–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slattery, D.A.; Neumann, I.D. Oxytocin and major depressive disorder: Experimental and clinical evidence for links to aetiology and possible treatment. Pharmaceuticals 2010, 3, 702–724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nandam, L.S.; Brazel, M.; Zhou, M.; Jhaveri, D.J. Cortisol and Major Depressive Disorder-Translating Findings from Humans to Animal Models and Back. Front. Psychiatry 2019, 10, 974. [Google Scholar] [CrossRef] [PubMed]
- Jain, F.A.; Connolly, C.G.; Reus, V.I.; Meyerhoff, D.J.; Yang, T.T.; Mellon, S.H.; Mackin, S.; Hough, C.M.; Morford, A.; Wolkowitz, O.M. Cortisol, moderated by age, is associated with antidepressant treatment outcome and memory improvement in Major Depressive Disorder: A retrospective analysis. Psychoneuroendocrinology 2019, 109, 104386. [Google Scholar] [CrossRef]
- Slavich, G.M.; Irwin, M.R. From stress to inflammation and major depressive disorder: A social signal transduction theory of depression. Psychol. Bull. 2014, 140, 774–815. [Google Scholar] [CrossRef]
- Crnković, D.; Buljan, D.; Karlović, D.; Krmek, M. Connection between inflammatory markers, antidepressants and depression. Acta Clin. Croat. 2012, 51, 25–33. [Google Scholar]
- Strawbridge, R.; Arnone, D.; Danese, A.; Papadopoulos, A.; Herane Vives, A.; Cleare, A.J. Inflammation and clinical response to treatment in depression: A meta-analysis. Eur. Neuropsychopharmacol. 2015, 25, 1532–1543. [Google Scholar] [CrossRef]
- Wang, J.; Mann, F.; Lloyd-Evans, B.; Ma, R.; Johnson, S. Associations between loneliness and perceived social support and outcomes of mental health problems: A systematic review. BMC Psychiatry 2018, 18, 156. [Google Scholar] [CrossRef] [Green Version]
- Tsai, T.Y.; Tseng, H.H.; Chi, M.H.; Chang, H.H.; Wu, C.K.; Yang, Y.K.; Chen, P.S. The interaction of oxytocin and social support, loneliness, and cortisol level in major depression. Clin. Psychopharmacol. Neurosci. 2019, 17, 487–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicolucci, A.; Haxhi, J.; D’Errico, V.; Sacchetti, M.; Orlando, G.; Cardelli, P.; Vitale, M.; Bollanti, L.; Conti, F.; Zanuso, S. Effect of a behavioural intervention for adoption and maintenance of a physically active lifestyle on psychological well-being and quality of life in patients with type 2 diabetes: The ides_2 randomized clinical trial. Sports Med. 2021, 52, 643–654. [Google Scholar] [CrossRef] [PubMed]
- Tang, A.L.; Thomas, S.J.; Larkin, T. Cortisol, oxytocin, and quality of life in major depressive disorder. Qual. Life Res. 2019, 28, 2919–2928. [Google Scholar] [CrossRef] [PubMed]
- IsHak, W.W.; Greenberg, J.M.; Balayan, K.; Kapitanski, N.; Jeffrey, J.; Fathy, H.; Fakhry, H.; Rapaport, M.H. Quality of life: The ultimate outcome measure of interventions in major depressive disorder. Harv. Rev. Psychiatry 2011, 19, 229–239. [Google Scholar] [CrossRef]
- Lee, K.W.; Ching, S.M.; Ramachandran, V.; Tusimin, M.; Mohd Nordin, N.; Chong, S.C.; Hoo, F.K. Association analysis of 14 candidate gene polymorphism with depression and stress among gestational diabetes mellitus. Genes 2019, 10, 988. [Google Scholar] [CrossRef] [Green Version]
- Chang, W.H.; Lee, I.H.; Chen, K.C.; Chi, M.H.; Chiu, N.T.; Yao, W.J.; Lu, R.B.; Yang, Y.K.; Chen, P.S. Oxytocin receptor gene rs53576 polymorphism modulates oxytocin-dopamine interaction and neuroticism traits--a SPECT study. Psychoneuroendocrinology 2014, 47, 212–220. [Google Scholar] [CrossRef]
- Wang, J.; Qin, W.; Liu, B.; Zhou, Y.; Wang, D.; Zhang, Y.; Jiang, T.; Yu, C. Neural mechanisms of oxytocin receptor gene mediating anxiety-related temperament. Brain Struct. Funct. 2014, 219, 1543–1554. [Google Scholar] [CrossRef]
- Feldman, R.; Monakhov, M.; Pratt, M.; Ebstein, R.P. Oxytocin pathway genes: Evolutionary ancient system impacting on human affiliation, sociality, and psychopathology. Biol. Psychiatry 2016, 79, 174–184. [Google Scholar] [CrossRef]
- Gao, S.; Calhoun, V.D.; Sui, J. Machine learning in major depression: From classification to treatment outcome prediction. CNS Neurosci. Ther. 2018, 24, 1037–1052. [Google Scholar] [CrossRef] [Green Version]
- Xue, L.; Pei, C.; Wang, X.; Wang, H.; Tian, S.; Yao, Z.; Lu, Q. Predicting neuroimaging biomarkers for antidepressant selection in early treatment of depression. J. Magn. Reson. Imaging 2021, 54, 551–559. [Google Scholar] [CrossRef]
- Zhao, W. Research on the deep learning of the small sample data based on transfer learning. In AIP Conference Proceedings; AIP Publishing LLC: Melville, NY, USA, 2017; Volume 1864, p. 020018. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Park, Y.; Ho, J.C. Tackling overfitting in boosting for noisy healthcare data. IEEE Trans. Knowl. Data Eng. 2021, 33, 2995–3006. [Google Scholar] [CrossRef]
- Sun, Q.; Liu, Y.; Chua, T.-S.; Schiele, B. Meta-transfer learning for few-shot learning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 403–412. Available online: https://www.computer.org/csdl/proceedings/cvpr/2019/1gyr6w5YIIU (accessed on 29 March 2022).
- Xu, Y.; Huang, H.; Heidari, A.A.; Gui, W.; Ye, X.; Chen, Y.; Chen, H.; Pan, Z.M. Feature: Towards high performance evolutionary tools for feature selection. Expert Syst. Appl. 2021, 186, 115655. [Google Scholar] [CrossRef]
- Tang, J.; Alelyani, S.; Liu, H. Feature selection for classification: A review. Data Classif. Algorithms Appl. 2014, 37, 1–29. [Google Scholar]
- Leeuwenberg, A.; van Smeden, M.; Langendijk, J.; Schaaf, A.; Mauer, M.; Moons, K.; Reitsma, J.; Schuit, E. Comparing methods addressing multi-collinearity when developing prediction models. arXiv 2021, arXiv:2101.01603. [Google Scholar]
- Howley, T.; Madden, M.G.; O’Connell, M.-L.; Ryder, A.G. The effect of principal component analysis on machine learning accuracy with high-dimensional spectral data. Knowl.-Based Syst. 2006, 19, 363–370. [Google Scholar] [CrossRef] [Green Version]
- Cai, Y.; Chow, M.Y.; Lu, W.; Li, L. Statistical feature selection from massive data in distribution fault diagnosis. IEEE Trans. Power Syst. 2010, 25, 642–648. [Google Scholar] [CrossRef]
- Sharma, A.; Paliwal, K.K. Linear discriminant analysis for the small sample size problem: An overview. Int. J. Mach. Learn. Cybern. 2015, 6, 443–454. [Google Scholar] [CrossRef] [Green Version]

| Characteristics | Remission | Nonremission | Comparison | |
|---|---|---|---|---|
| (N = 25) | (N = 45) | |||
| Mean ± SD | Mean ± SD | t/U/χ2 | p | |
| Clinical features | ||||
| Age, years | 40.8 ± 15.6 | 39.1 ± 12.0 | 536.5 | 0.754 |
| Gender, male (%) | 36.0% | 22.2% | 0.925 | 0.336 |
| HDRS scores of baseline | 22.9 ± 5.4 | 24.5 ± 5.6 | 1.184 | 0.242 |
| Peripheral biochemistry | ||||
| BH, cm | 161.1 ± 7.2 | 160.2 ± 8.0 | 420.5 | 0.360 |
| BW, kg | 58.98 ± 11.56 | 55.33 ± 11.99 | 402.0 | 0.242 |
| BMI, kg/m2 | 22.69 ± 3.95 | 21.48 ± 3.75 | −1.226 | 0.226 |
| SBP, mmHg | 117.6 ± 19.2 | 113.2 ± 16.8 | −0.952 | 0.346 |
| DBP, mmHg | 76.4 ± 9.6 | 74.2 ± 9.7 | −0.921 | 0.361 |
| Sugar profiles | ||||
| AC sugar, mg/dL | 96.1 ± 12.7 | 92.2 ± 11.7 | 428.5 | 0.204 |
| Insulin, µIn/mL | 6.77 ± 6.97 | 7.87 ± 9.44 | 611.5 | 0.446 |
| HbA1c (%) | 5.67 ± 0.44 | 5.56 ± 0.30 | 442.0 | 0.398 |
| HOMA-IR | 1.72 ± 1.90 | 1.95 ± 2.72 | 549.0 | 0.794 |
| HOMA-β (%) | 76.83 ± 69.15 | 88.79 ± 68.51 | 615.0 | 0.269 |
| Lipid profiles | ||||
| Cholesterol, mg/dL | 202.88 ± 45.84 | 189.02 ± 40.78 | 416.5 | 0.154 |
| TG, mg/dL | 113.83 ± 65.07 | 103.77 ± 70.81 | 458.5 | 0.376 |
| HDL, mg/dL | 56.57 ± 13.54 | 55.55 ± 14.05 | 505.5 | 1.000 |
| LDL, mg/dL | 126.35 ± 41.94 | 112.27 ± 38.83 | 413.5 | 0.224 |
| LDL/HDL | 2.32 ± 0.86 | 2.19 ± 1.07 | 423.0 | 0.276 |
| Other biochemical indices | ||||
| C-peptide, ng/mL | 1.98 ± 1.30 | 1.92 ± 1.83 | 484.5 | 0.416 |
| Cortisol, µg/dL | 17.2 ± 6.4 | 13.0 ± 6.8 | 348.0 | 0.011 * |
| Leptin, ng/mL | 8.78 ± 6.89 | 10.88 ± 14.36 | 480.0 | 0.388 |
| Oxytocin, pg/mL | 35.9 ± 25.4 | 26.5 ± 11.7 | 448.0 | 0.039 * |
| hsCRP, pg/mL | 287,440.0 ± 311,763.7 | 261,172.3 ± 357,027.6 | 511.0 | 0.721 |
| Questionnaire | Remission | Nonremission | Comparison | |
|---|---|---|---|---|
| (N = 25) | (N = 45) | |||
| Mean ± SD | Mean ± SD | t/U | p | |
| WHOQoL | ||||
| Overall | 5.6 ± 1.3 | 4.5 ± 1.7 | 238.5 | 0.015 * |
| Physical health | 18.7 ± 3.8 | 15.6 ± 5.3 | 213.5 | 0.005 * |
| Psychological | 15.0 ± 3.2 | 14.0 ± 4.1 | −0.909 | 0.368 |
| Social relationship | 13.6 ± 3.5 | 12.4 ± 3.6 | 308.0 | 0.207 |
| Environment | 34.1 ± 6.3 | 31.1 ± 5.8 | −1.857 | 0.070 |
| Social support scale | ||||
| Perceived crisis social support | 24.6 ± 4.6 | 20.6 ± 6.0 | 196.0 | 0.026 * |
| Received crisis social support | 30.3 ± 4.6 | 24.5 ± 7.7 | −3.476 | 0.001 * |
| Perceived routine social support | 23.4 ± 5.4 | 19.8 ± 6.6 | 229.0 | 0.047 * |
| Received routine social support | 26.5 ± 4.7 | 21.5 ± 6.5 | 190.5 | 0.007 * |
| Life event score | ||||
| Total score | 9.5 ± 8.0 | 10.6 ± 10.6 | 329.5 | 1.000 |
| Cognitive Function | Remission | Nonremission | Comparison | |
|---|---|---|---|---|
| (N = 25) | (N = 45) | |||
| Mean ± SD | Mean ± SD | U | p | |
| Finger-Tapping Test | ||||
| Dominant finger | 38.4 ± 11.1 | 36.8 ± 11.4 | 412.5 | 0.584 |
| Nondominant finger | 38.0 ± 11.2 | 35.5 ± 8.2 | 398.0 | 0.364 |
| Wisconsin Card-Sorting Test | ||||
| Perseverative errors | 18.8 ± 14.5 | 16.0 ± 12.6 | 436.0 | 0.718 |
| Completed categories | 1.3 ± 1.6 | 1.9 ± 1.6 | 568.0 | 0.125 |
| Continuous Performance test | ||||
| Unmasked | 3.83 ± 1.38 | 3.73 ± 1.08 | 367.0 | 0.226 |
| Masked | 3.07 ± 1.46 | 2.74 ± 1.26 | 328.0 | 0.264 |
| SNP | Related Gene | Chromosome | Reference Allele | Remission | Non-Remission | Comparison | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| (N = 25) | (N = 45) | |||||||||
| % | % | p | ||||||||
| rs6265 | BDNF | 11 | C | CC | CT | TT | CC | CT | TT | 0.772 |
| 40.0 | 32.0 | 28.0 | 37.8 | 40.0 | 22.2 | |||||
| rs5443 | GNB3 | 12 | C | CC | CT | TT | CC | CT | TT | 0.459 |
| 28.0 | 40.0 | 32.00 | 15.6 | 46.7 | 37.7 | |||||
| rs6313 | HTR2A | 13 | G | AA | AG | GG | AA | AG | GG | 0.949 |
| 32.0 | 48.0 | 20.0 | 35.6 | 44.4 | 20.0 | |||||
| rs6295 | HTR1A | 5 | G | CC | CG | GG | CC | CG | GG | 0.828 |
| 8.0 | 36.0 | 56.0 | 8.9 | 28.9 | 62.2 | |||||
| rs16944 | IL1B | 2 | A | AA | AG | GG | AA | AG | GG | 0.446 |
| 12.0 | 36.0 | 52.0 | 24.4 | 33.3 | 42.3 | |||||
| rs1800532 | TPH1 | 11 | G | TT | GT | GG | TT | GT | GG | 0.143 |
| 36.0 | 36.0 | 28.0 | 15.6 | 51.1 | 33.3 | |||||
| rs25533 | SLC6A4 | 17 | A | AA | AG | GG | AA | AG | GG | 0.302 |
| 76.0 | 24.0 | 0.0 | 71.1 | 20.0 | 8.9 | |||||
| rs53576 | OXTR | 3 | G | AA | AG | GG | AA | AG | GG | 0.014 * |
| 40.0 | 36.0 | 24.0 | 53.3 | 44.4 | 2.3 | |||||
| Model (No.) | Number of Markers | Accuracy (Mean ± SD) | AUC (Mean ± SD) |
|---|---|---|---|
| Age, sex, HDRS, clinical and peripheral biochemistry (1) | 23 | 64.286 ± 7.143% | 0.690 ± 0.281 |
| Age, sex, HDRS, questionnaire (2) | 15 | 64.286 ± 7.143% | 0.770 ± 0.154 |
| Age, sex, HDRS, cognitive function (3) | 9 | 64.286 ± 7.143% | 0.700 ± 0.152 |
| Age, sex, HDRS, SNP (4) | 11 | 65.714 ± 6.999% | 0.612 ± 0.177 |
| Age, sex, HDRS, clinical and peripheral biochemistry, questionnaire (5) | 35 | 70.000 ± 10.000% | 0.722 ± 0.160 |
| Age, sex, HDRS, clinical and peripheral biochemistry, cognitive function (6) | 29 | 65.714 ± 9.476% | 0.698 ± 0.238 |
| Age, sex, HDRS, clinical and peripheral biochemistry, SNP (7) | 31 | 62.857 ± 6.999% | 0.650 ± 0.203 |
| Age, sex, HDRS, questionnaire, cognitive function (8) | 21 | 64.286 ± 7.143% | 0.762 ± 0.184 |
| Age, sex, HDRS, questionnaire, SNP (9) | 23 | 67.143 ± 11.158% | 0.717 ± 0.123 |
| Age, sex, HDRS, cognitive function, SNP (10) | 17 | 64.286 ± 9.583% | 0.662 ± 0.188 |
| Age, sex, HDRS, clinical and peripheral biochemistry, questionnaire, cognitive function (11) | 41 | 67.143 ± 9.147% | 0.737 ± 0.232 |
| Age, sex, HDRS, questionnaire, cognitive function, SNP (12) | 29 | 65.714 ± 13.093% | 0.720 ± 0.195 |
| Age, sex, HDRS, clinical and peripheral biochemistry, cognitive function, SNP (13) | 37 | 70.000 ± 10.000% | 0.633 ± 0.243 |
| Age, sex, HDRS, clinical and peripheral biochemistry, questionnaire, SNP (14) | 43 | 67.143 ± 12.857% | 0.692 ± 0.163 |
| Age, sex, HDRS, clinical and peripheral biochemistry, questionnaire, cognitive function, SNP (15) | 49 | 68.571 ± 10.690% | 0.753 ± 0.154 |
| Model (No.) | Number of Markers | Accuracy (Mean ± SD) | AUC (Mean ± SD) |
|---|---|---|---|
| Age, sex, HDRS, clinical and peripheral biochemistry (1S) | 4 | 64.286 ± 7.143% | 0.707 ± 0.201 |
| Age, sex, HDRS, questionnaire (2S) | 11 | 62.857 ± 6.998% | 0.763 ± 0.124 |
| Age, sex, HDRS, SNP (3S) | 4 | 64.286 ± 7.143% | 0.757 ± 0.199 |
| Age, sex, HDRS, clinical and peripheral biochemistry, questionnaire (4S) | 13 | 64.286 ± 7.143% | 0.815 ± 0.184 |
| Age, sex, HDRS, clinical and peripheral biochemistry, SNP (5S) | 5 | 67.143 ± 9.147% | 0.763 ± 0.196 |
| Age, sex, HDRS, questionnaire, SNP (6S) | 12 | 65.714 ± 11.429% | 0.815 ± 0.137 |
| Age, sex, HDRS, clinical and peripheral biochemistry, questionnaire, SNP (7S) | 13 | 68.571 ± 12.454% | 0.825 ± 0.109 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsai, P.-L.; Chang, H.H.; Chen, P.S. Predicting the Treatment Outcomes of Antidepressants Using a Deep Neural Network of Deep Learning in Drug-Naïve Major Depressive Patients. J. Pers. Med. 2022, 12, 693. https://doi.org/10.3390/jpm12050693
Tsai P-L, Chang HH, Chen PS. Predicting the Treatment Outcomes of Antidepressants Using a Deep Neural Network of Deep Learning in Drug-Naïve Major Depressive Patients. Journal of Personalized Medicine. 2022; 12(5):693. https://doi.org/10.3390/jpm12050693
Chicago/Turabian StyleTsai, Ping-Lin, Hui Hua Chang, and Po See Chen. 2022. "Predicting the Treatment Outcomes of Antidepressants Using a Deep Neural Network of Deep Learning in Drug-Naïve Major Depressive Patients" Journal of Personalized Medicine 12, no. 5: 693. https://doi.org/10.3390/jpm12050693
APA StyleTsai, P.-L., Chang, H. H., & Chen, P. S. (2022). Predicting the Treatment Outcomes of Antidepressants Using a Deep Neural Network of Deep Learning in Drug-Naïve Major Depressive Patients. Journal of Personalized Medicine, 12(5), 693. https://doi.org/10.3390/jpm12050693








