Application of Patient-Derived Cancer Organoids to Personalized Medicine
Abstract
:1. Introduction
2. Three-Dimensional Culture
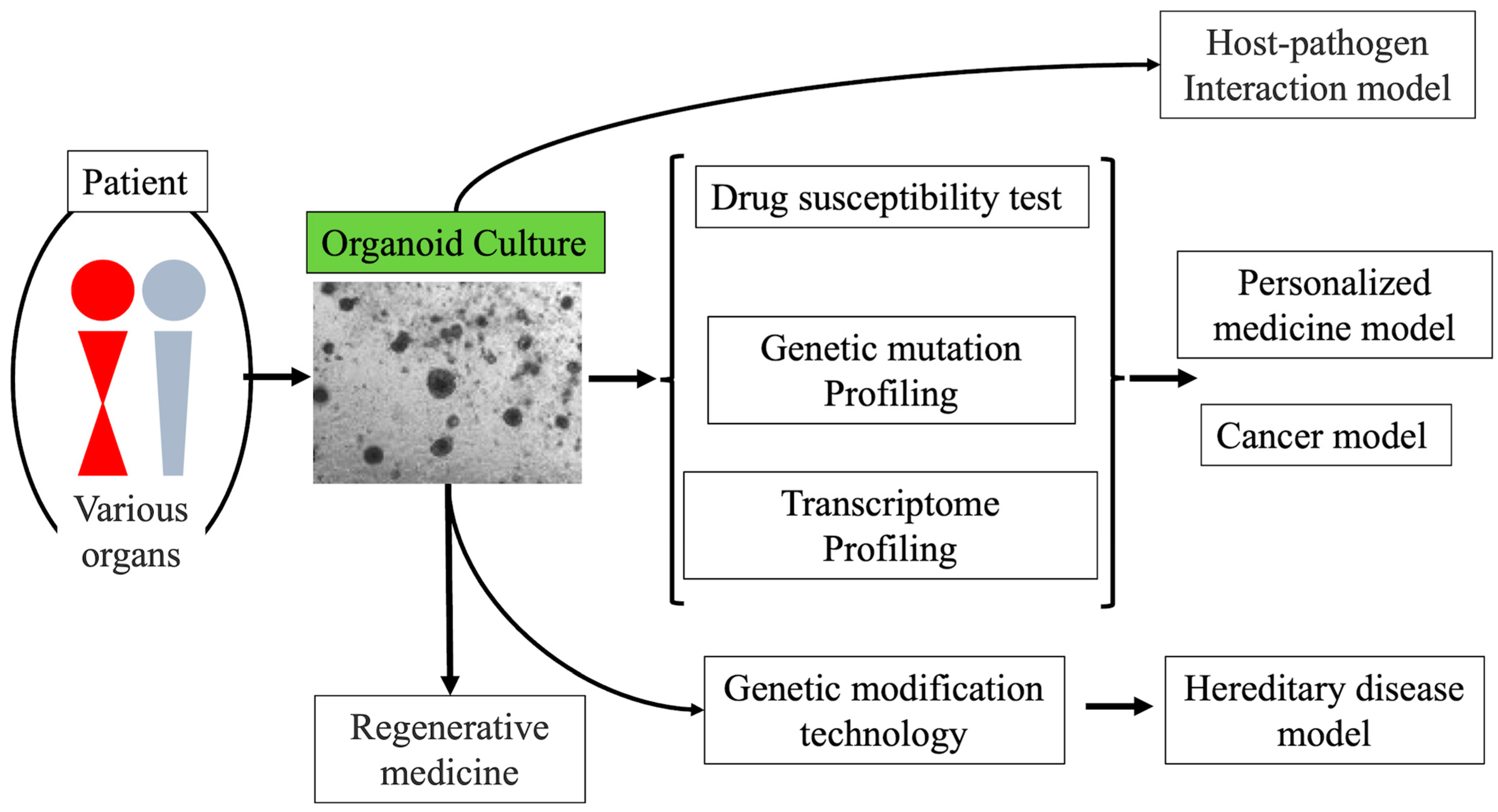
3. Organoid Culture
4. Cancer Organoid
5. General View of Pancreatobiliary Cancer
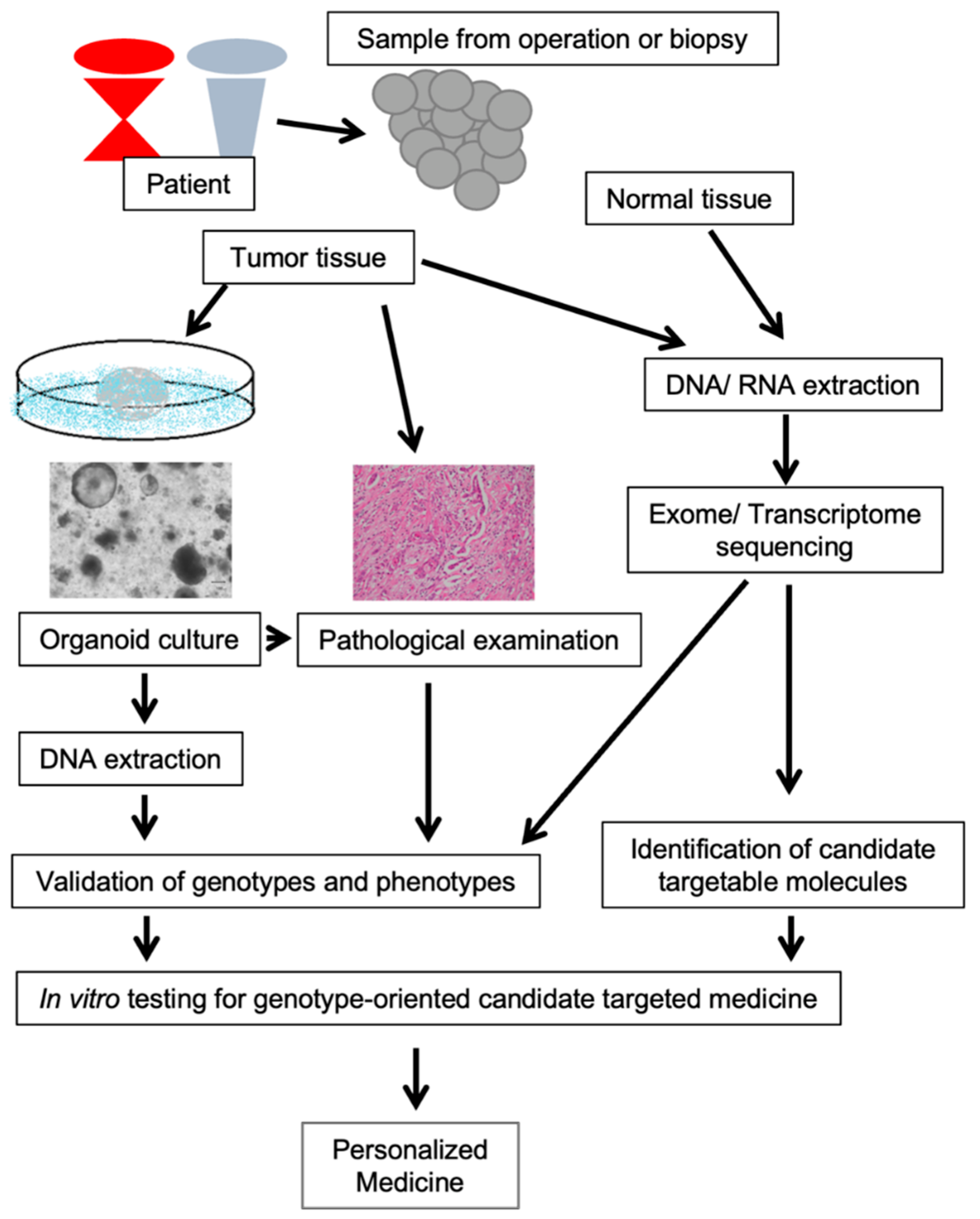
6. Pancreatobiliary Cancer Organoid
7. Personalized Medicine
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Dowden, H.; Munro, J. Trends in clinical success rates and therapeutic focus. Nat. Rev. Drug Discov. 2019, 18, 495–496. [Google Scholar] [CrossRef] [PubMed]
- Hwang, T.J.; Carpenter, D.; Lauffenburger, J.C.; Wang, B.; Franklin, J.M.; Kesselheim, A.S. Failure of Investigational Drugs in Late-Stage Clinical Development and Publication of Trial Results. JAMA Intern. Med. 2016, 176, 1826–1833. [Google Scholar] [CrossRef] [PubMed]
- Ben-David, U.; Siranosian, B.; Ha, G.; Tang, H.; Oren, Y.; Hinohara, K.; Strathdee, C.A.; Dempster, J.; Lyons, N.J.; Burns, R.; et al. Genetic and transcriptional evolution alters cancer cell line drug response. Nature 2018, 560, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Li, M.L.; Aggeler, J.; Farson, D.A.; Hatier, C.; Hassell, J.; Bissell, M.J. Influence of a reconstituted basement membrane and its components on casein gene expression and secretion in mouse mammary epithelial cells. Proc. Natl. Acad. Sci. USA 1987, 84, 136–140. [Google Scholar] [CrossRef] [Green Version]
- Shannon, J.M.; Mason, R.J.; Jennings, S.D. Functional differentiation of alveolar type II epithelial cells in vitro: Effects of cell shape, cell-matrix interactions and cell-cell interactions. Biochim. Biophys. Acta 1987, 931, 143–156. [Google Scholar] [CrossRef]
- Assawachananont, J.; Mandai, M.; Okamoto, S.; Yamada, C.; Eiraku, M.; Yonemura, S.; Sasai, Y.; Takahashi, M. Transplantation of embryonic and induced pluripotent stem cell-derived 3D retinal sheets into retinal degenerative mice. Stem Cell Rep. 2014, 2, 662–674. [Google Scholar] [CrossRef] [Green Version]
- McCracken, K.W.; Catá, E.M.; Crawford, C.M.; Sinagoga, K.L.; Schumacher, M.; Rockich, B.E.; Tsai, Y.H.; Mayhew, C.N.; Spence, J.R.; Zavros, Y.; et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 2014, 516, 400–404. [Google Scholar] [CrossRef] [Green Version]
- Spence, J.R.; Mayhew, C.N.; Rankin, S.A.; Kuhar, M.F.; Vallance, J.E.; Tolle, K.; Hoskins, E.E.; Kalinichenko, V.V.; Wells, S.I.; Zorn, A.M.; et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 2011, 470, 105–109. [Google Scholar] [CrossRef] [Green Version]
- Korinek, V.; Barker, N.; Willert, K.; Molenaar, M.; Roose, J.; Wagenaar, G.; Markman, M.; Lamers, W.; Destree, O.; Clevers, H. Two members of the Tcf family implicated in Wnt/beta-catenin signaling during embryogenesis in the mouse. Mol. Cell. Biol. 1998, 18, 1248–1256. [Google Scholar] [CrossRef] [Green Version]
- Dignass, A.U.; Sturm, A. Peptide growth factors in the intestine. Eur. J. Gastroenterol. Hepatol. 2001, 13, 763–770. [Google Scholar] [CrossRef]
- Haramis, A.P.; Begthel, H.; van den Born, M.; van Es, J.; Jonkheer, S.; Offerhaus, G.J.; Clevers, H. De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science 2004, 303, 1684–1686. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.A.; Kakitani, M.; Zhao, J.; Oshima, T.; Tang, T.; Binnerts, M.; Liu, Y.; Boyle, B.; Park, E.; Emtage, P.; et al. Mitogenic influence of human R-spondin1 on the intestinal epithelium. Science 2005, 309, 1256–1259. [Google Scholar] [CrossRef]
- Sato, T.; Stange, D.E.; Ferrante, M.; Vries, R.G.; Van Es, J.H.; Van den Brink, S.; Van Houdt, W.J.; Pronk, A.; Van Gorp, J.; Siersema, P.D.; et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 2011, 141, 1762–1772. [Google Scholar] [CrossRef]
- Jung, P.; Sato, T.; Merlos-Suárez, A.; Barriga, F.M.; Iglesias, M.; Rossell, D.; Auer, H.; Gallardo, M.; Blasco, M.A.; Sancho, E.; et al. Isolation and in vitro expansion of human colonic stem cells. Nat. Med. 2011, 17, 1225–1227. [Google Scholar] [CrossRef]
- Barker, N.; Huch, M.; Kujala, P.; van de Wetering, M.; Snippert, H.J.; van Es, J.H.; Sato, T.; Stange, D.E.; Begthel, H.; van den Born, M.; et al. Lgr5+ve stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 2010, 6, 25–36. [Google Scholar] [CrossRef] [Green Version]
- Broutier, L.; Andersson-Rolf, A.; Hindley, C.J.; Boj, S.F.; Clevers, H.; Koo, B.K.; Huch, M. Culture and establishment of self-renewing human and mouse adult liver and pancreas 3D organoids and their genetic manipulation. Nat. Protoc. 2016, 11, 1724–1743. [Google Scholar] [CrossRef]
- Kessler, M.; Hoffmann, K.; Brinkmann, V.; Thieck, O.; Jackisch, S.; Toelle, B.; Berger, H.; Mollenkopf, H.J.; Mangler, M.; Sehouli, J.; et al. The Notch and Wnt pathways regulate stemness and differentiation in human fallopian tube organoids. Nat. Commun. 2015, 6, 8989. [Google Scholar] [CrossRef]
- Karthaus, W.R.; Iaquinta, P.J.; Drost, J.; Gracanin, A.; van Boxtel, R.; Wongvipat, J.; Dowling, C.M.; Gao, D.; Begthel, H.; Sachs, N.; et al. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell 2014, 159, 163–175. [Google Scholar] [CrossRef] [Green Version]
- Maimets, M.; Rocchi, C.; Bron, R.; Pringle, S.; Kuipers, J.; Giepmans, B.N.; Vries, R.G.; Clevers, H.; de Haan, G.; van Os, R.; et al. Long-Term In Vitro Expansion of Salivary Gland Stem Cells Driven by Wnt Signals. Stem Cell Rep. 2016, 6, 150–162. [Google Scholar] [CrossRef] [Green Version]
- Dontu, G.; Abdallah, W.M.; Foley, J.M.; Jackson, K.W.; Clarke, M.F.; Kawamura, M.J.; Wicha, M.S. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003, 17, 1253–1270. [Google Scholar] [CrossRef] [Green Version]
- Kale, S.; Biermann, S.; Edwards, C.; Tarnowski, C.; Morris, M.; Long, M.W. Three-dimensional cellular development is essential for ex vivo formation of human bone. Nat. Biotechnol. 2000, 18, 954–958. [Google Scholar] [CrossRef]
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef]
- Ootani, A.; Li, X.; Sangiorgi, E.; Ho, Q.T.; Ueno, H.; Toda, S.; Sugihara, H.; Fujimoto, K.; Weissman, I.L.; Capecchi, M.R.; et al. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat. Med. 2009, 15, 701–706. [Google Scholar] [CrossRef] [Green Version]
- Stange, D.E.; Koo, B.K.; Huch, M.; Sibbel, G.; Basak, O.; Lyubimova, A.; Kujala, P.; Bartfeld, S.; Koster, J.; Geahlen, J.H.; et al. Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell 2013, 155, 357–368. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Nadauld, L.; Ootani, A.; Corney, D.C.; Pai, R.K.; Gevaert, O.; Cantrell, M.A.; Rack, P.G.; Neal, J.T.; Chan, C.W.; et al. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat. Med. 2014, 20, 769–777. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Bhang, D.H.; Beede, A.; Huang, T.L.; Stripp, B.R.; Bloch, K.D.; Wagers, A.J.; Tseng, Y.H.; Ryeom, S.; Kim, C.F. Lung stem cell differentiation in mice directed by endothelial cells via a BMP4-NFATc1-thrombospondin-1 axis. Cell 2014, 156, 440–455. [Google Scholar] [CrossRef] [Green Version]
- Huch, M.; Dorrell, C.; Boj, S.F.; van Es, J.H.; Li, V.S.; van de Wetering, M.; Sato, T.; Hamer, K.; Sasaki, N.; Finegold, M.J.; et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 2013, 494, 247–250. [Google Scholar] [CrossRef] [Green Version]
- Huch, M.; Koo, B.K. Modeling mouse and human development using organoid cultures. Development 2015, 142, 3113–3125. [Google Scholar] [CrossRef] [Green Version]
- Boj, S.F.; Hwang, C.I.; Baker, L.A.; Chio, I.I.C.; Engle, D.D.; Corbo, V.; Jager, M.; Ponz-Sarvise, M.; Tiriac, H.; Spector, M.S.; et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 2015, 160, 324–338. [Google Scholar] [CrossRef] [Green Version]
- Gao, D.; Vela, I.; Sboner, A.; Iaquinta, P.J.; Karthaus, W.R.; Gopalan, A.; Dowling, C.; Wanjala, J.N.; Undvall, E.A.; Arora, V.K.; et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 2014, 159, 176–187. [Google Scholar] [CrossRef] [Green Version]
- Nanduri, L.S.; Baanstra, M.; Faber, H.; Rocchi, C.; Zwart, E.; de Haan, G.; van Os, R.; Coppes, R.P. Purification and ex vivo expansion of fully functional salivary gland stem cells. Stem Cell Rep. 2014, 3, 957–964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hisha, H.; Tanaka, T.; Kanno, S.; Tokuyama, Y.; Komai, Y.; Ohe, S.; Yanai, H.; Omachi, T.; Ueno, H. Establishment of a novel lingual organoid culture system: Generation of organoids having mature keratinized epithelium from adult epithelial stem cells. Sci. Rep. 2013, 3, 3224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, K.G.; Zhong, P.; Zheng, W.; Beekman, J.M. Pharmacological analysis of CFTR variants of cystic fibrosis using stem cell-derived organoids. Drug Discov. Today 2019, 24, 2126–2138. [Google Scholar] [CrossRef] [PubMed]
- Nozaki, K.; Mochizuki, W.; Matsumoto, Y.; Matsumoto, T.; Fukuda, M.; Mizutani, T.; Watanabe, M.; Nakamura, T. Co-culture with intestinal epithelial organoids allows efficient expansion and motility analysis of intraepithelial lymphocytes. J. Gastroenterol. 2016, 51, 206–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben-David, U.; Ha, G.; Tseng, Y.Y.; Greenwald, N.F.; Oh, C.; Shih, J.; McFarland, J.M.; Wong, B.; Boehm, J.S.; Beroukhim, R.; et al. Patient-derived xenografts undergo mouse-specific tumor evolution. Nat. Genet. 2017, 49, 1567–1575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broutier, L.; Mastrogiovanni, G.; Verstegen, M.M.; Francies, H.E.; Gavarró, L.M.; Bradshaw, C.R.; Allen, G.E.; Arnes-Benito, R.; Sidorova, O.; Gaspersz, M.P.; et al. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat. Med. 2017, 23, 1424–1435. [Google Scholar] [CrossRef]
- Saito, Y.; Muramatsu, T.; Kanai, Y.; Ojima, H.; Sukeda, A.; Hiraoka, N.; Arai, E.; Sugiyama, Y.; Matsuzaki, J.; Uchida, R.; et al. Establishment of Patient-Derived Organoids and Drug Screening for Biliary Tract Carcinoma. Cell Rep. 2019, 27, 1265–1276.e1264. [Google Scholar] [CrossRef] [Green Version]
- Shimokawa, M.; Ohta, Y.; Nishikori, S.; Matano, M.; Takano, A.; Fujii, M.; Date, S.; Sugimoto, S.; Kanai, T.; Sato, T. Visualization and targeting of LGR5(+) human colon cancer stem cells. Nature 2017, 545, 187–192. [Google Scholar] [CrossRef]
- Koikawa, K.; Ohuchida, K.; Ando, Y.; Kibe, S.; Nakayama, H.; Takesue, S.; Endo, S.; Abe, T.; Okumura, T.; Iwamoto, C.; et al. Basement membrane destruction by pancreatic stellate cells leads to local invasion in pancreatic ductal adenocarcinoma. Cancer Lett. 2018, 425, 65–77. [Google Scholar] [CrossRef]
- van de Wetering, M.; Francies, H.E.; Francis, J.M.; Bounova, G.; Iorio, F.; Pronk, A.; van Houdt, W.; van Gorp, J.; Taylor-Weiner, A.; Kester, L.; et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015, 161, 933–945. [Google Scholar] [CrossRef] [Green Version]
- Sachs, N.; de Ligt, J.; Kopper, O.; Gogola, E.; Bounova, G.; Weeber, F.; Balgobind, A.V.; Wind, K.; Gracanin, A.; Begthel, H.; et al. A Living Biobank of Breast Cancer Organoids Captures Disease Heterogeneity. Cell 2018, 172, 373–386.e310. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Mun, H.; Sung, C.O.; Cho, E.J.; Jeon, H.J.; Chun, S.M.; Jung, D.J.; Shin, T.H.; Jeong, G.S.; Kim, D.K.; et al. Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat. Commun. 2019, 10, 3991. [Google Scholar] [CrossRef]
- Shiihara, M.; Ishikawa, T.; Saiki, Y.; Omori, Y.; Hirose, K.; Fukushige, S.; Ikari, N.; Higuchi, R.; Yamamoto, M.; Morikawa, T.; et al. Development of a system combining comprehensive genotyping and organoid cultures for identifying and testing genotype-oriented personalised medicine for pancreatobiliary cancers. Eur. J. Cancer 2021, 148, 239–250. [Google Scholar] [CrossRef]
- Huang, B.; Trujillo, M.A.; Fujikura, K.; Qiu, M.; Chen, F.; Felsenstein, M.; Zhou, C.; Skaro, M.; Gauthier, C.; Macgregor-Das, A.; et al. Molecular characterization of organoids derived from pancreatic intraductal papillary mucinous neoplasms. J. Pathol. 2020, 252, 252–262. [Google Scholar] [CrossRef]
- Kopper, O.; de Witte, C.J.; Lõhmussaar, K.; Valle-Inclan, J.E.; Hami, N.; Kester, L.; Balgobind, A.V.; Korving, J.; Proost, N.; Begthel, H.; et al. An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Nat. Med. 2019, 25, 838–849. [Google Scholar] [CrossRef]
- Kawasaki, K.; Fujii, M.; Sugimoto, S.; Ishikawa, K.; Matano, M.; Ohta, Y.; Toshimitsu, K.; Takahashi, S.; Hosoe, N.; Sekine, S.; et al. Chromosome Engineering of Human Colon-Derived Organoids to Develop a Model of Traditional Serrated Adenoma. Gastroenterology 2020, 158, 638–651.e638. [Google Scholar] [CrossRef] [Green Version]
- Patel, T. Worldwide trends in mortality from biliary tract malignancies. BMC Cancer 2002, 2, 10. [Google Scholar] [CrossRef] [Green Version]
- Khan, S.A.; Taylor-Robinson, S.D.; Toledano, M.B.; Beck, A.; Elliott, P.; Thomas, H.C. Changing international trends in mortality rates for liver, biliary and pancreatic tumours. J. Hepatol. 2002, 37, 806–813. [Google Scholar] [CrossRef]
- Carriaga, M.T.; Henson, D.E. Liver, gallbladder, extrahepatic bile ducts, and pancreas. Cancer 1995, 75, 171–190. [Google Scholar] [CrossRef]
- Valle, J.; Wasan, H.; Palmer, D.H.; Cunningham, D.; Anthoney, A.; Maraveyas, A.; Madhusudan, S.; Iveson, T.; Hughes, S.; Pereira, S.P.; et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N. Engl. J. Med. 2010, 362, 1273–1281. [Google Scholar] [CrossRef] [Green Version]
- Uesaka, K.; Boku, N.; Fukutomi, A.; Okamura, Y.; Konishi, M.; Matsumoto, I.; Kaneoka, Y.; Shimizu, Y.; Nakamori, S.; Sakamoto, H.; et al. Adjuvant chemotherapy of S-1 versus gemcitabine for resected pancreatic cancer: A phase 3, open-label, randomised, non-inferiority trial (JASPAC 01). Lancet 2016, 388, 248–257. [Google Scholar] [CrossRef]
- Abou-Alfa, G.K.; Macarulla, T.; Javle, M.M.; Kelley, R.K.; Lubner, S.J.; Adeva, J.; Cleary, J.M.; Catenacci, D.V.; Borad, M.J.; Bridgewater, J.; et al. Ivosidenib in IDH1-mutant, chemotherapy-refractory cholangiocarcinoma (ClarIDHy): A multicentre, randomised, double-blind, placebo-controlled, phase 3 study. Lancet Oncol. 2020, 21, 796–807. [Google Scholar] [CrossRef]
- Unno, M.; Hata, T.; Motoi, F. Long-term outcome following neoadjuvant therapy for resectable and borderline resectable pancreatic cancer compared to upfront surgery: A meta-analysis of comparative studies by intention-to-treat analysis. Surg. Today 2019, 49, 295–299. [Google Scholar] [CrossRef]
- Biankin, A.V.; Waddell, N.; Kassahn, K.S.; Gingras, M.C.; Muthuswamy, L.B.; Johns, A.L.; Miller, D.K.; Wilson, P.J.; Patch, A.M.; Wu, J.; et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012, 491, 399–405. [Google Scholar] [CrossRef]
- Jones, S.; Zhang, X.; Parsons, D.W.; Lin, J.C.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Kamiyama, H.; Jimeno, A.; et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 2008, 321, 1801–1806. [Google Scholar] [CrossRef] [Green Version]
- Jusakul, A.; Cutcutache, I.; Yong, C.H.; Lim, J.Q.; Huang, M.N.; Padmanabhan, N.; Nellore, V.; Kongpetch, S.; Ng, A.W.T.; Ng, L.M.; et al. Whole-Genome and Epigenomic Landscapes of Etiologically Distinct Subtypes of Cholangiocarcinoma. Cancer Discov. 2017, 7, 1116–1135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seino, T.; Kawasaki, S.; Shimokawa, M.; Tamagawa, H.; Toshimitsu, K.; Fujii, M.; Ohta, Y.; Matano, M.; Nanki, K.; Kawasaki, K.; et al. Human Pancreatic Tumor Organoids Reveal Loss of Stem Cell Niche Factor Dependence during Disease Progression. Cell Stem Cell 2018, 22, 454–467.e456. [Google Scholar] [CrossRef] [Green Version]
- Driehuis, E.; van Hoeck, A.; Moore, K.; Kolders, S.; Francies, H.E.; Gulersonmez, M.C.; Stigter, E.C.A.; Burgering, B.; Geurts, V.; Gracanin, A.; et al. Pancreatic cancer organoids recapitulate disease and allow personalized drug screening. Proc. Natl. Acad. Sci. USA 2019, 116, 26580–26590. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Shukla, S.K.; Vernucci, E.; He, C.; Wang, D.; King, R.J.; Jha, K.; Siddhanta, K.; Mullen, N.J.; Attri, K.S.; et al. Metabolic Rewiring by Loss of Sirt5 Promotes Kras-Induced Pancreatic Cancer Progression. Gastroenterology 2021, 161, 1584–1600. [Google Scholar] [CrossRef]
- Fujiwara, H.; Takahara, N.; Tateishi, K.; Tanaka, M.; Kanai, S.; Kato, H.; Nakatsuka, T.; Yamamoto, K.; Kogure, H.; Arita, J.; et al. 5-Aminolevulinic acid-mediated photodynamic activity in patient-derived cholangiocarcinoma organoids. Surg. Oncol. 2020, 35, 484–490. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.; McOlash, L.; Palen, K.; Johnson, B.; Duris, C.; Yang, Q.; Dwinell, M.B.; Hunt, B.; Evans, D.B.; Gershan, J.; et al. Development of primary human pancreatic cancer organoids, matched stromal and immune cells and 3D tumor microenvironment models. BMC Cancer 2018, 18, 335. [Google Scholar] [CrossRef]
- Öhlund, D.; Handly-Santana, A.; Biffi, G.; Elyada, E.; Almeida, A.S.; Ponz-Sarvise, M.; Corbo, V.; Oni, T.E.; Hearn, S.A.; Lee, E.J.; et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 2017, 214, 579–596. [Google Scholar] [CrossRef]
- Koikawa, K.; Kibe, S.; Suizu, F.; Sekino, N.; Kim, N.; Manz, T.D.; Pinch, B.J.; Akshinthala, D.; Verma, A.; Gaglia, G.; et al. Targeting Pin1 renders pancreatic cancer eradicable by synergizing with immunochemotherapy. Cell 2021, 184, 4753–4771.e4727. [Google Scholar] [CrossRef]
- Grandori, C.; Kemp, C.J. Personalized Cancer Models for Target Discovery and Precision Medicine. Trends Cancer 2018, 4, 634–642. [Google Scholar] [CrossRef]
- Nussinov, R.; Jang, H.; Tsai, C.J.; Cheng, F. Review: Precision medicine and driver mutations: Computational methods, functional assays and conformational principles for interpreting cancer drivers. PLoS Comput. Biol. 2019, 15, e1006658. [Google Scholar] [CrossRef]
- Pauli, C.; Hopkins, B.D.; Prandi, D.; Shaw, R.; Fedrizzi, T.; Sboner, A.; Sailer, V.; Augello, M.; Puca, L.; Rosati, R.; et al. Personalized In Vitro and In Vivo Cancer Models to Guide Precision Medicine. Cancer Discov. 2017, 7, 462–477. [Google Scholar] [CrossRef] [Green Version]
- Hou, S.; Tiriac, H.; Sridharan, B.P.; Scampavia, L.; Madoux, F.; Seldin, J.; Souza, G.R.; Watson, D.; Tuveson, D.; Spicer, T.P. Advanced Development of Primary Pancreatic Organoid Tumor Models for High-Throughput Phenotypic Drug Screening. SLAS Discov. 2018, 23, 574–584. [Google Scholar] [CrossRef] [Green Version]
- Maenhoudt, N.; Defraye, C.; Boretto, M.; Jan, Z.; Heremans, R.; Boeckx, B.; Hermans, F.; Arijs, I.; Cox, B.; Van Nieuwenhuysen, E.; et al. Developing Organoids from Ovarian Cancer as Experimental and Preclinical Models. Stem Cell Rep. 2020, 14, 717–729. [Google Scholar] [CrossRef]
- Narasimhan, V.; Wright, J.A.; Churchill, M.; Wang, T.; Rosati, R.; Lannagan, T.R.M.; Vrbanac, L.; Richardson, A.B.; Kobayashi, H.; Price, T.; et al. Medium-throughput Drug Screening of Patient-derived Organoids from Colorectal Peritoneal Metastases to Direct Personalized Therapy. Clin. Cancer Res. 2020, 26, 3662–3670. [Google Scholar] [CrossRef]
- Chen, J.H.; Chu, X.P.; Zhang, J.T.; Nie, Q.; Tang, W.F.; Su, J.; Yan, H.H.; Zheng, H.P.; Chen, Z.X.; Chen, X.; et al. Genomic characteristics and drug screening among organoids derived from non-small cell lung cancer patients. Thorac. Cancer 2020, 11, 2279–2290. [Google Scholar] [CrossRef]
- Li, Z.; Qian, Y.; Li, W.; Liu, L.; Yu, L.; Liu, X.; Wu, G.; Wang, Y.; Luo, W.; Fang, F.; et al. Human Lung Adenocarcinoma-Derived Organoid Models for Drug Screening. iScience 2020, 23, 101411. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Zhao, H.; Zhang, W.; Wang, J.; Liu, Y.; Cao, Y.; Zheng, H.; Hu, Z.; Wang, S.; Zhu, Y.; et al. An Automated Organoid Platform with Inter-organoid Homogeneity and Inter-patient Heterogeneity. Cell Rep. Med. 2020, 1, 100161. [Google Scholar] [CrossRef] [PubMed]
- Larsen, B.M.; Kannan, M.; Langer, L.F.; Leibowitz, B.D.; Bentaieb, A.; Cancino, A.; Dolgalev, I.; Drummond, B.E.; Dry, J.R.; Ho, C.S.; et al. A pan-cancer organoid platform for precision medicine. Cell Rep. 2021, 36, 109429. [Google Scholar] [CrossRef] [PubMed]
- Ramamoorthy, P.; Thomas, S.M.; Kaushik, G.; Subramaniam, D.; Chastain, K.M.; Dhar, A.; Tawfik, O.; Kasi, A.; Sun, W.; Ramalingam, S.; et al. Metastatic Tumor-in-a-Dish, a Novel Multicellular Organoid to Study Lung Colonization and Predict Therapeutic Response. Cancer Res. 2019, 79, 1681–1695. [Google Scholar] [CrossRef] [Green Version]
- Maier, C.F.; Zhu, L.; Nanduri, L.K.; Kühn, D.; Kochall, S.; Thepkaysone, M.L.; William, D.; Grützmann, K.; Klink, B.; Betge, J.; et al. Patient-Derived Organoids of Cholangiocarcinoma. Int. J. Mol. Sci. 2021, 22, 8675. [Google Scholar] [CrossRef]
- Cho, Y.W.; Min, D.W.; Kim, H.P.; An, Y.; Kim, S.; Youk, J.; Chun, J.; Im, J.P.; Song, S.H.; Ju, Y.S.; et al. Patient-derived organoids as a preclinical platform for precision medicine in colorectal cancer. Mol. Oncol. 2021, in press. [CrossRef]
- Yuan, B.; Zhao, X.; Wang, X.; Liu, E.; Liu, C.; Zong, Y.; Jiang, Y.; Hou, M.; Chen, Y.; Chen, L.; et al. Patient-derived organoids for personalized gallbladder cancer modelling and drug screening. Clin. Transl. Med. 2022, 12, e678. [Google Scholar] [CrossRef]
- Bi, J.; Newtson, A.M.; Zhang, Y.; Devor, E.J.; Samuelson, M.I.; Thiel, K.W.; Leslie, K.K. Successful Patient-Derived Organoid Culture of Gynecologic Cancers for Disease Modeling and Drug Sensitivity Testing. Cancers 2021, 13, 2901. [Google Scholar] [CrossRef]
- Gilles, M.E.; Hao, L.; Huang, L.; Rupaimoole, R.; Lopez-Casas, P.P.; Pulver, E.; Jeong, J.C.; Muthuswamy, S.K.; Hidalgo, M.; Bhatia, S.N.; et al. Personalized RNA Medicine for Pancreatic Cancer. Clin. Cancer Res. 2018, 24, 1734–1747. [Google Scholar] [CrossRef] [Green Version]
- Mazzocchi, A.R.; Rajan, S.A.P.; Votanopoulos, K.I.; Hall, A.R.; Skardal, A. In vitro patient-derived 3D mesothelioma tumor organoids facilitate patient-centric therapeutic screening. Sci. Rep. 2018, 8, 2886. [Google Scholar] [CrossRef] [Green Version]
- Tung, K.L.; Chen, K.Y.; Negrete, M.; Chen, T.; Safi, A.; Aljamal, A.A.; Song, L.; Crawford, G.E.; Ding, S.; Hsu, D.S.; et al. Integrated chromatin and transcriptomic profiling of patient-derived colon cancer organoids identifies personalized drug targets to overcome oxaliplatin resistance. Genes Dis. 2021, 8, 203–214. [Google Scholar] [CrossRef]
- Neal, J.T.; Li, X.; Zhu, J.; Giangarra, V.; Grzeskowiak, C.L.; Ju, J.; Liu, I.H.; Chiou, S.H.; Salahudeen, A.A.; Smith, A.R.; et al. Organoid Modeling of the Tumor Immune Microenvironment. Cell 2018, 175, 1972–1988.e1916. [Google Scholar] [CrossRef] [Green Version]
- Schnalzger, T.E.; de Groot, M.H.; Zhang, C.; Mosa, M.H.; Michels, B.E.; Röder, J.; Darvishi, T.; Wels, W.S.; Farin, H.F. 3D model for CAR-mediated cytotoxicity using patient-derived colorectal cancer organoids. EMBO J. 2019, 38, e100928. [Google Scholar] [CrossRef]
- Jacob, F.; Salinas, R.D.; Zhang, D.Y.; Nguyen, P.T.T.; Schnoll, J.G.; Wong, S.Z.H.; Thokala, R.; Sheikh, S.; Saxena, D.; Prokop, S.; et al. A Patient-Derived Glioblastoma Organoid Model and Biobank Recapitulates Inter- and Intra-tumoral Heterogeneity. Cell 2020, 180, 188–204.e122. [Google Scholar] [CrossRef]
- Forsythe, S.D.; Erali, R.A.; Sasikumar, S.; Laney, P.; Shelkey, E.; D’Agostino, R., Jr.; Miller, L.D.; Shen, P.; Levine, E.A.; Soker, S.; et al. Organoid Platform in Preclinical Investigation of Personalized Immunotherapy Efficacy in Appendiceal Cancer: Feasibility Study. Clin. Cancer Res. 2021, 27, 5141–5150. [Google Scholar] [CrossRef]
- Gong, Z.; Huang, L.; Tang, X.; Chen, K.; Wu, Z.; Zhang, L.; Sun, Y.; Xia, Y.; Chen, H.; Wei, Y.; et al. Acoustic Droplet Printing Tumor Organoids for Modeling Bladder Tumor Immune Microenvironment within a Week. Adv. Healthc. Mater. 2021, 10, e2101312. [Google Scholar] [CrossRef]
- Kholosy, W.M.; Derieppe, M.; van den Ham, F.; Ober, K.; Su, Y.; Custers, L.; Schild, L.; van Zogchel, L.M.J.; Wellens, L.M.; Ariese, H.R.; et al. Neuroblastoma and DIPG Organoid Coculture System for Personalized Assessment of Novel Anticancer Immunotherapies. J. Pers. Med. 2021, 11, 869. [Google Scholar] [CrossRef]
- Forsythe, S.D.; Sasikumar, S.; Moaven, O.; Sivakumar, H.; Shen, P.; Levine, E.A.; Soker, S.; Skardal, A.; Votanopoulos, K.I. Personalized Identification of Optimal HIPEC Perfusion Protocol in Patient-Derived Tumor Organoid Platform. Ann. Surg. Oncol. 2020, 27, 4950–4960. [Google Scholar] [CrossRef]
- Park, M.; Kwon, J.; Kong, J.; Moon, S.M.; Cho, S.; Yang, K.Y.; Jang, W.I.; Kim, M.S.; Kim, Y.; Shin, U.S. A Patient-Derived Organoid-Based Radiosensitivity Model for the Prediction of Radiation Responses in Patients with Rectal Cancer. Cancers 2021, 13, 3760. [Google Scholar] [CrossRef]
- Ding, R.B.; Chen, P.; Rajendran, B.K.; Lyu, X.; Wang, H.; Bao, J.; Zeng, J.; Hao, W.; Sun, H.; Wong, A.H.; et al. Molecular landscape and subtype-specific therapeutic response of nasopharyngeal carcinoma revealed by integrative pharmacogenomics. Nat. Commun. 2021, 12, 3046. [Google Scholar] [CrossRef]
- Votanopoulos, K.I.; Mazzocchi, A.; Sivakumar, H.; Forsythe, S.; Aleman, J.; Levine, E.A.; Skardal, A. Appendiceal Cancer Patient-Specific Tumor Organoid Model for Predicting Chemotherapy Efficacy Prior to Initiation of Treatment: A Feasibility Study. Ann. Surg. Oncol. 2019, 26, 139–147. [Google Scholar] [CrossRef]
- Nuciforo, S.; Fofana, I.; Matter, M.S.; Blumer, T.; Calabrese, D.; Boldanova, T.; Piscuoglio, S.; Wieland, S.; Ringnalda, F.; Schwank, G.; et al. Organoid Models of Human Liver Cancers Derived from Tumor Needle Biopsies. Cell Rep. 2018, 24, 1363–1376. [Google Scholar] [CrossRef] [Green Version]
- Pasch, C.A.; Favreau, P.F.; Yueh, A.E.; Babiarz, C.P.; Gillette, A.A.; Sharick, J.T.; Karim, M.R.; Nickel, K.P.; DeZeeuw, A.K.; Sprackling, C.M.; et al. Patient-Derived Cancer Organoid Cultures to Predict Sensitivity to Chemotherapy and Radiation. Clin. Cancer Res. 2019, 25, 5376–5387. [Google Scholar] [CrossRef]
| No | Author | Year | Precision Medicine | Organ |
|---|---|---|---|---|
| 1 | Pauli C, et al. | 2017 | Drug screening (High-throughput) | Various Cancer |
| 2 | Broutier L, et al. | 2018 | Drug screening (Genome-editing target therapy) | Liver |
| 3 | Sachs N, et al. | 2018 | Drug screening (Genome-editing target therapy) | Breast |
| 4 | Gilles ME, et al. | 2018 | Drug screening (Genome-editing target therapy) | Pancreas |
| 5 | Mazzocchi AR, et al. | 2019 | Drug screening (Genome-editing target therapy) | Mesothelioma |
| 6 | Hou S, et al. | 2019 | Drug screening (High-throughput) | Pancreas |
| 7 | Saito Y, et al. | 2019 | Drug screening (High-throughput) | Biliary |
| 8 | Kim M, et al. | 2019 | Drug screening (High-throughput) | Lung |
| 9 | Votanopoulos KI, et al. | 2019 | Drug screening | Appendiceal |
| 10 | Kijima T, et al. | 2019 | Drug screening | Oropharyngeal and Esophageal |
| 11 | Neal JT, et al. | 2019 | Immunotherapy | Melanoma, Renal Cell |
| 12 | Ramamoorthy P, et al. | 2019 | Drug screening (Clinical available drugs) | Lung |
| 13 | Li L, et al. | 2019 | Drug screening | Lung |
| 14 | Grassi L, et al. | 2019 | Drug screening | Renal Cell |
| 15 | Schnalzger TE, et al. | 2019 | Immunotherapy | Colorectal |
| 16 | Driehuis E, et al. | 2019 | Drug screening | Oral |
| 17 | Ubink I, et al. | 2019 | Drug screening (HIPEC) | Colon |
| 18 | Tung KL, et al. | 2019 | Drug screening (Genome-editing target therapy) | Colon |
| 19 | Driehuis E, et al. | 2019 | Drug screening (multiple) | Pancreas |
| 20 | Jacob F, et al. | 2020 | Immunotherapy | Glioblastoma |
| 21 | Maenhoudt N, et al. | 2020 | Drug screening (High-throughput) | Ovary |
| 22 | Narasimhan V, et al. | 2020 | Drug screening (High-throughput) | Colorectal |
| 23 | Forsythe SD, et al. | 2020 | Drug screening (HIPEC) | Colon, Appndiceal |
| 24 | Chen JH, et al. | 2020 | Drug screening (High-throughput) | Lung |
| 25 | Shiihara, et al. | 2021 | Drug screening (Genome-editing target therapy) | Pancreato-Biliary |
| 26 | Li Z, et al. | 2021 | Drug screening (High-throughput) | Lung |
| 27 | Luo X, et al. | 2021 | Drug screening | Colon |
| 28 | Jiang S, et al. | 2021 | Drug screening (High-throughput) | Various Cancer |
| 29 | Chen D, et al. | 2021 | Drug screening | Thyroid |
| 30 | Bie Y, et al. | 2021 | Drug screening (Genome-editing target therapy) | Lung |
| 31 | Park M, et al. | 2021 | Radiation treatment | Rectal |
| 32 | Ding RB, et al. | 2021 | Drug screening, Chemoradiation | Nasopharyngeal |
| 33 | Bi J, et al. | 2021 | Drug screening | Gynecologic |
| 34 | Forsythe SD, et al. | 2021 | Immunotherapy | Appendiceal |
| 35 | Larsen BM, et al. | 2021 | Drug screening (High-throughput) | Various Cancer |
| 36 | Bi J, et al. | 2021 | Drug screening | Endometrial |
| 37 | Maier CF, et al. | 2021 | Drug screening (Clinical available drugs) | Biliary |
| 38 | Kazama A, et al. | 2021 | Drug screening | Renal Cell |
| 39 | Kryeziu K, et al. | 2021 | Drug screening | Rectal |
| 40 | Gong Z, et al. | 2021 | Immunotherapy | Bladder |
| 41 | M Kholosy W, et al. | 2021 | Immunotherapy | Neuroblastoma |
| 42 | Boos SL, et al. | 2022 | Drug screening | Colorectal |
| 43 | Dong Y, et al. | 2022 | Drug screening | Hypopharyngeal |
| 44 | Grossman JE, et al. | 2022 | Drug screening | Pancreas |
| 45 | Cho YW, et al. | 2022 | Drug screening (Clinical available drugs) | Colorectal |
| 46 | Reed MR, et al. | 2022 | Drug screening | Glioblastoma |
| 47 | Yuan B, et al. | 2022 | Drug screening (Clinical available drugs) | Gallbladder |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shiihara, M.; Furukawa, T. Application of Patient-Derived Cancer Organoids to Personalized Medicine. J. Pers. Med. 2022, 12, 789. https://doi.org/10.3390/jpm12050789
Shiihara M, Furukawa T. Application of Patient-Derived Cancer Organoids to Personalized Medicine. Journal of Personalized Medicine. 2022; 12(5):789. https://doi.org/10.3390/jpm12050789
Chicago/Turabian StyleShiihara, Masahiro, and Toru Furukawa. 2022. "Application of Patient-Derived Cancer Organoids to Personalized Medicine" Journal of Personalized Medicine 12, no. 5: 789. https://doi.org/10.3390/jpm12050789
APA StyleShiihara, M., & Furukawa, T. (2022). Application of Patient-Derived Cancer Organoids to Personalized Medicine. Journal of Personalized Medicine, 12(5), 789. https://doi.org/10.3390/jpm12050789






