Immune Cell Networks Uncover Candidate Biomarkers of Melanoma Immunotherapy Response
Abstract
:1. Introduction
2. Results
2.1. IMMUNETS: Modelling Immune Cell Differentiation and Activation
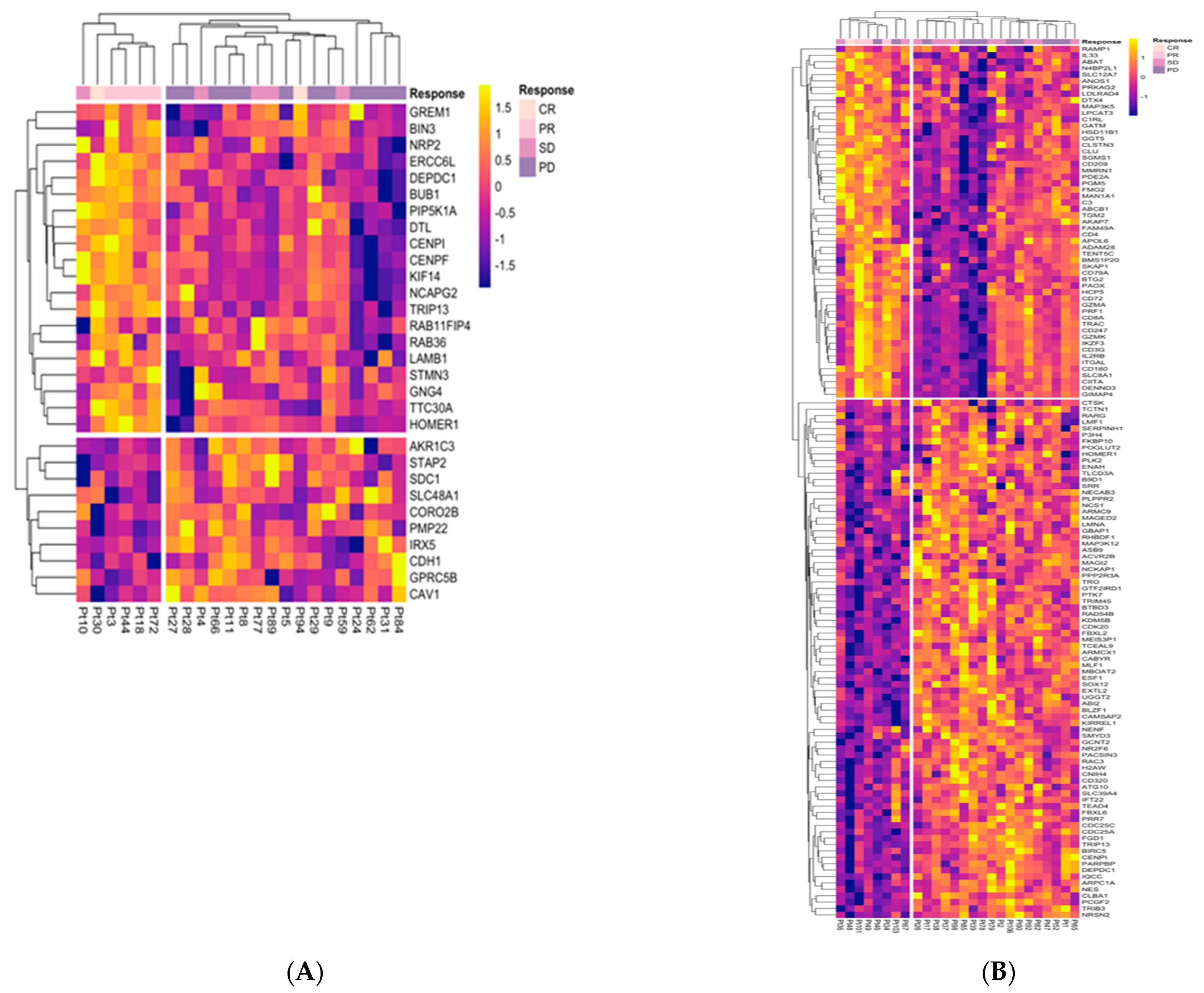
2.2. IMMUNETS Genes Stratify Melanoma Patients by Response to Nivolumab
2.3. Investigation of Candidate Nivolumab Response Biomarkers Expression in an Independent Cohort
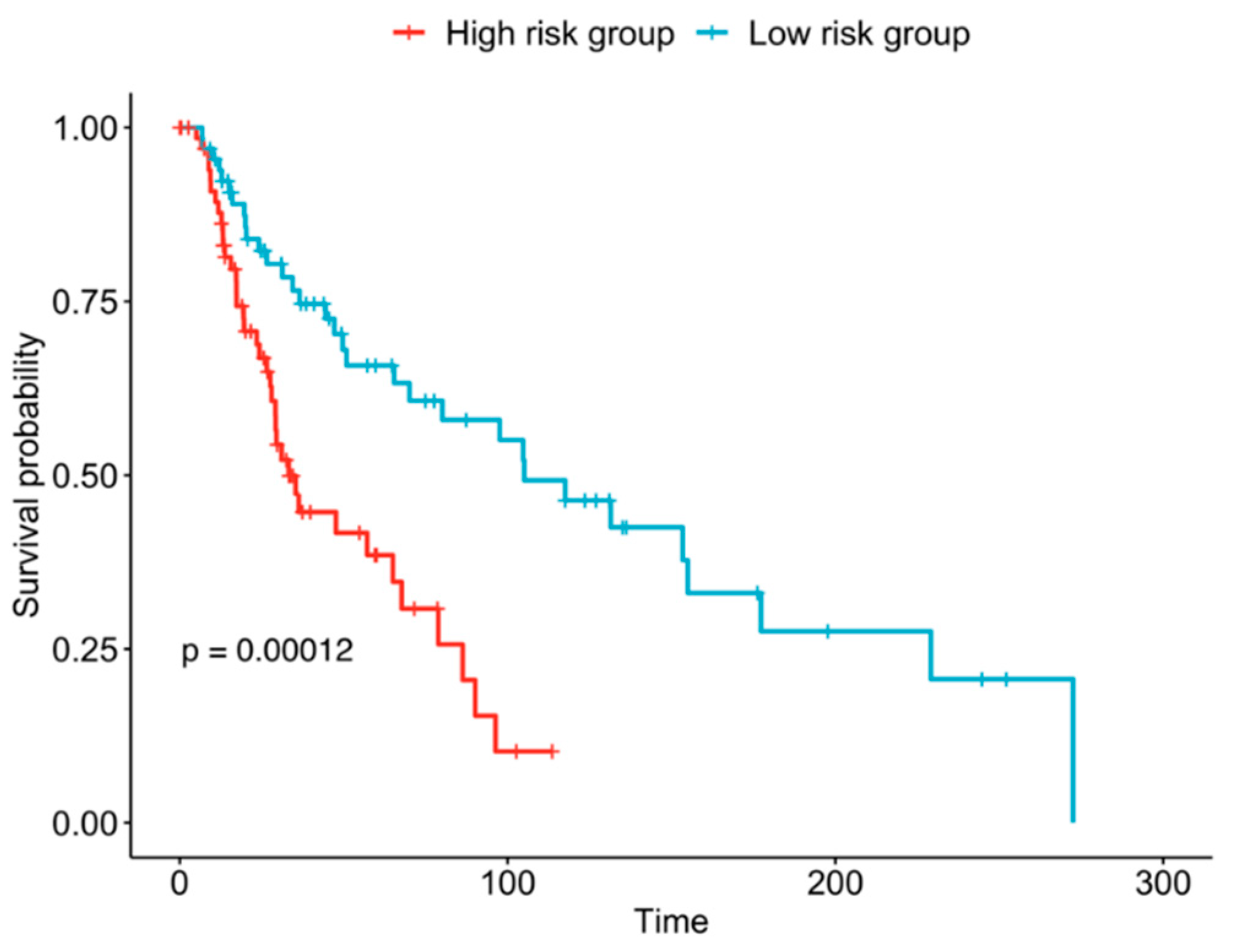
2.4. Candidate Immune Biomarkers Risk Stratify Melanoma by Overall Survival
3. Discussion
4. Methods
4.1. Co-Expression Gene Networks and Focus Network construction
4.2. Differential Expression Analysis of IMMUNETS Genes in Melanoma Treatment Response
4.3. Validation of Candidate Immunotherapy Response Genes in an Independent Cohort
4.4. Evaluation of Candidate Biomarkers for Risk Stratification of Melanoma
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- O’Donnell, J.S.; Teng, M.W.L.; Smyth, M.J. Cancer immunoediting and resistance to T cell-based immunotherapy. Nat. Rev. Clin. Oncol. 2019, 16, 151–167. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, H.; Hagerling, C.; Werb, Z. Roles of the immune system in cancer: From tumor initiation to metastatic progression. Genes Dev. 2018, 32, 1267–1284. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y. Cancer immunotherapy: Harnessing the immune system to battle cancer. J. Clin. Investig. 2015, 125, 3335–3337. [Google Scholar] [CrossRef] [PubMed]
- Farkona, S.; Diamandis, E.P.; Blasutig, I.M. Cancer immunotherapy: The beginning of the end of cancer? BMC Med. 2016, 14, 73. [Google Scholar] [CrossRef] [PubMed]
- Hoffner, B.; Zitella, L.J. Managing Side Effects of Cancer Patients Treated with Immunotherapy. J. Adv. Pract. Oncol. 2018, 9, 287. [Google Scholar]
- Sharma, P.; Hu-Lieskovan, S.; Wargo, J.A.; Ribas, A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 2017, 168, 707–723. [Google Scholar] [CrossRef]
- Cristescu, R.; Mogg, R.; Ayers, M.; Albright, A.; Murphy, E.; Yearley, J.; Sher, X.; Liu, X.Q.; Lu, H.; Nebozhyn, M.; et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 2018, 362, 6411. [Google Scholar] [CrossRef]
- Cabrita, R.; Lauss, M.; Sanna, A.; Donia, M.; Skaarup Larsen, M.; Mitra, S.; Johansson, I.; Phung, B.; Harbst, K.; Vallon-Christersson, J.; et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature 2020, 577, 561–565. [Google Scholar] [CrossRef]
- Mukherjee, S. Genomics-Guided Immunotherapy for Precision Medicine in Cancer. Cancer Biother. Radiopharm. 2019, 34, 487–497. [Google Scholar] [CrossRef]
- Emens, L.A.; Ascierto, P.A.; Darcy, P.K.; Demaria, S.; Eggermont, A.M.M.; Redmond, W.L.; Seliger, B.; Marincola, F.M. Cancer immunotherapy: Opportunities and challenges in the rapidly evolving clinical landscape. Eur. J. Cancer 2017, 81, 116–129. [Google Scholar] [CrossRef]
- Horn, L.A.; Fousek, K.; Palena, C. Tumor Plasticity and Resistance to Immunotherapy. Trends Cancer 2020, 6, 432–441. [Google Scholar] [CrossRef] [PubMed]
- Oldenhuis, C.N.A.M.; Oosting, S.F.; Gietema, J.A.; de Vries, E.G.E. Prognostic versus predictive value of biomarkers in oncology. Eur. J. Cancer 2008, 44, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Leonardi, G.C.; Falzone, L.; Salemi, R.; Zanghi, A.; Spandidos, D.A.; Mccubrey, J.A.; Candido, S.; Libra, M. Cutaneous melanoma: From pathogenesis to therapy (Review). Int. J. Oncol. 2018, 52, 1071–1080. [Google Scholar] [CrossRef]
- Hodi, F.S.; O’Day, S.J.; McDermott, D.F.; Weber, R.W.; Sosman, J.A.; Haanen, J.B.; Gonzalez, R.; Robert, C.; Schadendorf, D.; Hassel, J.C.; et al. Improved Survival with Ipilimumab in Patients with Metastatic Melanoma. N. Engl. J. Med. 2010, 363, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Schachter, J.; Ribas, A.; Long, G.V.; Arance, A.; Grob, J.J.; Mortier, L.; Daud, A.; Carlino, M.S.; McNeil, C.; Lotem, M.; et al. Pembrolizumab versus ipilimumab for advanced melanoma: Final overall survival results of a multicentre, randomised, open-label phase 3 study (KEYNOTE-006). Lancet 2017, 390, 1853–1862. [Google Scholar] [CrossRef]
- Robert, C.; Ribas, A.; Schachter, J.; Arance, A.; Grob, J.J.; Mortier, L.; Daud, A.; Carlino, M.S.; McNeil, C.M.; Lotem, M.; et al. Pembrolizumab versus ipilimumab in advanced melanoma (KEYNOTE-006): Post-hoc 5-year results from an open-label, multicentre, randomised, controlled, phase 3 study. Lancet Oncol. 2019, 20, 1239–1251. [Google Scholar] [CrossRef]
- Turanli, B.; Karagoz, K.; Gulfidan, G.; Sinha, R.; Mardinoglu, A.; Arga, K.Y. A Network-Based Cancer Drug Discovery: From Integrated Multi-Omics Approaches to Precision Medicine. Curr. Pharm. Des. 2019, 24, 3778–3790. [Google Scholar] [CrossRef]
- Demchak, B.; Kreisberg, J.F.; Bass, J.I.F. Theory and Application of Network Biology Toward Precision Medicine. J. Mol. Biol. 2018, 430, 2873–2874. [Google Scholar] [CrossRef]
- Abbas, A.R.; Baldwin, D.; Ma, Y.; Ouyang, W.; Gurney, A.; Martin, F.; Fong, S.; van Lookeren Campagne, M.; Godowski, P.; Williams, P.M.; et al. Immune response in silico (IRIS): Immune-specific genes identified from a compendium of microarray expression data. Genes Immun. 2005, 6, 319–331. [Google Scholar] [CrossRef]
- Riaz, N.; Havel, J.J.; Makarov, V.; Horak, C.E.; Weinhold, N.; Chan, T.A. Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab Article Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. Cell 2017, 171, 934–949. [Google Scholar] [CrossRef]
- Topalian, S.L.; Hodi, F.S.; Brahmer, J.R.; Geetinger, S.N.; Smith, D.C.; McDermott, D.F.; Powderly, J.D.; Cervajal, R.D.; Sosman, J.A.; Atkins, M.B.; et al. Safety, Activity, and Immune Correlates of Anti–PD-1 Antibody in Cancer. N. Engl. J. Med. 2012, 366, 2443–2454. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Schilling, B.; Liu, D.; Sucker, A.; Livingstone, E.; Jerby-Arnon, L.; Zimmer, L.; Gutzmer, R.; Satzger, I.; Loquai, C.; et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat. Med. 2019, 25, 1916–1927. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Network. Genomic Classification of Cutaneous Melanoma. Cell 2015, 161, 1681–1696. [Google Scholar] [CrossRef] [PubMed]
- Crinier, A.; Milpied, P.; Escaliere, B.; Piperoglou, C.; Galluso, J.; Balsamo, A.; Spinelli, L.; Cervera-Marzal, I.; Ebbo, M.; Girard-Madoux, M.; et al. High-Dimensional Single-Cell Analysis Identifies Organ-Specific Signatures and Conserved NK Cell Subsets in Humans and Mice. Immunity 2018, 49, 971.e5–986.e5. [Google Scholar] [CrossRef]
- Zheng, C.; Zheng, L.; Yoo, J.K.; Guo, H.; Zhang, Y.; Guo, X.; Kang, B.; Hu, R.; Huang, J.Y.; Zhang, Q.; et al. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell 2017, 169, 1342.e16–1356.e16. [Google Scholar] [CrossRef] [PubMed]
- Overton, I.M.; Sims, A.H.; Owen, J.A.; Heale, B.S.E.; Ford, M.J.; Lubbock, A.L.R.; Pairo-Castineira, E.; Essafi, A. Functional Transcription Factor Target Networks Illuminate Control of Epithelial Remodelling. Cancers 2020, 12, 2823. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Blom, U.M.; Wang, P.I.; Shim, J.E.; Marcotte, E.M. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Res. 2011, 21, 1109–1121. [Google Scholar] [CrossRef]
- Theofilopoulos, A.N.; Dummer, W.; Kono, D.H. T cell homeostasis and systemic autoimmunity. J. Clin. Investig. 2001, 108, 335–340. [Google Scholar] [CrossRef]
- Raingeaud, J.; Whitmarsh, A.J.; Barrett, T.; Dérijard, B.; Davis, R.J. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol. Cell. Biol. 1996, 16, 1247–1255. [Google Scholar] [CrossRef]
- Dey, M.; Huff, W.X.; Kwon, J.H.; Henriquez, M.; Fetcko, K. The evolving role of CD8+ CD28− immunosenescent T cells in cancer immunology. Int. J. Mol. Sci. 2019, 20, 2810. [Google Scholar] [CrossRef]
- Aandahl, E.M.; Sandberg, J.K.; Beckerman, K.P.; Taskén, K.; Moretto, W.J.; Nixon, D.F. CD7 Is a Differentiation Marker That Identifies Multiple CD8 T Cell Effector Subsets. J. Immunol. 2003, 170, 2349–2355. [Google Scholar] [CrossRef] [PubMed]
- Tabbekh, M.; Mokrani-Hammani, M.; Bismuth, G.; Mami-Chouaib, F. T-cell modulatory properties of CD5 and its role in antitumor immune responses. OncoImmunology 2013, 2, 1. [Google Scholar] [CrossRef] [PubMed]
- Junttila, I.S. Tuning the cytokine responses: An update on interleukin (IL)-4 and IL-13 receptor complexes. Front. Immunol. 2018, 9, 888. [Google Scholar] [CrossRef]
- Kelly, B.L.; Locksley, R.M. Coordinate Regulation of the IL-4, IL-13, and IL-5 Cytokine Cluster in Th2 Clones Revealed by Allelic Expression Patterns. J. Immunol. 2000, 165, 2982–2986. [Google Scholar] [CrossRef]
- Ouyang, W.; Kolls, J.K.; Zheng, Y. The Biological Functions of T Helper 17 Cell Effector Cytokines in Inflammation. Immunity 2008, 28, 454–467. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Bechara, R.; Zhao, J.; McGeachy, M.J.; Gaffen, S.L. IL-17 receptor–based signaling and implications for disease. Nature Immunology 2019, 20, 1594–1602. [Google Scholar] [CrossRef]
- Minguet, S.; Swamy, M.; Alarcon, B.; Luescher, I.F.; Schamel, W.W.A. Full activation of the T cell receptor requires both clustering and conformational changes at CD3. Immunity 2007, 26, 43–54. [Google Scholar] [CrossRef]
- Hardy, I.R.; Anceriz, N.; Rousseau, F.; Seefeldt, M.B.; Hatterer, E.; Irla, M.; Buatois, V.; Chatel, L.E.; Getahun, A.; Fletcher, A.; et al. Anti-CD79 Antibody Induces B Cell Anergy That Protects against Autoimmunity. J. Immunol. 2014, 192, 1641–1650. [Google Scholar] [CrossRef]
- Wang, K.; Wei, G.; Liu, D. CD19: A biomarker for B cell development, lymphoma diagnosis and therapy. Exp. Hematol. Oncol. 2012, 1, 36. [Google Scholar] [CrossRef]
- Clark, E.A.; Giltiay, N.V. CD22: A Regulator of Innate and Adaptive B Cell Responses and Autoimmunity. Front. Immunol. 2018, 9, 2235. [Google Scholar] [CrossRef]
- Boross, P.; Leusen, J.H.W. Mechanisms of action of CD20 antibodies. Am. J. Cancer Res. 2012, 2, 676–690. [Google Scholar]
- Yu, Y.; Wang, J.; Khaled, W.; Burke, S.; Li, P.; Chen, X.; Yang, W.; Jenkins, N.A.; Copeland, N.G.; Zhang, S.; et al. Bcl11a is essential for lymphoid development and negatively regulates p53. J. Exp. Med. 2012, 209, 2467–2483. [Google Scholar] [CrossRef]
- Cobaleda, C.; Schebesta, A.; Delogu, A.; Busslinger, M. Pax5: The guardian of B cell identity and function. Nat. Immunol. 2007, 8, 463–470. [Google Scholar] [CrossRef]
- Rutz, S.; Wang, X.; Ouyang, W. The IL-20 subfamily of cytokines-from host defence to tissue homeostasis. Nat. Rev. Immunol. 2014, 14, 783–795. [Google Scholar] [CrossRef]
- Barone, F.; Nayar, S.; Campos, J.; Cloake, T.; Withers, D.R.; Toellner, K.M.; Zhang, Y.; Fouser, L.; Fisher, B.; Bowman, S.; et al. IL-22 regulates lymphoid chemokine production and assembly of tertiary lymphoid organs. Proc. Natl. Acad. Sci. USA 2015, 112, 11024–11029. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, Z.; Graner, M.W.; Yang, L.; Liao, M.; Yang, Q.; Gou, J.; Zhu, Y.; Wu, C.; Liu, H.; et al. B cell infiltration is associated with the increased IL-17 and IL-22 expression in the lungs of patients with tuberculosis. Cell. Immunol. 2011, 270, 217–223. [Google Scholar] [CrossRef]
- He, Y.; Yang, Y.; Xu, J.; Liao, Y.; Liu, L.; Deng, L.; Xiong, X. IL22 drives cutaneous melanoma cell proliferation, migration and invasion through activation of miR-181/STAT3/AKT axis. J. Cancer 2020, 11, 2679–2687. [Google Scholar] [CrossRef]
- ClinicalTrials.gov. PH1 Biomarker Study of Nivolumab and Ipilimumab and Nivolumab in Combination with Ipilimumab in Advanced Melanoma (PD-1). Available online: https://clinicaltrials.gov/ct2/show/NCT01621490 (accessed on 1 December 2020).
- Francisco, L.M.; Sage, P.T.; Sharpe, A.H. The PD-1 pathway in tolerance and autoimmunity. Immunol. Rev. 2010, 236, 219–242. [Google Scholar] [CrossRef] [PubMed]
- Taube, J.M.; Klein, A.; Brahmer, J.R.; Xu, H.; Pan, X.; Kim, J.H.; Chen, L.; Pardoll, D.M.; Topalian, S.L.; Anders, R.A. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin. Cancer Res. 2014, 20, 5064–5074. [Google Scholar] [CrossRef]
- Liu, X.; Guo, C.Y.; Tou, F.F.; Wen, X.M.; Kuang, Y.K.; Zhu, Q.; Hu, H. Association of PD-L1 expression status with the efficacy of PD-1/PD-L1 inhibitors and overall survival in solid tumours: A systematic review and meta-analysis. Int. J. Cancer 2020, 147, 116–127. [Google Scholar] [CrossRef]
- Okada, M.; Cheeseman, I.M.; Hori, T.; Okawa, K.; McLeod, I.X.; Yates, J.R., 3rd; Desai, A.; Fukagawa, T. The cenp-H–I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 2006, 8, 446–457. [Google Scholar] [CrossRef]
- Ding, N.; Li, R.; Shi, W.; He, C. CENPI is overexpressed in colorectal cancer and regulates cell migration and invasion. Gene 2018, 674, 80–86. [Google Scholar] [CrossRef]
- Reinherz, E.L.; Schlossman, S.F. The differentiation and function of human T lymphocytes. Cell 1980, 19, 821–827. [Google Scholar] [CrossRef]
- Espinosa, E.; Márquez-Rodas, I.; Soria, A.; Berrocal, A.; Manzano, J.L.; Gonzalez-Cao, M.; Martin-Algarra, S.; Spanish Melanoma Group (GEM). Predictive factors of response to immunotherapy-a review from the Spanish Melanoma Group (GEM). Ann. Transl. Med. 2017, 5, 6. [Google Scholar] [CrossRef]
- Porciello, N.; Kunkl, M.; Viola, A.; Tuosto, L. Phosphatidylinositol 4-phosphate 5-kinases in the regulation of T cell activation. Front. Immunol. 2016, 7, 186. [Google Scholar] [CrossRef]
- van Vlodrop, I.J.; Baldewijns, M.M.; Smits, K.M.; Schouten, L.J.; van Neste, L.; van Criekinge, W.; van Poppel, H.; Lerut, E.; Schuebel, K.E.; Ahuja, N.; et al. Prognostic significance of Gremlin1 (GREM1) promoter CpG island hypermethylation in clear cell renal cell carcinoma. Am. J. Pathol. 2010, 176, 575–584. [Google Scholar] [CrossRef]
- Kovalova, N.; Nault, R.; Crawford, R.; Zacharewski, T.R.; Kaminski, N.E. Comparative analysis of TCDD-induced AhR-mediated gene expression in human, mouse and rat primary B cells. Toxicol. Appl. Pharmacol. 2017, 316, 95–106. [Google Scholar] [CrossRef]
- Lu, E.; Cyster, J.G. G-protein coupled receptors and ligands that organize humoral immune responses. Immunol. Rev. 2019, 289, 158–172. [Google Scholar] [CrossRef]
- Roy, S.; Bag, A.K.; Singh, R.K.; Talmadge, J.E.; Batra, S.K.; Datta, K. Multifaceted role of neuropilins in the immune system: Potential targets for immunotherapy. Front. Immunol. 2017, 8, 1228. [Google Scholar] [CrossRef]
- Ngoenkam, J.; Schamel, W.W.; Pongcharoen, S. Selected signalling proteins recruited to the T-cell receptor-CD3 complex. Immunology 2018, 153, 42–50. [Google Scholar] [CrossRef]
- Schmidt, T.; Samaras, P.; Frejno, M.; Gessulat, S.; Barnert, M.; Kienegger, H.; Krcmar, H.; Schlegl, J.; Ehrlich, H.C.; Aiche, S.; et al. ProteomicsDB. Nucleic Acids Res. 2018, 46, D1271–D1281. [Google Scholar] [CrossRef]
- Goodall, K.J.; Nguyen, A.; McKenzie, C.; Eckle, S.; Sullivan, L.C.; Andrews, D.M. The murine CD94/NKG2 ligand, Qa-1 b, is a high-affinity, functional ligand for the CD8αα homodimer. J. Biol. Chem. 2020, 295, 3239–3246. [Google Scholar] [CrossRef]
- McGinn, O.J.; English, W.R.; Roberts, S.; Ager, A.; Newham, P.; Murphy, G. Modulation of integrin α4β1 by ADAM28 promotes lymphocyte adhesion and transendothelial migration. Cell Biol. Int. 2011, 35, 1043–1053. [Google Scholar] [CrossRef]
- Chapman, K.E.; Coutinho, A.E.; Gray, M.; Gilmour, J.S.; Savill, J.S.; Seckl, J.R. The role and regulation of 11β-hydroxysteroid dehydrogenase type 1 in the inflammatory response. Mol. Cell. Endocrinol. 2009, 301, 123–131. [Google Scholar] [CrossRef]
- Liu, N.; Jiang, Y.; Chen, J.; Nan, H.; Zhao, Y.; Chu, X.; Wang, A.; Wang, D.; Qin, T.; Gao, S.; et al. IL-33 drives the antitumor effects of dendritic cells via the induction of Tc9 cells. Cell. Mol. Immunol. 2019, 16, 644–651. [Google Scholar] [CrossRef]
- Liew, F.Y.; Pitman, N.I.; McInnes, I.B. Disease-associated functions of IL-33: The new kid in the IL-1 family. Nat. Rev. Immunol. 2010, 10, 103–110. [Google Scholar] [CrossRef]
- Gérard, C.; Hubeau, C.; Carnet, O.; Bellefroid, M.; Sounni, N.E.; Blacher, S.; Bendavid, G.; Moser, M.; Fässler, R.; Noel, A.; et al. Microenvironment-derived ADAM28 prevents cancer dissemination. Oncotarget 2018, 9, 37185–37199. [Google Scholar] [CrossRef]
- Hubeau, C.; Rocks, N.; Cataldo, D. ADAM28: Another ambivalent protease in cancer. Cancer Lett. 2020, 494, 18–26. [Google Scholar] [CrossRef]
- Huang, G.N.; Huso, D.L.; Bouyain, S.; Tu, J.; McCorkell, K.A.; May, M.J.; Zhu, Y.; Lutz, M.; Collins, S.; Dehoff, M.; et al. NFAT binding and regulation of T cell activation by the cytoplasmic scaffolding homer proteins. Science 2008, 319, 476–481. [Google Scholar] [CrossRef]
- Fagni, L.; Worley, P.F.; Ango, F. Homer as both a scaffold and transduction molecule. Sci. STKE Signal Transduct. Knowl. Environ. 2002, 137, re8. [Google Scholar] [CrossRef]
- Liu, S.-T.; Hittle, J.C.; Jablonski, S.A.; Campbell, M.S.; Yoda, K.; Yen, T.J. Human CENP-I specifies localization of CENP-F, MAD1 and MAD2 to kinetochores and is essential for mitosis. Nat. Cell Biol. 2003, 5, 341–345. [Google Scholar] [CrossRef]
- Feng, X.; Zhang, C.; Zhu, L.; Zhang, L.; Li, H.; He, L.; Mi, Y.; Wang, Y.; Zhu, J.; Bu, Y. DEPDC1 is required for cell cycle progression and motility in nasopharyngeal carcinoma. Oncotarget 2017, 8, 63605–63619. [Google Scholar] [CrossRef]
- Lu, S.; Qian, J.; Guo, M.; Gu, C.; Yang, Y. Insights into a Crucial Role of TRIP13 in Human Cancer. Comput. Struct. Biotechnol. J. 2019, 17, 854–861. [Google Scholar] [CrossRef]
- Brakeman, P.R.; Lanahan, A.A.; O’Brien, R.; Roche, K.; Barnes, C.A.; Huganir, R.L.; Worley, P.F. Homer: A protein that selectively binds metabotropic glutamate receptors. Nature 1997, 386, 284–288. [Google Scholar] [CrossRef]
- Duncan, R.S.; Hwang, S.Y.; Koulen, P. Effects of Vesl/Homer proteins on intracellular signaling. Exp. Biol. Med. 2005, 230, 527–535. [Google Scholar] [CrossRef]
- Lominac, K.D.; Oleson, E.B.; Pava, M.; Klugmann, M.; Schwarz, M.K.; Seeburg, P.H.; During, M.J.; Worley, P.F.; Kalivas, P.W.; Szumlinski, K.K. Distinct Roles for Different Homer1 Isoforms in Behaviors and Associated Prefrontal Cortex Function. J. Neurosci. 2005, 25, 11586–11594. [Google Scholar] [CrossRef]
- Bilska, A.; Kusio-Kobiałka, M.; Krawczyk, P.S.; Gewartowska, O.; Tarkowski, B.; Kobyłecki, K.; Nowis, D.; Golab, J.; Gruchota, J.; Borsuk, E.; et al. Immunoglobulin expression and the humoral immune response is regulated by the non-canonical poly(A) polymerase TENT5C. Nat. Commun. 2020, 11, 1–17. [Google Scholar] [CrossRef]
- Luisiri, P.; Lee, Y.J.; Eisfelder, B.J.; Clark, M.R. Cooperativity and Segregation of Function within the Ig-α/β Heterodimer of the B Cell Antigen Receptor Complex (∗). J. Biol. Chem. 1996, 271, 5158–5163. [Google Scholar] [CrossRef]
- Chang, C.H.; Fontes, J.D.; Peterlin, M.; Flavell, R.A. Class II transactivator (CIITA) is sufficient for the inducible expression of major histocompatibility complex class II genes. J. Exp. Med. 1994, 180, 1367–1374. [Google Scholar] [CrossRef]
- Martin, B.K.; Chin, K.C.; Olsen, J.C.; Skinner, C.A.; Dey, A.; Ozato, K.; Ting, J.P. Induction of MHC Class I Expression by the MHC Class II Transactivator CIITA. Immunity 1997, 6, 591–600. [Google Scholar] [CrossRef]
- Wang, J.H.; Avitahl, N.; Cariappa, A.; Friedrich, C.; Ikeda, T.; Renold, A.; Andrikopoulos, K.; Liang, L.; Pillai, S.; Morgan, B.A.; et al. Aiolos Regulates B Cell Activation and Maturation to Effector State. Immunity 1998, 9, 543–553. [Google Scholar] [CrossRef]
- Fernandez, I.Z.; Baxter, R.M.; Garcia-Perez, J.E.; Vendrame, E.; Ranganath, T.; Kong, D.S.; Lundquist, K.; Nguyen, T.; Ogolla, S.; Black, J.; et al. A novel human IL2RB mutation results in T and NK cell–driven immune dysregulation. J. Exp. Med. 2019, 216, 1255–1267. [Google Scholar] [CrossRef] [PubMed]
- Barber, E.K.; Dasgupta, J.D.; Schlossman, S.F.; Trevillyan, J.M.; Rudd, C.E. The CD4 and CD8 antigens are coupled to a protein-tyrosine kinase (p56lck) that phosphorylates the CD3 complex. Proc. Natl. Acad. Sci. USA 1989, 86, 3277–3281. [Google Scholar] [CrossRef]
- Doyle, C.; Strominger, J.L. Interaction between CD4 and class II MHC molecules mediates cell adhesion. Nature 1987, 330, 256–259. [Google Scholar] [CrossRef]
- Uhlen, M.; Karlsson, M.J.; Zhong, W.; Tebani, A.; Pou, C.; Mikes, J.; Lakshmikanth, T.; Forsström, B.; Edfors, F.; Odeberg, J.; et al. A genome-wide transcriptomic analysis of protein-coding genes in human blood cells. Science 2019, 366, 6472. [Google Scholar] [CrossRef]
- Balbas, M.D.; Burgess, M.R.; Murali, R.; Wongvipat, J.; Skaggs, B.J.; Mundel, P.; Weins, A.; Sawyers, C.L. MAGI-2 scaffold protein is critical for kidney barrier function. Proc. Natl. Acad. Sci. USA 2014, 111, 14876–14881. [Google Scholar] [CrossRef]
- Wu, X.; Hepner, K.; Castelino-Prabhu, S.; Do, D.; Kaye, M.B.; Yuan, X.J.; Wood, J.; Ross, C.; Sawyers, C.L.; Whang, Y.E. Evidence for regulation of the PTEN tumor suppressor by a membrane-localized multi-PDZ domain containing scaffold protein MAGI-2. Proc. Natl. Acad. Sci. USA 2000, 97, 4233–4238. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate—A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Freeman, S.S.; Sade-Feldman, M.; Kim, J.; Stewart, C.; Gonye, A.; Ravi, A.; Arniella, M.B.; Gushterova, I.; LaSalle, T.J.; Blaum, E.M.; et al. Combined tumor and immune signals from genomes or transcriptomes predict outcomes of checkpoint inhibition in melanoma. Cell Rep. Med. 2022, 3, 100500. [Google Scholar] [CrossRef]
- Van Allen, E.M.; Miao, D.; Schilling, B.; Shukla, S.A.; Blank, C.; Zimmer, L.; Sucker, A.; Hillen, U.; Foppen, M.; Goldinger, S.M.; et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015, 350, 207–211. [Google Scholar] [CrossRef]
- Hugo, W.; Zaretsky, J.M.; Sun, L.; Song, C.; Moreno, B.H.; Hu-Lieskovan, S.; Berent-Maoz, B.; Pang, J.; Chmielowski, B.; Cherry, G.; et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell 2016, 165, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Gide, T.N.; Quek, C.; Menzies, A.M.; Tasker, A.T.; Shang, P.; Holst, J.; Madore, J.; Lim, S.Y.; Velickovic, R.; Wongchenko, M.; et al. Distinct Immune Cell Populations Define Response to Anti-PD-1 Monotherapy and Anti-PD-1/Anti-CTLA-4 Combined Therapy. Cancer Cell 2019, 35, 238.e6–255.e6. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.R. Regression Models and Life-Tables. J. R. Stat. Society. Ser. B Methodol. 1972, 34, 187–220. [Google Scholar] [CrossRef]
- Akaike, H. Information Theory and an Extension of the Maximum Likelihood Principle; Akademiai: Budapest, Hungary, 1998; pp. 199–213. [Google Scholar] [CrossRef]
- Machado, J.A.L.; Steimle, V. The MHC Class II Transactivator CIITA: Not (Quite) the Odd-One-Out Anymore among NLR Proteins. Int. J. Mol. Sci. 2021, 22, 1074. [Google Scholar] [CrossRef] [PubMed]
- Morgan, B.; Sun, L.; Avitahl, N.; Andrikopoulos, K.; Ikeda, T.; Gonzales, E.; Wu, P.; Neben, S.; Georgopoulos, K. Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. EMBO J. 1997, 16, 2004–2013. [Google Scholar] [CrossRef]
- Romero, F.; Martinez, C.; Camonis, J.; Rebollo, A. Aiolos transcription factor controls cell death in T cells by regulating Bcl-2 expression and its cellular localization. EMBO J. 1999, 18, 3419–3430. [Google Scholar] [CrossRef]
- Ma, S.; Pathak, S.; Mandal, M.; Trinh, L.; Clark, M.R.; Lu, R. Ikaros and Aiolos inhibit pre-B-cell proliferation by directly suppressing c-Myc expression. Mol. Cell Biol. 2010, 30, 4149–4158. [Google Scholar] [CrossRef]
- Holmes, M.L.; Huntington, N.D.; Thong, R.P.; Brady, J.; Hayakawa, Y.; Andoniou, C.E.; Fleming, P.; Shi, W.; Smyth, G.K.; Degli-Esposti, M.A.; et al. Peripheral natural killer cell maturation depends on the transcription factor Aiolos. EMBO J. 2014, 33, 2721–2734. [Google Scholar] [CrossRef]
- Davis, M.M.; Tato, C.M.; Furman, D. Systems immunology: Just getting started. Nat. Immunol. 2017, 18, 725–732. [Google Scholar] [CrossRef]
- Nirmal, A.J.; Regan, T.; Shih, B.B.; Hume, D.A.; Sims, A.H.; Freeman, T.C. Immune Cell Gene Signatures for Profiling the Microenvironment of Solid Tumors. Cancer Immunol. Res. 2018, 6, 1388–1400. [Google Scholar] [CrossRef]
- Neagu, M.; Constantin, C.; Tanase, C. Immune-related biomarkers for diagnosis/prognosis and therapy monitoring of cutaneous melanoma. Expert review of molecular diagnostics. 2010, 10, 897–919. [Google Scholar] [CrossRef] [PubMed]
- Steimle, V.; Siegrist, C.A.; Mottet, A.; Lisowska-Grospierre, B.; Mach, B. Regulation of MHC class II expression by interferon-gamma mediated by the transactivator gene CIITA. Science. 1994, 265, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Mkrtichyan, M.; Najjar, Y.G.; Raulfs, E.C.; Abdalla, M.Y.; Samara, R.; Rotem-Yehudar, R.; Cook, L.; Khleif, S.N. Anti-PD-1 synergizes with cyclophosphamide to induce potent anti-tumor vaccine effects through novel mechanisms. Eur. J. Immunol. 2011, 41, 2977–2986. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.A.; Patel, V.G. The role of PD-L1 expression as a predictive biomarker: An analysis of all US food and drug administration (FDA) approvals of immune checkpoint inhibitors. J. ImmunoTher. Cancer 2019, 7, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Müller, T.; Braun, M.; Dietrich, D.; Aktekin, S.; Höft, S.; Kristiansen, G.; Göke, F.; Schröck, A.; Brägelmann, J.; Held, S.; et al. PD-L1: A novel prognostic biomarker in head and neck squamous cell carcinoma. Oncotarget 2017, 32, 52889. [Google Scholar] [CrossRef]
- Baptista, M.Z.; Sarian, L.O.; Derchain, S.F.M.; Pinto, G.A.; Vassallo, J. Prognostic significance of PD-L1 and PD-L2 in breast cancer. Hum. Pathol. 2016, 47, 78–84. [Google Scholar] [CrossRef]
- Gelman, A. Analysis of Variance. Ann. Stat. 2005, 33, 1–53. [Google Scholar] [CrossRef]
- Glazko, G.; Mushegian, A. Measuring gene expression divergence: The distance to keep. Biol. Direct 2010, 5, 1–10. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2001, 13, 2498–2504. [Google Scholar] [CrossRef]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Sha, Y.; Phan, J.H.; Wang, M.D. Effect of low-expression gene filtering on detection of differentially expressed genes in RNA-seq data. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2015, 2015, 6461. [Google Scholar] [CrossRef]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef] [PubMed]
- Blom, G. Statistical Estimates and Transformed Beta Variables. Inc. Stat. 1960, 10, 53. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Collins, J.R.; Alvord, W.G.; Roayaei, J.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. The DAVID Gene Functional Classification Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007, 8, 1–16. [Google Scholar] [CrossRef]
- Lubbock, A.L.R.; Katz, E.; Harrison, D.J.; Overton, I.M. TMA Navigator: Network inference, patient stratification and survival analysis with tissue microarray data. Nucleic Acids Res. 2013, 41, W562. [Google Scholar] [CrossRef]
- Amin, M.B.; Edge, S.; Greene, F.; Byrd, D.R.; Brookland, R.K.; Washington, M.K.; Gershenwald, J.E.; Compton, C.C.; Hess, K.R.; Sullivan, D.C.; et al. AJCC Cancer Staging Manual, 8th ed.; Springer International Publishing: New York, NY, USA, 2017. [Google Scholar]
- Grambsch, P.M.; Therneau, T.M. Proportional Hazards Tests and Diagnostics Based on Weighted Residuals. Biometrika 1994, 81, 515. [Google Scholar] [CrossRef]



| T Cell | NK Cell | B Cell | Monocyte | Dendritic Cell | |
|---|---|---|---|---|---|
| T cell | 233 | - | - | - | - |
| NK cell | 357 | 935 | - | - | - |
| B cell | 131 | 535 | 1022 | - | - |
| Monocyte | 58 | 171 | 120 | 233 | - |
| Dendritic cell | 143 | 750 | 333 | 222 | 839 |
| Dataset | Biological Descriptor (s) | Score | Genes |
|---|---|---|---|
| MEL_NAI | Cell cycle, Cell division, Mitosis | 1.90 | BUB1, ERCC6L, CENPF, CENPI, NCAPG2, KIF14, BIN3, DTL, DEPDC1, PIP5K1A, CDH1, AKR1C3, STAP2 |
| MEL_PROG | Immunoreceptor signaling, ITAM | 2.17 | CD247, CD79A, CD3G, CD72, CD4 |
| MEL_PROG | Immunity, Adaptive Immunity | 1.81 | CD180, CD79A, SKAP1, CD4, CD209, C1RL, MAP3K5, CD8A, C3, CLU |
| MEL_PROG | Regulation of immune response, including T cell receptor signaling | 1.79 | CD247, SKAP1, CD4, CD8A, CD3G, PRF1, ITGAL, TRAC, C3, CD72, CD79A, KIRREL1, PTK7 |
| MEL_PROG | Immunity, Innate Immunity | 1.44 | CD180, CD79A, SKAP1, CD4, CD209, C1RL, MAP3K5, CD8A, C3, CLU |
| MEL_PROG | Complement pathway | 1.38 | C1RL, C3, CLU, CD180, CD209, MAP3K5 |
| MEL_PROG | Antigen processing and presentation | 1.33 | CIITA, CD79A, CD4, CD8A, GZMA, C3 |
| Gene | MEL_TCGA | MI × ED_ICI |
|---|---|---|
| ADAM28 | 1.161 × 10−4 * | 1.230 × 10−2 * |
| TGM2 | 2.368 × 10−3 * | 4.607 × 10−1 |
| CD247 | 8.566 × 10−5* | 3.450 × 10−2 * |
| CD4 | 1.161 × 10−4 * | 3.757 × 10−1 |
| IKZF3 | 4.098 × 10−6 * | 9.800 × 10−4 * |
| TENT5C | 2.593 × 10−4 * | 2.015 × 10−1 |
| BTG2 | 4.684 × 10−2 * | 4.607 × 10−1 |
| HOMER1 | 9.643 × 10−2 | 1.055 × 10−1 |
| CIITA | 2.150 × 10−4 * | 6.800 × 10−4 * |
| CABYR | 5.233 × 10−1 | 2.015 × 10−1 |
| CD79A | 1.617 × 10−3 * | 6.800 × 10−4 * |
| IL2RB | 1.161 × 10−4 * | 2.670 × 10−3 * |
| MAGI2 | 1.018 × 10−1 | 5.875 × 10−1 |
| Prognostic Factor | p-Value | Hazard Ratio | 95% Confidence Interval |
|---|---|---|---|
| Age | 0.00492 * | 1.0189 | 1.00–1.03 |
| Tumour stage | 0.017 * | 1.3205 | 1.05–1.66 |
| CIITA | 0.012 * | 0.8602 | 0.76–0.97 |
| IKZF3 | 0.976 | 1.0015 | 0.91–1.11 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vo, D.H.T.; McGleave, G.; Overton, I.M. Immune Cell Networks Uncover Candidate Biomarkers of Melanoma Immunotherapy Response. J. Pers. Med. 2022, 12, 958. https://doi.org/10.3390/jpm12060958
Vo DHT, McGleave G, Overton IM. Immune Cell Networks Uncover Candidate Biomarkers of Melanoma Immunotherapy Response. Journal of Personalized Medicine. 2022; 12(6):958. https://doi.org/10.3390/jpm12060958
Chicago/Turabian StyleVo, Duong H. T., Gerard McGleave, and Ian M. Overton. 2022. "Immune Cell Networks Uncover Candidate Biomarkers of Melanoma Immunotherapy Response" Journal of Personalized Medicine 12, no. 6: 958. https://doi.org/10.3390/jpm12060958
APA StyleVo, D. H. T., McGleave, G., & Overton, I. M. (2022). Immune Cell Networks Uncover Candidate Biomarkers of Melanoma Immunotherapy Response. Journal of Personalized Medicine, 12(6), 958. https://doi.org/10.3390/jpm12060958







