Gut Microbiota Dysbiosis in Childhood Vasculitis: A Perspective Comparative Pilot Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design, Patients and Sample Collection
2.2. Microbial DNA Extraction, Library Preparation and Sequencing
2.3. Bioinformatics and Statistics
3. Results
3.1. Study Cohort Description
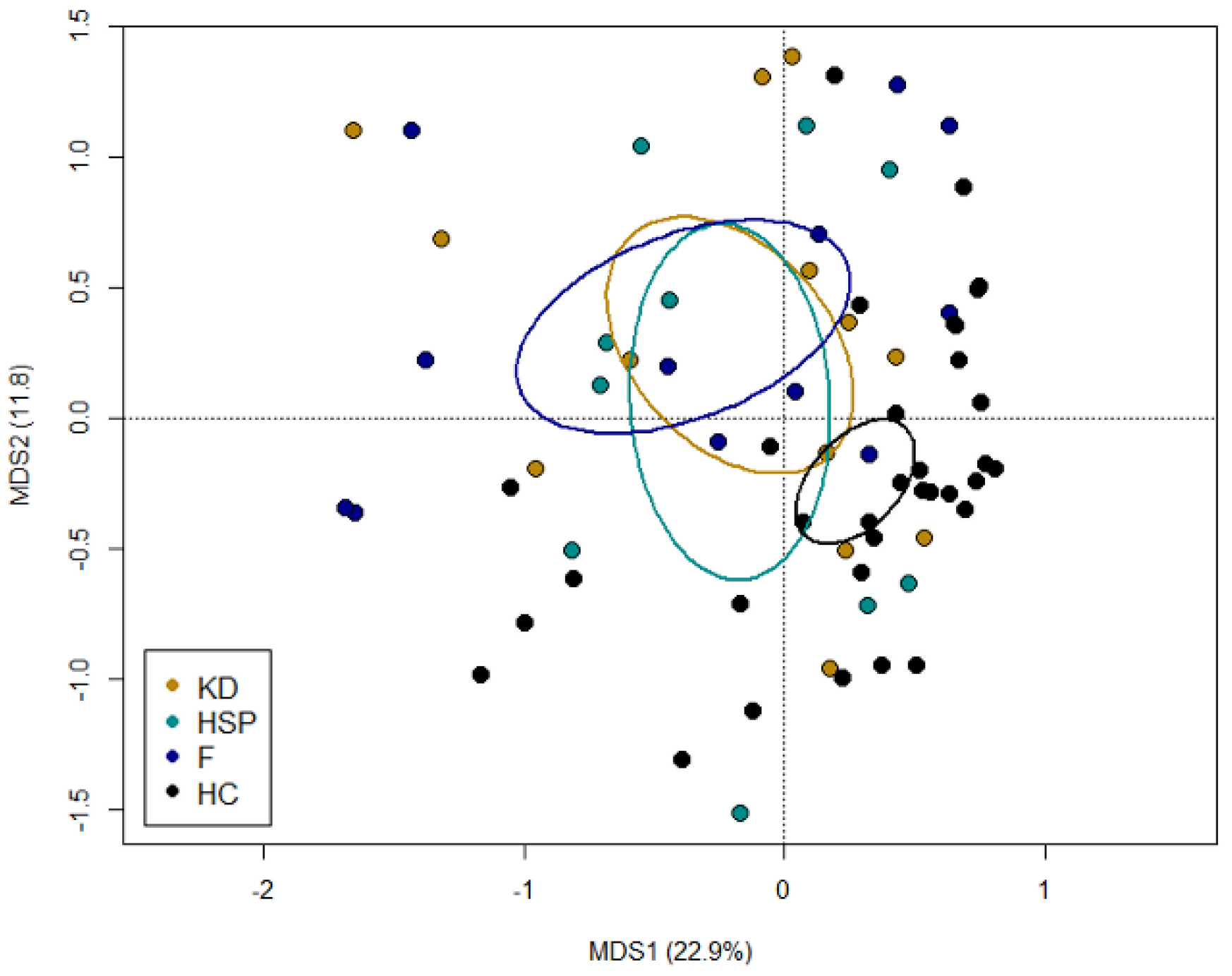
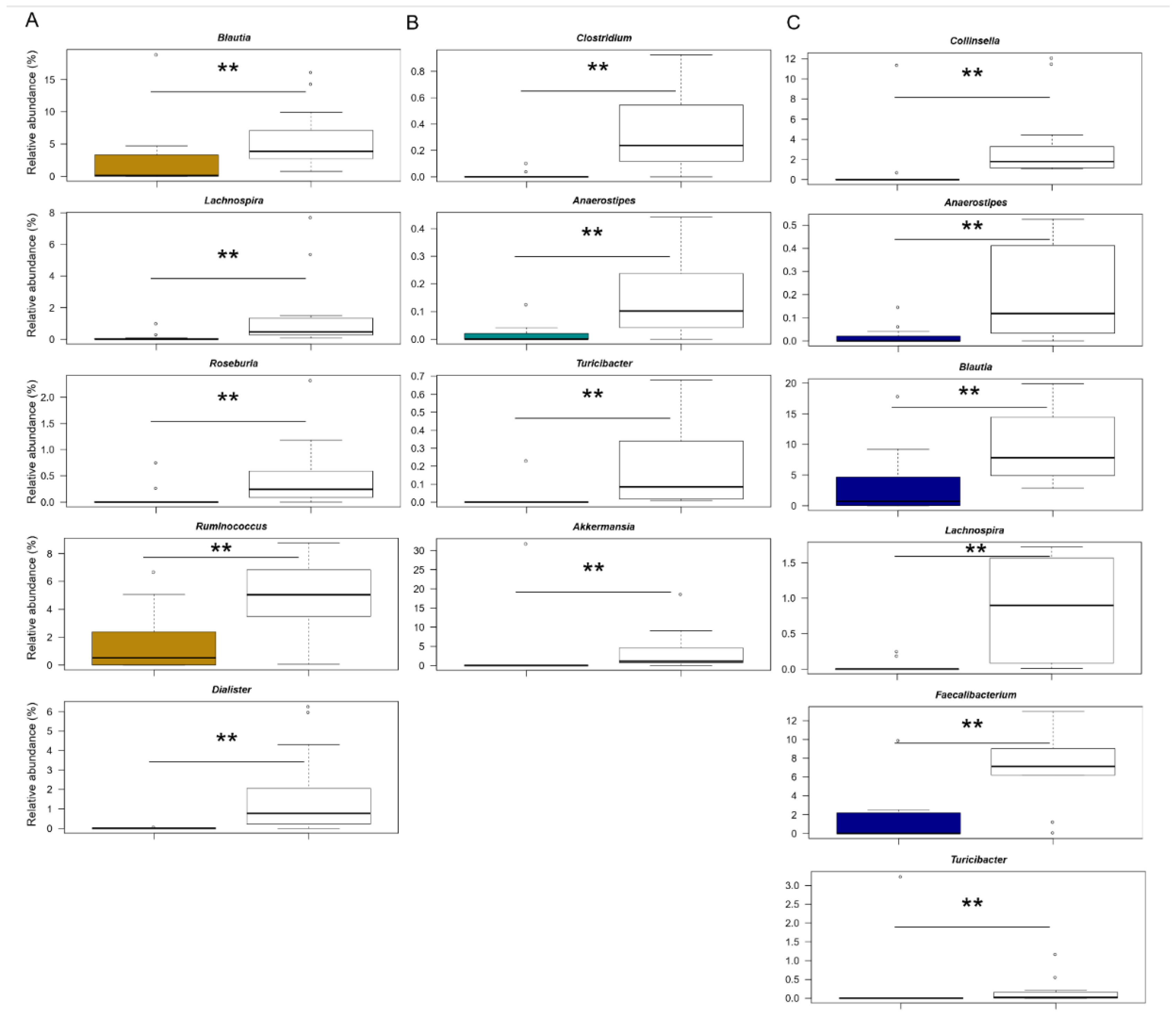
3.2. The Gut Microbiota Dysbiosis of KD, HSP and F Children
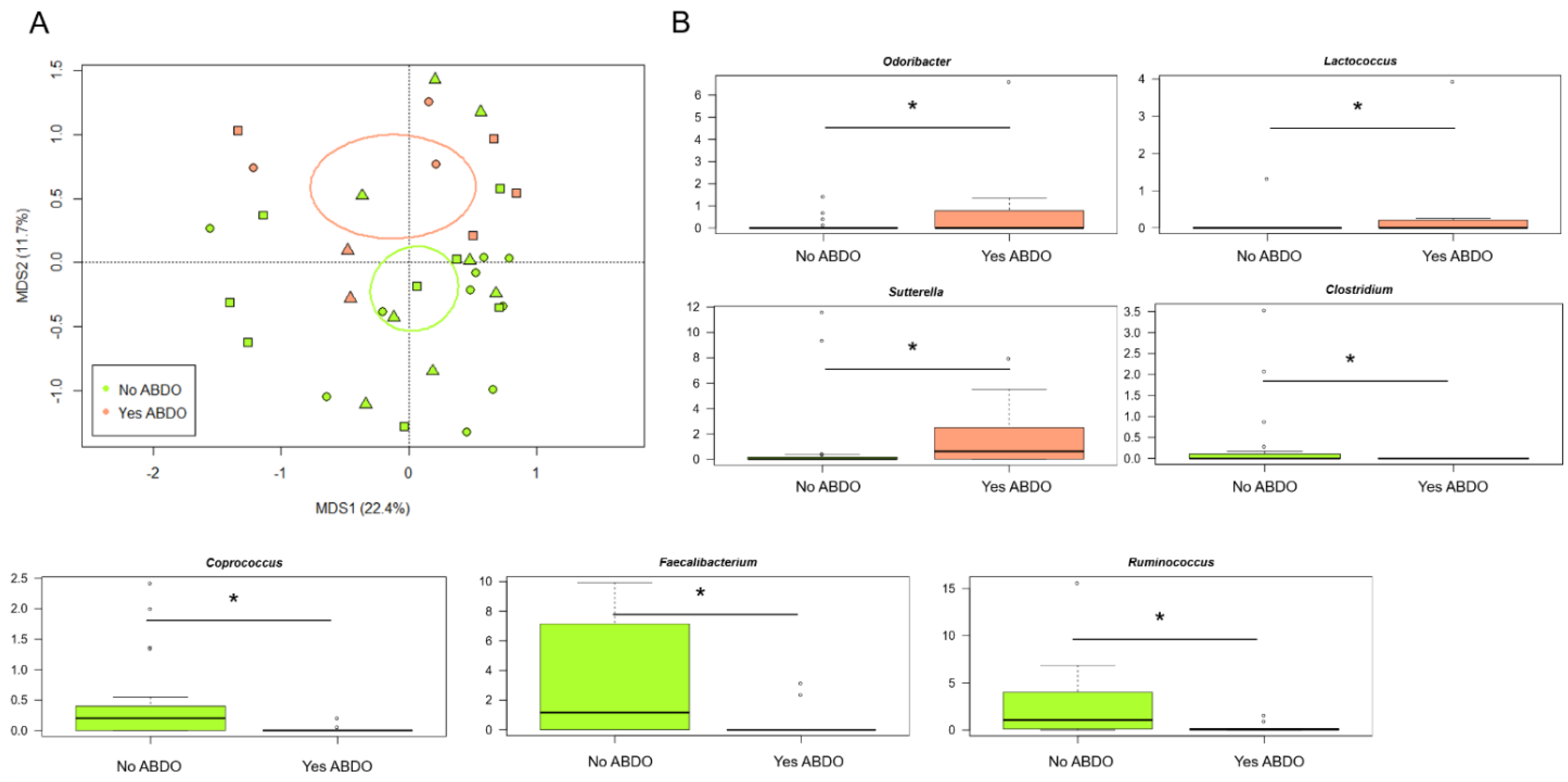
3.3. The Gut Microbiota Profiles of KD, HSP and F Children Stratify by Gastrointestinal Involvement
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Weiss, P.F. Pediatric vasculitis. Pediatrics Clin. N. Am. 2012, 59, 407–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowyer, S.; Roettcher, P. Pediatric rheumatology clinic populations in the United States: Results of a 3 year survey. J. Rheumatol. 1996, 23, 1968–1974. [Google Scholar] [PubMed]
- Hetland, L.E.; Susrud, K.S.; Lindahl, K.H.; Bygum, A. Henoch-Schönlein Purpura: A Literature Review. Acta Derm. Venereol. 2017, 97, 1160–1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozen, S.; Pistorio, A.; Iusan, S.M.; Bakkaloglu, A.; Herlin, T.; Brik, R.; Buoncompagni, A.; Lazar, C.; Bilge, I.; Uziel, Y.; et al. EULAR/PRINTO/PRES criteria for Henoch–Schönlein purpura, childhood polyarteritis nodosa, childhood Wegener granulomatosis and childhood Takayasu arteritis: Ankara 2008. Part II: Final classification criteria. Ann. Rheum. Dis. 2010, 69, 798–806. [Google Scholar] [CrossRef] [Green Version]
- McCrindle, B.W.; Rowley, A.H.; Newburger, J.W.; Burns, J.C.; Bolger, A.F.; Gewitz, M.; Baker, A.L.; Jackson, M.A.; Takahashi, M.; Shah, P.B.; et al. Diagnosis, treatment, and long-term management of Kawasaki disease: A scientific statement for health professionals from the American Heart Association. Circulation 2017, 135, e927–e999. [Google Scholar] [CrossRef]
- Rowley, A.; Baker, S.C.; Arrollo, D.; Gruen, L.J.; Bodnar, T.; Innocentini, N.; Hackbart, M.; Pulido, Y.C.; Wylie, K.M.; A Kim, K.-Y.; et al. A protein epitope targeted by the antibody response to Kawasaki disease. J. Infect. Dis. 2020, 222, 158–168. [Google Scholar] [CrossRef]
- Rowley, A.H.; Baker, S.C.; Shulman, S.T.; Rand, K.H.; Tretiakova, M.S.; Perlman, E.; Garcia, F.L.; Tajuddin, N.F.; Fox, L.M.; Huang, J.H.; et al. Ultrastructural, immunofluorescence, and RNA evidence support the hypothesis of a “new” virus associated with Kawasaki disease. J. Infect. Dis. 2011, 203, 1021–1030. [Google Scholar] [CrossRef]
- Rowley, A.H.; Baker, S.C.; Shulman, S.T.; Garcia, F.L.; Fox, L.M.; Kos, I.M.; Crawford, S.E.; Russo, P.A.; Hammadeh, R.; Takahashi, K.; et al. RNA-containing cytoplasmic inclusion bodies in ciliated bronchial epithelium months to years after acute Kawasaki disease. PLoS ONE 2008, 3, e1582. [Google Scholar] [CrossRef]
- Yim, D.; Curtis, N.; Cheung, M.; Burgner, D. Update on Kawasaki disease: Epidemiology, aetiology and pathogenesis. J. Paediatr. Child. Health 2013, 49, 704–708. [Google Scholar] [CrossRef]
- Takeshita, S.; Nakatani, K.; Kawase, H.; Seki, S.; Yamamoto, M.; Sekine, I.; Yoshioka, S. The role of bacterial lipopolysaccharide-bound neutrophils in the pathogenesis of Kawasaki disease. J. Infect. Dis. 1999, 179, 508–512. [Google Scholar] [CrossRef]
- Kim, G.B. Reality of Kawasaki disease epidemiology. Korean J. Pediatrics 2019, 62, 292–296. [Google Scholar] [CrossRef] [PubMed]
- Makino, N.; Nakamura, Y.; Yashiro, M.; Kosami, K.; Matsubara, Y.; Ae, R.; Aoyama, Y.; Yanagawa, H. Nationwide epidemiologic survey of Kawasaki disease in Japan, 2015–2016. Pediatrics Int. 2019, 61, 397–403. [Google Scholar] [CrossRef]
- Mauro, A.; Fabi, M.; Da Frè, M.; Guastaroba, P.; Corinaldesi, E.; Calabri, G.B.; Giani, T.; Simonini, G.; Rusconi, F.; Cimaz, R. Kawasaki disease: An epidemiological study in central Italy. Pediatrics Rheumatol. Online J. 2016, 14, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carmona, E.G.; García-Giménez, J.A.; López-Mejías, R.; Khor, C.C.; Lee, J.K.; Taskiran, E.; Ozen, S.; Hocevar, A.; Liu, L.; Gorenjak, M.; et al. Identification of a shared genetic risk locus for Kawasaki disease and IgA vasculitis by a cross-phenotype meta-analysis. Rheumatology 2021, 61, 1204–1210. [Google Scholar] [CrossRef] [PubMed]
- Fabi, M.; Corinaldesi, E.; Pierantoni, L.; Mazzoni, E.; Landini, C.; Bigucci, B.; Ancora, G.; Malaigia, L.; Bodnar, T.; Di Fazzio, G.; et al. Gastrointestinal presentation of Kawasaki disease: A red flag for severe disease? PLoS ONE 2018, 13, e0202658. [Google Scholar] [CrossRef] [PubMed]
- Turroni, S.; Brigidi, P.; Cavalli, A.; Candela, M. Microbiota-host transgenomic metabolism, bioactive molecules from the inside: Miniperspective. J. Med. Chem. 2018, 61, 47–61. [Google Scholar] [CrossRef]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef]
- Ling, Z.; Li, Z.; Liu, X.; Cheng, Y.; Luo, Y.; Tong, X.; Yuan, L.; Wang, Y.; Sun, J.; Li, L.; et al. Altered Fecal Microbiota Composition Associated with Food Allergy in Infants. Appl. Environ. Microbiol. 2014, 80, 2546–2554. [Google Scholar] [CrossRef] [Green Version]
- Cheng, J.; Palva, A.M.; Vos, W.M.D.; Satokari, R. Contribution of the Intestinal Microbiota to Human Health: From Birth to 100 Years of Age. Curr. Top. Microbiol. Immunol. 2013, 358, 323–346. [Google Scholar] [CrossRef]
- Wu, H.J.; Wu, E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes 2012, 3, 4–14. [Google Scholar] [CrossRef] [Green Version]
- Berer, K.; Mues, M.; Koutrolos, M.; Al Rasbi, Z.; Boziki, M.; Johner, C.; Wekerle, H.; Krishnamoorthy, G. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature 2011, 479, 538–541. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Ding, Y.; Yang, Z.; Zhang, X.; Zhao, M. Effects of changes on gut microbiota in children with acute Kawasaki disease. PeerJ 2020, 8, e9698. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Yue, Y.; Wang, L.; Deng, Z.; Yuan, Y.; Zhao, M.; Yuan, Z.; Tan, C.; Cao, Y. Altered gut microbiota correlated with systemic inflammation in children with Kawasaki disease. Sci. Rep. 2020, 10, 14525. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, L.; Wang, Y.; Liu, X.; Zhang, H.; Liu, Y.; Shen, N.; Yang, J.; Gai, Z. Gut microbiota dysbiosis is associated with Henoch-Schönlein Purpura in children. Int. Immunopharmacol. 2018, 58, 1–8. [Google Scholar] [CrossRef]
- Kinumaki, A.; Sekizuka, T.; Hamada, H.; Kato, K.; Yamashita, A.; Kuroda, M. Characterization of the gut microbiota of Kawasaki disease patients by metagenomic analysis. Front. Microbiol. 2015, 6, 824. [Google Scholar] [CrossRef] [Green Version]
- Nagelkerke, S.Q.; Tacke, C.E.; Breunis, W.B.; Tanck, M.W.T.; Geissler, J.; Png, E.; Hoang, L.T.; van der Heijden, J.; Naim, A.N.M.; Yeung, R.S.M.; et al. Extensive Ethnic Variation and Linkage Disequilibrium at the FCGR2/3 Locus: Different Genetic Associations Revealed in Kawasaki Disease. Front. Immunol. 2019, 10, 185. [Google Scholar] [CrossRef] [Green Version]
- Deschasaux, M.; Bouter, K.E.; Prodan, A.; Levin, E.; Groen, A.K.; Herrema, H.; Tremaroli, V.; Bakker, G.J.; Attaye, I.; Pinto-Sietsma, S.-J.; et al. Depicting the composition of gut microbiota in a population with varied ethnic origins but shared geography. Nat. Med. 2018, 24, 1526–1531. [Google Scholar] [CrossRef]
- Cancello, R.; Turroni, S.; Rampelli, S.; Cattaldo, S.; Candela, M.; Cattani, L.; Mai, S.; Vietti, R.; Scacchi, M.; Brigidi, P.; et al. Effect of short-term dietary intervention and probiotic mix supplementation on the gut microbiota of elderly obese women. Nutrients 2019, 11, 3011. [Google Scholar] [CrossRef] [Green Version]
- D’Amico, F.; Biagi, E.; Rampelli, S.; Fiori, J.; Zama, D.; Soverini, M.; Barone, M.; Leardini, D.; Muratore, E.; Prete, A.; et al. Enteral nutrition in pediatric patients undergoing hematopoietic SCT promotes the recovery of gut microbiome homeostasis. Nutrients 2019, 11, 2958. [Google Scholar] [CrossRef] [Green Version]
- Masella, A.P.; Bartram, A.K.; Truszkowski, J.M.; Brown, D.G.; Neufeld, J.D. PANDAseq: Paired-end assembler for illumina sequences. BMC Bioinform. 2012, 13, 31. [Google Scholar] [CrossRef] [Green Version]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. QIIME 2: Reproducible, interactive, scalable, and extensible microbiome data science. Nat. Biotechnol. 2018, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Rampelli, S.; Guenther, K.; Turroni, S.; Wolters, M.; Veidebaum, T.; Kourides, Y.; Molnár, D.; Lissner, L.; Benitez-Paez, A.; Sanz, Y.; et al. Pre-obese children’s dysbiotic gut microbiome and unhealthy diets may predict the development of obesity. Commun. Biol. 2018, 1, 222. [Google Scholar] [CrossRef] [PubMed]
- Muleviciene, A.; D’Amico, F.; Turroni, S.; Candela, M.; Jankauskiene, A. Iron deficiency anemia-related gut microbiota dysbiosis in infants and young children: A pilot study. Acta Microbiol. Immunol. Hung. 2018, 65, 551–564. [Google Scholar] [CrossRef] [Green Version]
- Biagi, E.; Franceschi, C.; Rampelli, S.; Severgnini, M.; Ostan, R.; Turroni, S.; Consolandi, C.; Quercia, S.; Scurti, M.; Monti, D.; et al. Gut microbiota and extreme longevity. Curr. Biol. 2016, 26, 1480–1485. [Google Scholar] [CrossRef] [Green Version]
- Culhane, A.C.; Thioulouse, J.; Perrière, G.; Higgins, D.G. MADE4: An R package for multivariate analysis of gene expression data. Bioinformatics 2005, 21, 2789–2790. [Google Scholar] [CrossRef] [Green Version]
- Derrien, M.; Alvarez, A.S.; de Vos, W.M. The gut microbiota in the first decade of life. Trends Microbiol. 2019, 27, 997–1010. [Google Scholar] [CrossRef] [Green Version]
- Vujkovic-Cvijin, I.; Sklar, J.; Jiang, L.; Natarajan, L.; Knight, R.; Belkaid, Y. Host variables confound gut microbiota studies of human disease. Nature 2020, 587, 448–454. [Google Scholar] [CrossRef]
- Duvallet, C.; Gibbons, S.; Gurry, T.; Irizarry, R.; Alm, E. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat. Commun. 2017, 8, 1784. [Google Scholar] [CrossRef] [Green Version]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From dietary fiber to host physiology: Short-chain fatty acids as key bacterial metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaneko, K.; Akagawa, S.; Akagawa, Y.; Kimata, T.; Tsuji, S. Our evolving understanding of Kawasaki disease pathogenesis: Role of the gut microbiota. Front. Immunol. 2020, 11, 1616. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.; Ikeda, K.; Hamaoka, K. Aetiological significance of infectious stimuli in Kawasaki disease. Front. Pediatrics 2019, 7, 244. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef] [Green Version]
- Milani, C.; Duranti, S.; Bottacini, F.; Casey, E.; Turroni, F.; Mahony, J.; Belzer, C.; Delgado Palacio, S.; Arboleya Montes, S.; Mancabelli, L.; et al. The first microbial colonizers of the human gut: Composition, activities, and health implications of the infant gut microbiota. Microbiol. Mol. Biol. Rev. 2017, 81, e00036-17. [Google Scholar] [CrossRef] [Green Version]
- Stanislawski, M.A.; Dabelea, D.; Wagner, B.D.; Iszatt, N.; Dahl, C.; Sontag, M.K.; Knight, R.; Lozupone, C.A.; Eggesbø, M. Gut microbiota in the first 2 years of life and the association with body mass index at age 12 in a Norwegian birth cohort. mBio 2018, 9, e01751-18. [Google Scholar] [CrossRef] [Green Version]
- Ho, N.T.; Li, F.; Lee-Sarwar, K.A.; Tun, H.M.; Brown, B.; Pannaraj, P.S.; Bender, J.M.; Azad, M.B.; Thompson, A.L.; Weiss, S.T.; et al. Meta-analysis of effects of exclusive breastfeeding on infant gut microbiota across populations. Nat. Commun. 2018, 9, 4169. [Google Scholar] [CrossRef]
- Stokholm, J.; Blaser, M.J.; Thorsen, J.; Rasmussen, M.A.; Waage, J.; Vinding, R.K.; Schoos, A.-M.M.; Kunøe, A.; Fink, N.R.; Chawes, B.; et al. Maturation of the gut microbiome and risk of asthma in childhood. Nat. Commun. 2018, 9, 141. [Google Scholar] [CrossRef]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef] [Green Version]
- De Vos, W.M. Microbe Profile: Akkermansia muciniphila: A conserved intestinal symbiont that acts as the gatekeeper of our mucosa. Microbiology 2017, 163, 646–648. [Google Scholar] [CrossRef]
- Derrien, M.; Van Baarlen, P.; Hooiveld, G.; Norin, E.; Muller, M.; de Vos, W. Modulation of mucosal immune response, tolerance, and proliferation in mice colonized by the mucin-degrader Akkermansia muciniphila. Front. Microbiol. 2011, 2, 166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, B.L.; Hornig, M.; Parekh, T.; Lipkin, W.I. Application of novel PCR-based methods for detection, quantitation, and phylogenetic characterization of Sutterella species in intestinal biopsy samples from children with autism and gastrointestinal disturbances. mBio 2012, 3, e00261-11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, W.L.; Yang, Y.H.; Lin, Y.T.; Chiang, B.L. Gastrointestinal manifestations in Henoch-Schönlein purpura: A review of 261 patients. Acta Paediatrics 2004, 3, 1427–1431. [Google Scholar] [CrossRef]
- Mosli, M.; Zou, G.; Garg, S.K.; Feagan, S.G.; MacDonald, J.K.; Chande, N.; Sandborn, W.J.; Feagan, B.G. C-reactive protein, fecal calprotectin, and stool lactoferrin for detection of endoscopic activity in symptomatic inflammatory bowel disease patients: A systematic review and meta-analysis. Am. J. Gastroenterol. 2015, 110, 802–819. [Google Scholar] [CrossRef]
| KD (n = 13) | HSP (n = 10) | F (n = 12) | p | ||
|---|---|---|---|---|---|
| Ethnicity, n (%) | Caucasian | 10 (76.9%) | 8 (80.0%) | 8 (66.7%) | n.s. |
| Asian | 3 (23.1%) | 2 (20.0%) | 1 (8.3%) | ||
| Hispanic | 1 (8.3%) | ||||
| Mixed | 1 (8.3%) | ||||
| Black | 1 (8.3%) | ||||
| Sex, n (%) | Male | 10 (76.9%) | 5 (50.0%) | 7 (58.3%) | n.s. |
| Female | 3 (23.1%) | 5 (50.0%) | 5 (41.7%) | ||
| Age (months), median (IQR) | 31 (14.5–43) * | 62 (52.3–104.3) *§ | 30 (15–67.8) § | 0.005 | |
| Class, n (%) | Responder | 11 (84.6%) | - | - | - |
| Non-responder | 2 (15.4%) | ||||
| Clinical form, n (%) | Complete | 8 (61.5%) | - | - | - |
| Atypical/ incomplete | 5 (38.5%) | ||||
| Abdominal symptoms, n (%) | Yes | 3 (23.1%) | 2 (20.0%) | 4 (33.3%) | n.s. |
| No | 10 (76.9%) | 8 (80.0%) | 8 (66.7%) | ||
| White blood cells (×109/L), median (IQR) | 15.0 (12.3–20.0) | 11.5 (8.7–15.2) | 12.5 (10.8–13.7) | n.s. | |
| Neutrophils %, median (IQR) | 73.8 (59.7–80.3) | 68.5 (58.1–73.5) | 70 (59.9–76.6) | n.s. | |
| Lymphocytes %, median (IQR) | 19.0 (12.6–28.4) | 28.6 (19.6–35.6) | 20.6 (13.5–32.2) | n.s. | |
| Eosinophils %, median (IQR) | 1.4 (0.3–3.6) | 1.7 (0.6–3.7) | 0.5 (0.2–1.9) | n.s. | |
| Red blood cells (×1012/L), median (IQR) | 4.15 (3.90–4.46) | 4.68 (4.12–4.97) | 4.58 (4.01–5.79) | n.s. | |
| Haemoglobin (g/dL), median (IQR) | 10.7 (10.1–11.7) * | 12.9 (11.0–13.8) * | 11.2 (10.6–12.0) | 0.034 | |
| Platelets count (×109/L), median (IQR) | 377 (317–536) ° | 358 (319–402) | 293 (229–318) ° | 0.010 | |
| C-reactive protein (mg/dL), mean (SD) | 9.68 (4.89) * | 2.74 (3.11) *§ | 6.96 (4.29) § | 0.002 | |
| Aspartate aminotransferase (IU/L), median (IQR) | 32 (26–55) | 29 (23–33) | 36 (28–74) | n.s. | |
| Alanine aminotransferase (IU/L), median (IQR) | 27 (18–94) * | 14 (9–16) * | 16 (10–25) | 0.011 | |
| Calprotectin (mcg/g), median (IQR) | 706 (117–1445) | - | 175 (38–501) | n.s. | |
| TNF-alpha (pg/mL), median (IQR) | 2.5 (0–24.3) | - | 1.0 (0–2.8) | n.s. | |
| IL6 (pg/mL), median (IQR) | 132.0 (58.4–1747.0) | - | 18.1 (8.6–104.5) | 0.025 | |
| IL8 (pg/mL), median (IQR) | 214 (62–14910) | - | 123 (18–4502) | n.s. | |
| IL12p70 (pg/mL), median (IQR) | 0 (0–5) | - | 0 (0–1.25) | n.s. | |
| IL10 (pg/mL), median (IQR) | 7.0 (1.8–30.3) | - | 6.0 (3.3–16.0) | n.s. | |
| IgG (mg/dL), median (IQR) | 729 (664–861) | 1044 (850–1237) | 944 (674–1299) | n.s. | |
| IgM (mg/dL), mean (SD) | 96.6 (48.2) | 102.0 (44.2) | 111.6 (29.5) | n.s. | |
| IgA (mg/dL), mean (SD) | 101.5 (80.3) * | 170.2 (65.2) *§ | 96.3 (51.6) § | 0.031 | |
| Cardiac non-coronary involvement, n (%) | Yes | 7 (53.8%) | - | - | - |
| No | 6 (46.2%) | ||||
| Coronary involvement, n (%) | Yes | 7 (53.8%) | - | - | - |
| No | 6 (46.2%) | ||||
| Coronary artery lesions, n (%) | No involvement | 6 (46.2%) | - | - | - |
| Dilations | 2 (15.4%) | ||||
| Aneurysms | 5 (38.5%) | ||||
| Total days of fever, median (IQR) | 8 (7–10) | - | 8 (6–10) | n.s. | |
| Delayed therapy, n (%) | Yes | 2 (15.4%) | - | - | - |
| No | 10 (76.9%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fabi, M.; D’Amico, F.; Turroni, S.; Andreozzi, L.; Filice, E.; Brigidi, P.; Lanari, M. Gut Microbiota Dysbiosis in Childhood Vasculitis: A Perspective Comparative Pilot Study. J. Pers. Med. 2022, 12, 973. https://doi.org/10.3390/jpm12060973
Fabi M, D’Amico F, Turroni S, Andreozzi L, Filice E, Brigidi P, Lanari M. Gut Microbiota Dysbiosis in Childhood Vasculitis: A Perspective Comparative Pilot Study. Journal of Personalized Medicine. 2022; 12(6):973. https://doi.org/10.3390/jpm12060973
Chicago/Turabian StyleFabi, Marianna, Federica D’Amico, Silvia Turroni, Laura Andreozzi, Emanuele Filice, Patrizia Brigidi, and Marcello Lanari. 2022. "Gut Microbiota Dysbiosis in Childhood Vasculitis: A Perspective Comparative Pilot Study" Journal of Personalized Medicine 12, no. 6: 973. https://doi.org/10.3390/jpm12060973
APA StyleFabi, M., D’Amico, F., Turroni, S., Andreozzi, L., Filice, E., Brigidi, P., & Lanari, M. (2022). Gut Microbiota Dysbiosis in Childhood Vasculitis: A Perspective Comparative Pilot Study. Journal of Personalized Medicine, 12(6), 973. https://doi.org/10.3390/jpm12060973











