Prediction of All-Cause Mortality Following Percutaneous Coronary Intervention in Bifurcation Lesions Using Machine Learning Algorithms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Model Development
2.3. Statistical Analysis
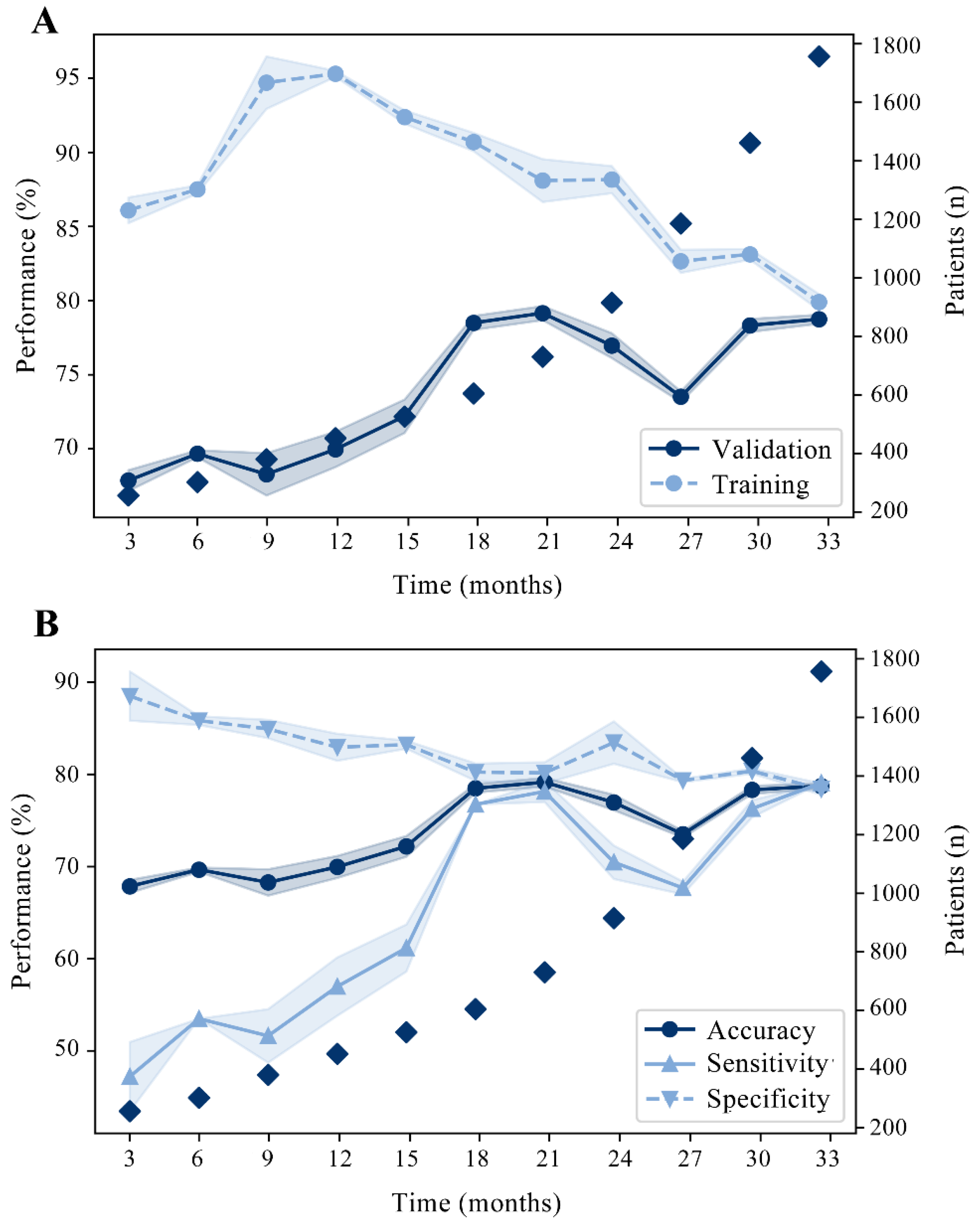
3. Results
3.1. Characteristics of the Discovery Cohort
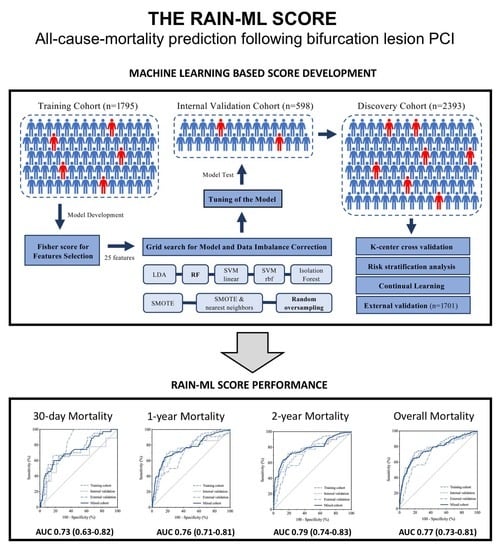
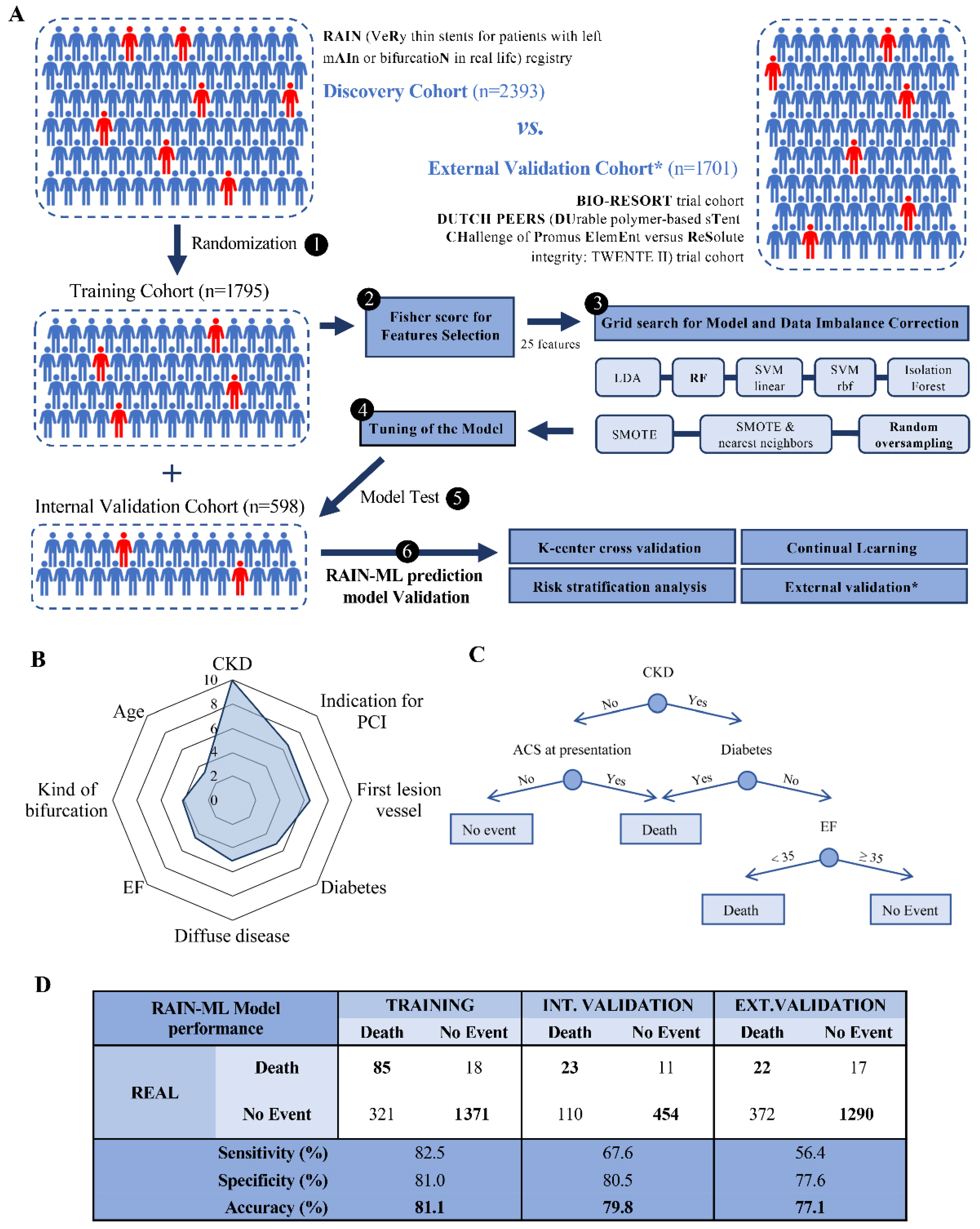
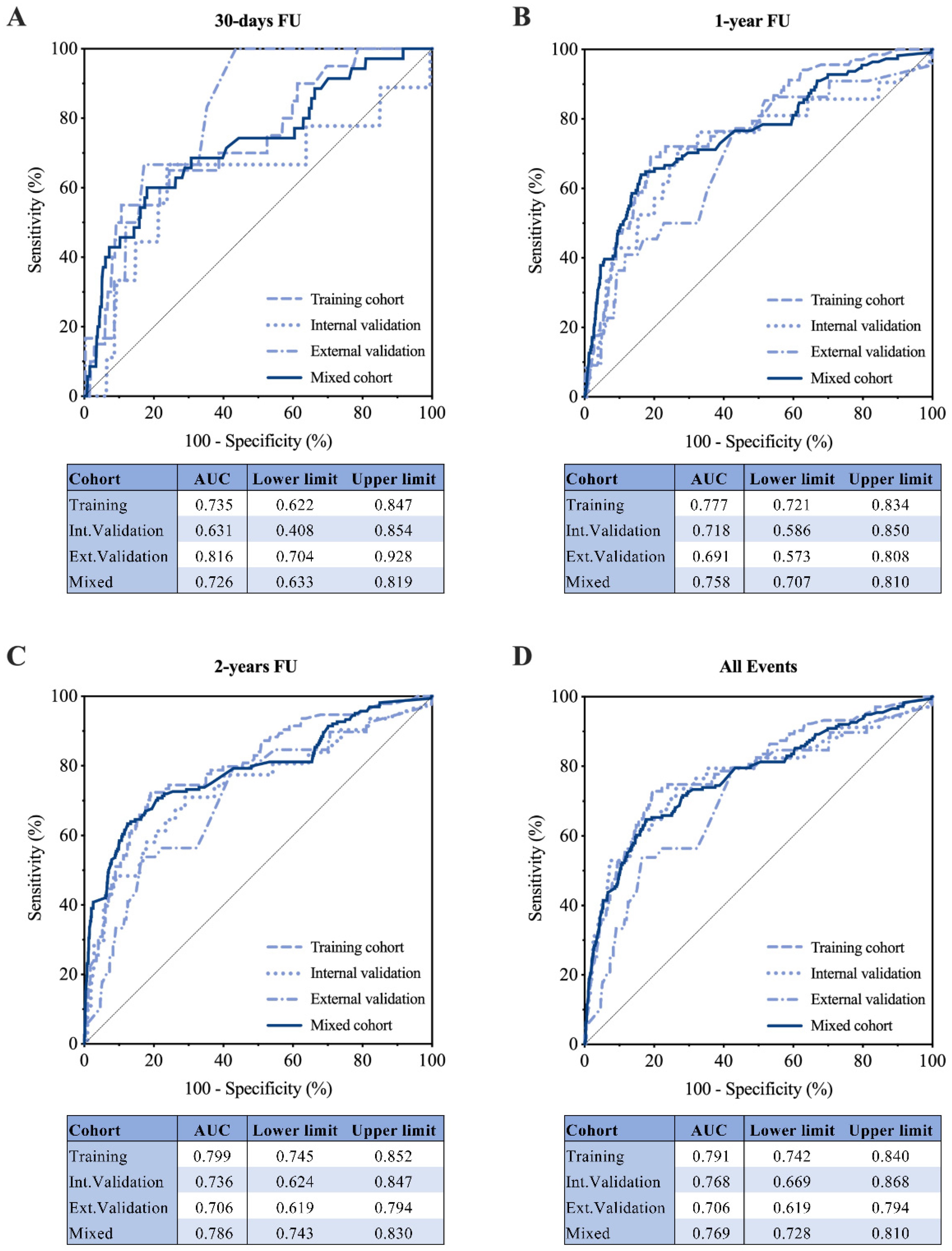
3.2. Development and Internal Validation of the RAIN-ML Prediction Model
3.3. External Validation of the RAIN-ML Model
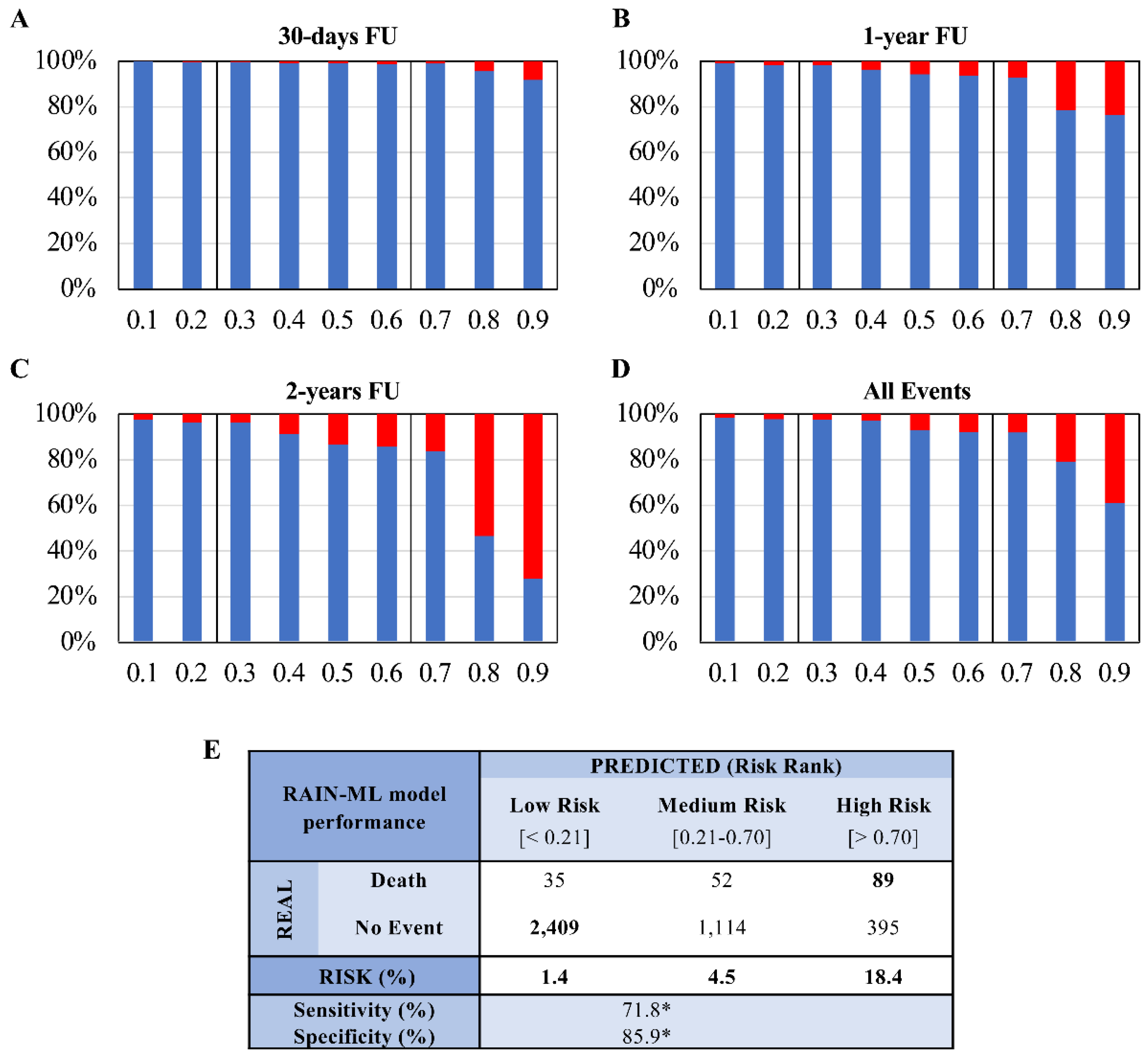
3.4. Risk Stratification Analysis
4. Discussion
4.1. Rationale of the Study and Related Work
4.2. Perspectives
4.3. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bangalore, S.; Kumar, S.; Fusaro, M.; Amoroso, N.; Attubato, M.; Feit, F.; Bhatt, D.L.; Slater, J. Short-and long-term outcomes with drug-eluting and bare-metal coronary stents: A mixed-treatment comparison analysis of 117 762 patient-years of follow-up from randomized trials. Circulation 2012, 125, 2873–2891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- von Birgelen, C.; Kok, M.M.; van der Heijden, L.C.; Danse, P.W.; E Schotborgh, C.; Scholte, M.; Gin, R.M.T.J.; Somi, S.; van Houwelingen, K.G.; Stoel, M.G.; et al. Very thin strut biodegradable polymer everolimus-eluting and sirolimus-eluting stents versus durable polymer zotarolimus-eluting stents in allcomers with coronary artery disease (BIO-RESORT): A three-arm, randomised, non-inferiority trial. Lancet 2016, 388, 2607–2617. [Google Scholar] [CrossRef]
- Biolè, C.; Huczek, Z.; Nuñez-Gil, I.; Boccuzzi, G.; Autelli, M.; Montefusco, A.; Trabattoni, D.; Ryan, N.; Venuti, G.; Imori, Y.; et al. Daily risk of adverse outcomes in patients undergoing complex lesions revascularization: A subgroup analysis from the RAIN-CARDIOGROUP VII study (veRy thin stents for patients with left mAIn or bifurcatioN in real life). Int. J. Cardiol. 2019, 290, 64–69. [Google Scholar] [CrossRef] [PubMed]
- D’Ascenzo, F.; Omedè, P.; De Filippo, O.; Cerrato, E.; Autelli, M.; Trabattoni, D.; Ryan, N.; Venuti, G.; Muscoli, S.; Montabone, A.; et al. Impact of final kissing balloon and of Imaging on patients treated on unprotected left main coronary artery with thin-strut stents (from the RAIN-CARDIOGROUP VII study). Am. J. Cardiol. 2019, 123, 1610–1619. [Google Scholar] [CrossRef]
- Gaido, L.; D’Ascenzo, F.; Imori, Y.; Wojakowski, W.; Saglietto, A.; Figini, F.; Mattesini, A.; Trabattoni, D.; Rognoni, A.; Tomassini, F.; et al. Impact of kissing balloon in patients treated with ultrathin stents for left main lesions and bifurcations: An analysis from the RAIN-CARDIOGROUP VII study. Circ. Cardiovasc. Interv. 2020, 13, e008325. [Google Scholar] [CrossRef]
- D’Ascenzo, F.; Chieffo, A.; Cerrato, E.; Ugo, F.; Pavani, M.; Kawamoto, H.; di Summa, R.; Varbella, F.; Boccuzzi, G.; Omedè, P.; et al. Incidence and Management of Restenosis After Treatment of Unprotected Left Main Disease with Second-Generation Drug-Eluting Stents (from Failure in Left Main Study with 2nd Generation Stents–Cardiogroup III Study). Am. J. Cardiol. 2017, 119, 978–982. [Google Scholar] [CrossRef]
- Naganuma, T.; Chieffo, A.; Meliga, E.; Capodanno, D.; Park, S.-J.; Onuma, Y.; Valgimigli, M.; Jegere, S.; Makkar, R.R.; Palacios, I.F.; et al. Long-term clinical outcomes after percutaneous coronary intervention for ostial/mid-shaft lesions versus distal bifurcation lesions in unprotected left main coronary artery: The DELTA Registry (Drug-Eluting Stent for Left Main Coronary Artery Disease). JACC Cardiovasc. Interv. 2013, 6, 1242–1249. [Google Scholar] [CrossRef] [Green Version]
- Baber, U.; Mehran, R.; Giustino, G.; Cohen, D.J.; Henry, T.D.; Sartori, S.; Ariti, C.; Litherland, C.; Dangas, G.; Gibson, C.M.; et al. Coronary Thrombosis and Major Bleeding after PCI with Drug-Eluting Stents Risk Scores from Paris. J. Am. Coll. Cardiol. 2016, 67, 2224–2234. [Google Scholar] [CrossRef]
- Mehran, R.; Baber, U.; Steg, P.G.; Ariti, C.; Weisz, G.; Witzenbichler, B.; Henry, T.D.; Kini, A.S.; Stuckey, T.; Cohen, D.J.; et al. Cessation of dual antiplatelet treatment and cardiac events after percutaneous coronary intervention (PARIS): 2 years results from a prospective observational study. Lancet 2013, 382, 1714–1722. [Google Scholar] [CrossRef]
- Moretti, C.; D’Ascenzo, F.; Omedè, P.; Sciuto, F.; Presutti, D.G.; Di Cuia, M.; Colaci, C.; Giusto, F.; Ballocca, F.; Cerrato, E.; et al. Thirty-day readmission rates after PCI in a metropolitan center in Europe: Incidence and impact on prognosis. J. Cardiovasc. Med. 2015, 16, 238–245. [Google Scholar] [CrossRef]
- Giustino, G.; Chieffo, A.; Palmerini, T.; Valgimigli, M.; Feres, F.; Abizaid, A.; Costa, R.A.; Hong, M.-K.; Kim, B.-K.; Jang, Y.; et al. Efficacy and Safety of Dual Antiplatelet Therapy After Complex PCI. J. Am. Coll. Cardiol. 2016, 68, 1851–1864. [Google Scholar] [CrossRef] [PubMed]
- Raposeiras-Roubín, S.; Caneiro Queija, B.; D’Ascenzo, F.; Kinnaird, T.; Ariza-Solé, A.; Manzano-Fernández, S.; Templin, C.; Velicki, L.; Xanthopoulou, I.; Cerrato, E.; et al. Usefulness of the PARIS Score to Evaluate the Ischemic-hemorrhagic Net Benefit with Ticagrelor and Prasugrel After an Acute Coronary Syndrome. Rev. Esp. Cardiol. 2019, 72, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Burzotta, F.; Annone, U.; Paraggio, L.; D’Ascenzo, F.; Biondi-Zoccai, G.; Aurigemma, C.; Romagnoli, E.; Verdirosi, D.; Trani, C.; Crea, F. Clinical outcome after percutaneous coronary intervention with drug-eluting stent in bifurcation and nonbifurcation lesions: A meta-analysis of 23 981 patients. Coron. Artery Dis. 2020, 31, 438–445. [Google Scholar] [CrossRef] [PubMed]
- Van Der Giessen, A.G.; Wentzel, J.J.; Meijboom, W.B.; Mollet, N.R.; Van Der Steen, A.F.W.; Van De Vosse, F.N.; De Feyter, P.J.; Gijsen, F.J.H. Plaque and shear stress distribution in human coronary bifurcations: A multislice computed tomography study. EuroIntervention 2009, 4, 654–661. [Google Scholar] [CrossRef]
- Lassen, J.F.; Burzotta, F.; Banning, A.P.; Lefèvre, T.; Darremont, O.; Hildick-Smith, D.; Chieffo, A.; Pan, M.; Holm, N.; Louvard, Y.; et al. Percutaneous coronary intervention for the left main stem and other bifurcation lesions: 12th consensus document from the European Bifurcation Club. EuroIntervention 2018, 13, 1540–1553. [Google Scholar] [CrossRef] [Green Version]
- Behan, M.W.; Holm, N.R.; De Belder, A.J.; Cockburn, J.; Erglis, A.; Curzen, N.P.; Niemelä, M.; Oldroyd, K.G.; Kervinen, K.; Kumsars, I.; et al. Coronary bifurcation lesions treated with simple or complex stenting: 5-year survival from patient-level pooled analysis of the Nordic Bifurcation Study and the British Bifurcation Coronary Study. Eur. Heart J. 2016, 37, 1923–1928. [Google Scholar] [CrossRef]
- Ambati, L.S.; El-Gayar, O.; Nawar, N. Design principles for multiple sclerosis mobile self-management applications: A patient-centric perspective. In Proceedings of the 27th Annual Americas Conference on Information Systems, AMCIS 2021, Online, 9–13 August 2021. [Google Scholar]
- Ambati, L.S.; El-Gayar, O.F.; Nawar, N. Influence of The Digital Divide and Socio-Economic Factors on Prevalence of Diabetes. Issues Inf. Syst. 2020, 21, 103–113. [Google Scholar]
- Costa, F.; Van Klaveren, D.; Feres, F.; James, S.; Räber, L.; Pilgrim, T.; Hong, M.-K.; Kim, H.-S.; Colombo, A.; Steg, P.G.; et al. Dual Antiplatelet Therapy Duration Based on Ischemic and Bleeding Risks After Coronary Stenting. J. Am. Coll. Cardiol. 2019, 73, 741–754. [Google Scholar] [CrossRef]
- Gallone, G.; Bruno, F.; D’ascenzo, F.; De Ferrari, G. What will we ask to artificial intelligence for cardiovascular medicine in the next decade? Minerva Cardiol. Angiol. 2022, 70, 92–101. [Google Scholar] [CrossRef]
- Dey, D.; Slomka, P.J.; Leeson, P.; Comaniciu, D.; Shrestha, S.; Sengupta, P.P.; Marwick, T.H. Artificial Intelligence in Cardiovascular Imaging: JACC State-of-the-Art Review. J. Am. Coll. Cardiol. 2019, 73, 1317–1335. [Google Scholar] [CrossRef]
- Motwani, M.; Dey, D.; Berman, D.S.; Germano, G.; Achenbach, S.; Al-Mallah, M.; Andreini, D.; Budoff, M.J.; Cademartiri, F.; Callister, T.Q.; et al. Machine learning for prediction of all-cause mortality in patients with suspected coronary artery disease: A 5-year multicentre prospective registry analysis. Eur. Heart J. 2016, 38, 500–507. [Google Scholar] [CrossRef] [PubMed]
- Tokodi, M.; Schwertner, W.R.; Kovács, A.; Tősér, Z.; Staub, L.; Sárkány, A.; Lakatos, B.K.; Behon, A.; Boros, A.M.; Perge, P.; et al. Machine learning-based mortality prediction of patients undergoing cardiac resynchronization therapy: The SEMMELWEIS-CRT score. Eur. Heart J. 2020, 41, 1747–1756. [Google Scholar] [CrossRef] [PubMed]
- D’Ascenzo, F.; Filippo, O.; De Gallone, G.; Mittone, G.; Deriu, M.A.; Iannaccone, M.; Ariza-Solé, A.; Liebetrau, C.; Manzano-Fernández, S.; Quadri, G.; et al. Machine learning-based prediction of adverse events following an acute coronary syndrome (PRAISE): A modelling study of pooled datasets. Lancet 2021, 397, 199–207. [Google Scholar] [CrossRef]
- Von Birgelen, C.; Sen, H.; Lam, M.K.; Danse, P.W.; Jessurun, G.A.J.; Hautvast, R.W.M.; van Houwelingen, G.K.; Schramm, A.R.; Gin, R.M.T.J.; Louwerenburg, J.W.; et al. Third-generation zotarolimus-eluting and everolimus-eluting stents in all-comer patients requiring a percutaneous coronary intervention (DUTCH PEERS): A randomised, single-blind, multicentre, non-inferiority trial. Lancet 2014, 383, 413–423. [Google Scholar] [CrossRef]
- Zocca, P.; Kok, M.M.; Tandjung, K.; Danse, P.W.; Jessurun, G.A.; Hautvast, R.W.; van Houwelingen, K.G.; Stoel, M.G.; Schramm, A.R.; Gin, R.M.T.J.; et al. 5-Year Outcome Following Randomized Treatment of All-Comers with Zotarolimus-Eluting Resolute Integrity and Everolimus-Eluting PROMUS Element Coronary Stents: Final Report of the DUTCH PEERS (TWENTE II) Trial. JACC Cardiovasc. Interv. 2018, 11, 462–469. [Google Scholar] [CrossRef]
- D’Ascenzo, F.; De Filippo, O.; Elia, E.; Doronzo, M.; Montefusco, A.; Pennone, M.; Angelini, F.; Franchin, L.; Bruno, F.; Salizzoni, S.; et al. Percutaneous vs. Surgical Revascularization for Patients with Unprotected Left Main Stenosis: A Meta-Analysis of 5 Years Follow-Up RCTs. Eur. Heart J. Qual. Care Clin. Outcomes 2020, 7, 476–485. [Google Scholar] [CrossRef]
- Gallone, G.; D’Ascenzo, F.; Conrotto, F.; Costa, F.; Capodanno, D.; Muscoli, S.; Chieffo, A.; Yoichi, I.; Pennacchi, M.; Quadri, G.; et al. Accuracy of the PARIS score and PCI complexity to predict ischemic events in patients treated with very thin stents in unprotected left main or coronary bifurcations. Catheter. Cardiovasc. Interv. 2020, 97, E227–E236. [Google Scholar] [CrossRef]
- Christodoulou, E.; Ma, J.; Collins, G.S.; Steyerberg, E.W.; Verbakel, J.Y.; Van Calster, B. A systematic review shows no performance benefit of machine learning over logistic regression for clinical prediction models. J. Clin. Epidemiol. 2019, 110, 12–22. [Google Scholar] [CrossRef]
- Gupta, V.; Mittal, M.; Mittal, V.; Gupta, A. ECG signal analysis using CWT, spectrogram and autoregressive technique. Iran J. Comput. Sci. 2021, 4, 265–280. [Google Scholar] [CrossRef]
- Parameswari, C.; Siva Ranjani, S. Prediction of atherosclerosis pathology in retinal fundal images with machine learning approaches. J. Ambient Intell. Humaniz. Comput. 2020, 12, 6701–6711. [Google Scholar] [CrossRef]
- Talaat, F.M.; Gamel, S.A. RL based hyper-parameters optimization algorithm (ROA) for convolutional neural network. J. Ambient Intell. Humaniz. Comput. 2022, 1–11. [Google Scholar] [CrossRef]
- Elavarasan, D.; Vincent, P.M.D.R. A reinforced random forest model for enhanced crop yield prediction by integrating agrarian parameters. J. Ambient Intell. Humaniz. Comput. 2021, 12, 10009–10022. [Google Scholar] [CrossRef]
- Jayavadivel, R.; Prabaharan, P. Investigation on automated surveillance monitoring for human identification and recognition using face and iris biometric. J. Ambient Intell. Humaniz. Comput. 2021, 12, 10197–10208. [Google Scholar] [CrossRef]
- Usharani, R.; Shanthini, A. Neuropathic complications: Type II diabetes mellitus and other risky parameters using machine learning algorithms. J. Ambient Intell. Humaniz. Comput. 2021, 12, 0123456789. [Google Scholar] [CrossRef]
- Saravanan, S.; Karthigaivel, R.; Magudeeswaran, V. A brain tumor image segmentation technique in image processing using ICA-LDA algorithm with ARHE model. J. Ambient Intell. Humaniz. Comput. 2021, 12, 4727–4735. [Google Scholar] [CrossRef]
- González, I.; Fontecha, J.; Bravo, J. Relationship between stride interval variability and aging: Use of linear and non-linear estimators for gait variability assessment in assisted living environments. J. Ambient Intell. Humaniz. Comput. 2019, 10, 2095–2109. [Google Scholar] [CrossRef]
| Variable | Training Cohort (n = 1795) | Internal Validation Cohort (n = 598) | External Validation Cohort (n = 1701) | |
|---|---|---|---|---|
| Patient parameters | Age (years) | 70 [61; 77] | 69 [61; 78] | 65 [57; 72] |
| Sex (ref. male; n, %) | 1379 (76.9) | 440 (73.6) | 1329 (78.1) | |
| Hypertension (ref. yes; n, %) | 1331 (74.2) | 460 (76.9) | 820 (48.2) | |
| Hyperlipidemia (ref. yes; n, %) | 1074 (59.8) | 377 (63.0) | N.A. | |
| Diabetes (ref. yes; n, %) | 605 (33.7) | 200 (33.4) | 319 (18.8) | |
| Smoker | N.A. | |||
| No (n, %) | 872 (48.6) | 291 (48.6) | ||
| Previous (n, %) | 548 (30.5) | 181 (30.3) | ||
| Current (n, %) | 375 (20.9) | 126 (21.1) | ||
| CKD (ref. GFR < 60 mL/min; n, %) | 391 (21.8) | 147 (24.6) | 50 (2.9) | |
| Previous PCI (ref. yes; n, %) | 605 (33.7) | 187 (31.3) | 301 (17.7) | |
| Previous CABG (ref. yes; n, %) | 97 (5.4) | 29 (4.8) | 120 (7.1) | |
| Previous MI (ref. yes; n, %) | 554 (30.9) | 183 (30.6) | 339 (19.9) | |
| EF at echo (%) | 55 [55; 60] | 55 [55; 65] | 50 [50; 50] * | |
| PCI indication | ||||
| STEMI (n, %) | 305 (17.0) | 114 (19.1) | 383 (22.5) | |
| NSTEMI (n, %) | 447 (24.9) | 133 (22.2) | 388 (22.8) | |
| Unstable angina (n, %) | 257 (14.3) | 90 (15.1) | 296 (17.4) | |
| Stable angina (n, %) | 440 (24.6) | 147 (24.5) | 634 (37.3) | |
| Positive stress test (n, %) | 227 (12.6) | 82 (13.7) | 0 (0.0) | |
| Planned angiography (n, %) | 119 (6.6) | 32 (5.4) | 0 (0.0) | |
| ACS at presentation (ref. yes; n, %) | 1007 (56.1) | 337 (56.4) | 1067 (62.7) | |
| STEMI at presentation (ref. yes; n, %) | 305 (17.0) | 114 (91.1) | 383 (22.5) | |
| Kind of DAT (aspirin plus) | ||||
| Clopidogrel (n, %) | 1170 (65.2) | 391 (65.4) | 1131 (66.4) | |
| Ticagrelor (n, %) | 479 (26.7) | 162 (27.1) | 513 (30.2) | |
| Prasugrel (n, %) | 146 (8.1) | 45 (7.5) | 57 (3.4) | |
| Predilatation (ref. yes; n, %) | First lesion vessel | |||
| LM (n, %) | 435 (24.2) | 160 (26.8) | 179 (10.5) | |
| LAD (n, %) | 876 (48.9) | 296 (49.5) | 1044 (61.5) | |
| Cx/MO (n, %) | 320 (17.8) | 98 (16.4) | 332 (19.5) | |
| RCA (n, %) | 133 (7.4) | 33 (5.5) | 142 (8.3) | |
| RI (n, %) | 31 (1.7) | 11 (1.8) | 4 (0.2) | |
| Lesion localization | ||||
| Ostial (n, %) | 64 (3.6) | 21 (3.5) | 63 (3.7) | |
| Proximal (n, %) | 545 (30.4) | 187 (31.3) | 964 (56.7) | |
| Middle (n, %) | 642 (35.7) | 209 (34.9) | 401 (23.6) | |
| Distal (n, %) | 544 (30.3) | 181 (30.3) | 273 (16.0) | |
| Type C lesion (ref. yes; n, %) | 685 (38.2) | 212 (35.5) | N.A. | |
| Severe calcification (ref. yes; n, %) | 261 (14.5) | 88 (14.7) | 397 (23.3) | |
| Diffuse disease (ref. yes; n, %) | 700 (39.0) | 238 (39.8) | N.A. | |
| Kind of bifurcation | ||||
| Distal LM (n, %) | 481 (26.8) | 174 (29.1) | 179 (10.5) | |
| LAD/Diag (n, %) | 839 (46.7) | 284 (47.5) | 1045 (61.5) | |
| Cx/MO (n, %) | 341 (19.0) | 109 (18.2) | 334 (19.6) | |
| RCA/PL (n, %) | 134 (7.5) | 31 (5.2) | 143 (8.4) | |
| Medina class | ||||
| 1,1,1 (n, %) | 605 (33.8) | 205 (34.2) | 378 (22.2) | |
| 1,1,0 (n, %) | 587 (32.7) | 198 (33.0) | 640 (37.6) | |
| 1,0,1 (n, %) | 174 (9.7) | 47 (7.9) | 85 (5.0) | |
| 0,1,1 (n, %) | 87 (4.8) | 32 (5.4) | 80 (4.7) | |
| 1,0,0 (n, %) | 158 (8.8) | 41 (6.9) | 119 (7.0) | |
| 0,1,0 (n, %) | 92 (5.1) | 47 (7.9) | 258 (15.2) | |
| 0,0,1 (n, %) | 92 (5.1) | 28 (4.7) | 141 (8.3) | |
| Kind of strategy | ||||
| Provisional (n, %) | 1474 (82.1) | 490 (81.9) | 1447 (85.1) | |
| Two stents (n, %) | 321 (17.9) | 108 (18.1) | 254 (14.9) | |
| Use of imaging | ||||
| No (n, %) | 1186 (66.1) | 400 (66.9) | 1648 (96.9) | |
| IVUS (n, %) | 586 (32.6) | 194 (32.4) | 36 (2.1) | |
| OCT (n, %) | 23 (1.3) | 4 (0.7) | 17 (1.0) | |
| Predilatation (ref. yes; n, %) | 1559 (86.9) | 531 (88.8) | 1375 (80.8) | |
| Kind of balloon | ||||
| Conventional (n, %) | 1521 (97.5) | 518 (97.5) | 1666 (97.9) | |
| Angiosculpt (n, %) | 18 (1.2) | 4 (0.8) | 35 (2.1) | |
| Scoring (n, %) | 20 (1.5) | 9 (1.7) | 0 (0.0) | |
| Rotablator (ref. yes; n, %) | 106 (5.9) | 38 (6.4) | 25 (1.5) | |
| Kind of stent on MB | N.A. | |||
| Resolute Onyx (n, %) | 506 (28.6) | 186 (31.3) | ||
| Xience Alpine (n, %) | 454 (25.5) | 136 (22.9) | ||
| Synergy (n, %) | 374 (21.0) | 131 (22.1) | ||
| Ultimaster (n, %) | 165 (9.3) | 50 (8.4) | ||
| Biomatrix alpha (n, %) | 2 (0.1) | 2 (0.3) | ||
| Promus (n, %) | 276 (15.5) | 89 (15.0) | ||
| MB lesion diameter (mm) | 3.0 [2.8; 3.5] | 3.0 [2.8; 3.5] | N.A. | |
| MB lesion length (mm) | 22 [16; 28] | 23 [16; 28] | 15 [10; 22] | |
| MB atmospheres (atm) | 12 [12; 16] | 14 [12; 16] | N.A. | |
| Stent on SB (ref. yes; n, %) | 321 (17.9) | 108 (18.1) | 442 (26.0) | |
| SB lesion diameter (mm) | 2.3 [1.0; 2.8] | 2.5 [1.0; 2.8] | N.A. | |
| SB lesion length (mm) | 28 [20; 33] | 28 [20; 33] | N.A. | |
| SB atmospheres (atm) | 12 [10; 14] | 12 [12; 14] | N.A. | |
| POT (ref. yes; n, %) | 1384 (77.1) | 447 (74.7) | N.A. | |
| ATM Post (atm) | 20 [16; 22] | 20 [16; 20] | N.A. | |
| Final kissing balloon (ref. yes; n, %) | 746 (41.6) | 248 (41.5) | 366 (21.5) | |
| Outcome | Death (ref. yes; n, %) | 103 (5.7) | 34 (5.7) | 39 (2.3) |
| Median follow-up at the event (days) | 274 [61; 433] | 253 [23; 458] | 284 [65; 500] | |
| Death within 30 days (ref. yes; n, %) | 20 (1.1) | 9 (1.5) | 6 (0.4) | |
| Death within 1 year (ref. yes; n, %) | 68 (3.8) | 21 (3.5) | 22 (1.3) | |
| Death within 2 years (ref. yes; n, %) | 94 (5.2) | 31 (5.2) | 39 (2.3) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Burrello, J.; Gallone, G.; Burrello, A.; Jahier Pagliari, D.; Ploumen, E.H.; Iannaccone, M.; De Luca, L.; Zocca, P.; Patti, G.; Cerrato, E.; et al. Prediction of All-Cause Mortality Following Percutaneous Coronary Intervention in Bifurcation Lesions Using Machine Learning Algorithms. J. Pers. Med. 2022, 12, 990. https://doi.org/10.3390/jpm12060990
Burrello J, Gallone G, Burrello A, Jahier Pagliari D, Ploumen EH, Iannaccone M, De Luca L, Zocca P, Patti G, Cerrato E, et al. Prediction of All-Cause Mortality Following Percutaneous Coronary Intervention in Bifurcation Lesions Using Machine Learning Algorithms. Journal of Personalized Medicine. 2022; 12(6):990. https://doi.org/10.3390/jpm12060990
Chicago/Turabian StyleBurrello, Jacopo, Guglielmo Gallone, Alessio Burrello, Daniele Jahier Pagliari, Eline H. Ploumen, Mario Iannaccone, Leonardo De Luca, Paolo Zocca, Giuseppe Patti, Enrico Cerrato, and et al. 2022. "Prediction of All-Cause Mortality Following Percutaneous Coronary Intervention in Bifurcation Lesions Using Machine Learning Algorithms" Journal of Personalized Medicine 12, no. 6: 990. https://doi.org/10.3390/jpm12060990
APA StyleBurrello, J., Gallone, G., Burrello, A., Jahier Pagliari, D., Ploumen, E. H., Iannaccone, M., De Luca, L., Zocca, P., Patti, G., Cerrato, E., Wojakowski, W., Venuti, G., De Filippo, O., Mattesini, A., Ryan, N., Helft, G., Muscoli, S., Kan, J., Sheiban, I., ... D’Ascenzo, F. (2022). Prediction of All-Cause Mortality Following Percutaneous Coronary Intervention in Bifurcation Lesions Using Machine Learning Algorithms. Journal of Personalized Medicine, 12(6), 990. https://doi.org/10.3390/jpm12060990