An Integrated Approach for the Early Detection of Endometrial and Ovarian Cancers (Screenwide Study): Rationale, Study Design and Pilot Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Epidemiological and Clinical Questionnaires
2.2. Biological Samples and Histopathological Examination
2.3. Genomic Biomarkers in Minimally Invasive Sampling Methods—Pilot Study
2.4. Statistical and Cost-Effectiveness Analyses
2.5. Ethical Approval
3. Results
3.1. Overall Enrollment
3.2. Pilot Study to Detect Somatic Variants in Minimally Invasive Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today; International Agency for Research on Cancer: Lyon, France, 2018. [Google Scholar]
- Lortet-Tieulent, J.; Ferlay, J.; Bray, F.; Jemal, A. International Patterns and Trends in Endometrial Cancer Incidence, 1978–2013. JNCI J. Natl. Cancer Inst. 2018, 110, 354–361. [Google Scholar] [CrossRef]
- Hao, W.; Zhang, Y.; Li, Z.; Zhang, E.; Gao, S.; Yin, C.; Yue, W. International Trends in Ovarian Cancer Incidence from 1973 to 2012. Arch. Gynecol. Obstet. 2021, 303, 1589–1597. [Google Scholar] [CrossRef]
- Lheureux, S.; Gourley, C.; Vergote, I.; Oza, A.M. Epithelial Ovarian Cancer. Lancet 2019, 393, 1240–1253. [Google Scholar] [CrossRef] [Green Version]
- Crosbie, E.J.; Kitson, S.J.; McAlpine, J.N.; Mukhopadhyay, A.; Powell, M.E.; Singh, N. Endometrial Cancer. Lancet 2022, 399, 1412–1428. [Google Scholar] [CrossRef]
- Setiawan, V.W.; Yang, H.P.; Pike, M.C.; McCann, S.E.; Yu, H.; Xiang, Y.-B.; Wolk, A.; Wentzensen, N.; Weiss, N.S.; Webb, P.M.; et al. Type I and II Endometrial Cancers: Have They Different Risk Factors? J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2013, 31, 2607–2618. [Google Scholar] [CrossRef]
- Yang, H.P.; Cook, L.S.; Weiderpass, E.; Adami, H.-O.; Anderson, K.E.; Cai, H.; Cerhan, J.R.; Clendenen, T.V.; Felix, A.S.; Friedenreich, C.M.; et al. Infertility and Incident Endometrial Cancer Risk: A Pooled Analysis from the Epidemiology of Endometrial Cancer Consortium (E2C2). Br. J. Cancer 2015, 112, 925–933. [Google Scholar] [CrossRef] [Green Version]
- Setiawan, V.W.; Pike, M.C.; Karageorgi, S.; Deming, S.L.; Anderson, K.; Bernstein, L.; Brinton, L.A.; Cai, H.; Cerhan, J.R.; Cozen, W.; et al. Age at Last Birth in Relation to Risk of Endometrial Cancer: Pooled Analysis in the Epidemiology of Endometrial Cancer Consortium. Am. J. Epidemiol. 2012, 176, 269–278. [Google Scholar] [CrossRef]
- Raglan, O.; Kalliala, I.; Markozannes, G.; Cividini, S.; Gunter, M.J.; Nautiyal, J.; Gabra, H.; Paraskevaidis, E.; Martin-Hirsch, P.; Tsilidis, K.K.; et al. Risk Factors for Endometrial Cancer: An Umbrella Review of the Literature. Int. J. Cancer 2019, 145, 1719–1730. [Google Scholar] [CrossRef] [Green Version]
- Onstad, M.A.; Schmandt, R.E.; Lu, K.H. Addressing the Role of Obesity in Endometrial Cancer Risk, Prevention, and Treatment. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2016, 34, 4225–4230. [Google Scholar] [CrossRef]
- Tanha, K.; Mottaghi, A.; Nojomi, M.; Moradi, M.; Rajabzadeh, R.; Lotfi, S.; Janani, L. Investigation on Factors Associated with Ovarian Cancer: An Umbrella Review of Systematic Review and Meta-Analyses. J. Ovarian Res. 2021, 14, 153. [Google Scholar] [CrossRef]
- Papantoniou, K.; Pozo, O.J.; Espinosa, A.; Marcos, J.; Castaño-Vinyals, G.; Basagaña, X.; Juanola Pagès, E.; Mirabent, J.; Martín, J.; Such Faro, P.; et al. Increased and Mistimed Sex Hormone Production in Night Shift Workers. Cancer Epidemiol. Biomark. Prev. Publ. Am. Assoc. Cancer Res. Cosponsored Am. Soc. Prev. Oncol. 2015, 24, 854–863. [Google Scholar] [CrossRef] [Green Version]
- Viswanathan, A.N.; Hankinson, S.E.; Schernhammer, E.S. Night Shift Work and the Risk of Endometrial Cancer. Cancer Res. 2007, 67, 10618–10622. [Google Scholar] [CrossRef] [Green Version]
- Schwartzbaum, J.; Ahlbom, A.; Feychting, M. Cohort Study of Cancer Risk among Male and Female Shift Workers. Scand. J. Work. Environ. Health 2007, 33, 336–343. [Google Scholar] [CrossRef]
- Leung, L.; Grundy, A.; Siemiatycki, J.; Arseneau, J.; Gilbert, L.; Gotlieb, W.H.; Provencher, D.M.; Aronson, K.J.; Koushik, A. Shift Work Patterns, Chronotype, and Epithelial Ovarian Cancer Risk. Cancer Epidemiol. Biomark. Prev. Publ. Am. Assoc. Cancer Res. Cosponsored Am. Soc. Prev. Oncol. 2019, 28, 987–995. [Google Scholar] [CrossRef] [Green Version]
- Bhatti, P.; Cushing-Haugen, K.L.; Wicklund, K.G.; Doherty, J.A.; Rossing, M.A. Nightshift Work and Risk of Ovarian Cancer. Occup. Environ. Med. 2013, 70, 231–237. [Google Scholar] [CrossRef] [Green Version]
- Poole, E.M.; Schernhammer, E.S.; Tworoger, S.S. Rotating Night Shift Work and Risk of Ovarian Cancer. Cancer Epidemiol. Biomark. Prev. 2011, 20, 934–938. [Google Scholar] [CrossRef] [Green Version]
- Dun, A.; Zhao, X.; Jin, X.; Wei, T.; Gao, X.; Wang, Y.; Hou, H. Association Between Night-Shift Work and Cancer Risk: Updated Systematic Review and Meta-Analysis. Front. Oncol. 2020, 10, 1006. [Google Scholar] [CrossRef]
- Zheng, W.; Xiang, L.; Fadare, O.; Kong, B. A Proposed Model for Endometrial Serous Carcinogenesis. Am. J. Surg. Pathol. 2011, 35, e1–e14. [Google Scholar] [CrossRef]
- Fadare, O.; Zheng, W. Endometrial Glandular Dysplasia (EmGD): Morphologically and Biologically Distinctive Putative Precursor Lesions of Type II Endometrial Cancers. Diagn. Pathol. 2008, 3, 6. [Google Scholar] [CrossRef] [Green Version]
- Labidi-Galy, S.I.; Papp, E.; Hallberg, D.; Niknafs, N.; Adleff, V.; Noe, M.; Bhattacharya, R.; Novak, M.; Jones, S.; Phallen, J.; et al. High Grade Serous Ovarian Carcinomas Originate in the Fallopian Tube. Nat. Commun. 2017, 8, 1093. [Google Scholar] [CrossRef]
- Shih, I.-M.; Wang, Y.; Wang, T.-L. The Origin of Ovarian Cancer Species and Precancerous Landscape. Am. J. Pathol. 2021, 191, 26–39. [Google Scholar] [CrossRef] [PubMed]
- Maksem, J.A.; Robboy, S.J.; Bishop, J.W.; Meiers, I. Endometrial Precancer. In Endometrial Cytology with Tissue Correlations; Essentials in Cytopathology; Springer: Boston, MA, USA, 2009; pp. 189–230. ISBN 978-0-387-89909-1. [Google Scholar]
- Cancer Genome Atlas Research Network; Kandoth, C.; Schultz, N.; Cherniack, A.D.; Akbani, R.; Liu, Y.; Shen, H.; Robertson, A.G.; Pashtan, I.; Shen, R.; et al. Integrated Genomic Characterization of Endometrial Carcinoma. Nature 2013, 497, 67–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, L.; Liu, Y.; Yi, X.; Miron, A.; Crum, C.P.; Kong, B.; Zheng, W. Endometrial Glandular Dysplasia with Frequent P53 Gene Mutation: A Genetic Evidence Supporting Its Precancer Nature for Endometrial Serous Carcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2008, 14, 2263–2269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Putten, L.J.M.; van Hoof, R.; Tops, B.B.J.; Snijders, M.P.L.M.; van den Berg-van Erp, S.H.; van der Wurff, A.A.M.; Bulten, J.; Pijnenborg, J.M.A.; Massuger, L.F.A.G. Molecular Profiles of Benign and (Pre)Malignant Endometrial Lesions. Carcinogenesis 2017, 38, 329–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurman, R.J. WHO Classification of Tumours of Female Reproductive Organs. In World Health Organization Classification of Tumours, 4th ed.; International Agency for Research on Cancer, World Health Organization, Eds.; International Agency for Research on Cancer: Lyon, France, 2014; ISBN 978-92-832-2435-8. [Google Scholar]
- Jacobs, I.J.; Menon, U.; Ryan, A.; Gentry-Maharaj, A.; Burnell, M.; Kalsi, J.K.; Amso, N.N.; Apostolidou, S.; Benjamin, E.; Cruickshank, D.; et al. Ovarian Cancer Screening and Mortality in the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS): A Randomised Controlled Trial. Lancet Lond. Engl. 2016, 387, 945–956. [Google Scholar] [CrossRef] [Green Version]
- Costas, L.; Frias-Gomez, J.; Guardiola, M.; Benavente, Y.; Pineda, M.; Pavón, M.Á.; Martínez, J.M.; Climent, M.; Barahona, M.; Canet, J.; et al. New Perspectives on Screening and Early Detection of Endometrial Cancer. Int. J. Cancer 2019, 145, 3194–3206. [Google Scholar] [CrossRef]
- Nebgen, D.R.; Lu, K.H.; Bast, R.C. Novel Approaches to Ovarian Cancer Screening. Curr. Oncol. Rep. 2019, 21, 75. [Google Scholar] [CrossRef]
- Wang, Y.; Li, L.; Douville, C.; Cohen, J.D.; Yen, T.-T.; Kinde, I.; Sundfelt, K.; Kjær, S.K.; Hruban, R.H.; Shih, I.-M.; et al. Evaluation of Liquid from the Papanicolaou Test and Other Liquid Biopsies for the Detection of Endometrial and Ovarian Cancers. Sci. Transl. Med. 2018, 10, eaap8793. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.C.; Wang, H.C.; Liao, Y.P.; Chen, Y.C.; Weng, Y.C.; Yu, M.H.; Lai, H.C. The Feasibility of Detecting Endometrial and Ovarian Cancer Using DNA Methylation Biomarkers in Cervical Scrapings. J. Gynecol. Oncol. 2018, 29, e17. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Zhang, Z.-H.; Wang, S.; Lang, J.-H. Circulating Cell-Free DNA or Circulating Tumor DNA in the Management of Ovarian and Endometrial Cancer. OncoTargets Ther. 2019, 12, 11517–11530. [Google Scholar] [CrossRef] [Green Version]
- Reijnen, C.; Putten, L.J.M.; Bulten, J.; Snijders, M.P.L.M.; Küsters-Vandevelde, H.V.N.; Sweegers, S.; Vos, M.C.; Wurff, A.A.M.; Ligtenberg, M.J.L.; Massuger, L.F.A.G.; et al. Mutational Analysis of Cervical Cytology Improves Diagnosis of Endometrial Cancer: A Prospective Multicentre Cohort Study. Int. J. Cancer 2020, 146, 2628–2635. [Google Scholar] [CrossRef] [PubMed]
- Bakkum-Gamez, J.N.; Wentzensen, N.; Maurer, M.J.; Hawthorne, K.M.; Voss, J.S.; Kroneman, T.N.; Famuyide, A.O.; Clayton, A.C.; Halling, K.C.; Kerr, S.E.; et al. Detection of Endometrial Cancer via Molecular Analysis of DNA Collected with Vaginal Tampons. Gynecol. Oncol. 2015, 137, 14–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costas, L.; Palomero, L.; Benavente, Y.; Guardiola, M.; Frias-Gomez, J.; Pavón, M.Á.; Climent, M.; Martinez, J.M.; Barahona, M.; Salinas, M.; et al. Defining a Mutational Signature for Endometrial Cancer Screening and Early Detection. Cancer Epidemiol. 2019, 61, 129–132. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Jiménez, F.; Muiños, F.; Sentís, I.; Deu-Pons, J.; Reyes-Salazar, I.; Arnedo-Pac, C.; Mularoni, L.; Pich, O.; Bonet, J.; Kranas, H.; et al. A Compendium of Mutational Cancer Driver Genes. Nat. Rev. Cancer 2020, 20, 555–572. [Google Scholar] [CrossRef]
- Sullivan, L.M.; Massaro, J.M.; D’Agostino, R.B. Presentation of Multivariate Data for Clinical Use: The Framingham Study Risk Score Functions. Stat. Med. 2004, 23, 1631–1660. [Google Scholar] [CrossRef]
- Slattery, M.L.; Edwards, S.L.; Caan, B.J.; Kerber, R.A.; Potter, J.D. Response Rates among Control Subjects in Case-Control Studies. Ann. Epidemiol. 1995, 5, 245–249. [Google Scholar] [CrossRef]
- Frias-Gomez, J.; Tovar, E.; Vidal, A.; Murgui, L.; Ibáñez, R.; Peremiquel-Trillas, P.; Paytubi, S.; Baixeras, N.; Zanca, A.; Ponce, J.; et al. Sensitivity of Cervical Cytology in Endometrial Cancer Detection in a Tertiary Hospital in Spain. Cancer Med. 2021, 10, 6762–6766. [Google Scholar] [CrossRef]
- Lac, V.; Nazeran, T.M.; Tessier-Cloutier, B.; Aguirre-Hernandez, R.; Albert, A.; Lum, A.; Khattra, J.; Praetorius, T.; Mason, M.; Chiu, D.; et al. Oncogenic Mutations in Histologically Normal Endometrium: The New Normal? J. Pathol. 2019, 249, 173–181. [Google Scholar] [CrossRef]
- Ketelaars, P.J.W.; Bosgraaf, R.P.; Siebers, A.G.; Massuger, L.F.a.G.; van der Linden, J.C.; Wauters, C.a.P.; Rahamat-Langendoen, J.C.; van den Brule, A.J.C.; IntHout, J.; Melchers, W.J.G.; et al. High-Risk Human Papillomavirus Detection in Self-Sampling Compared to Physician-Taken Smear in a Responder Population of the Dutch Cervical Screening: Results of the VERA Study. Prev. Med. 2017, 101, 96–101. [Google Scholar] [CrossRef]
- Mota, A.; Colás, E.; García-Sanz, P.; Campoy, I.; Rojo-Sebastián, A.; Gatius, S.; García, Á.; Chiva, L.; Alonso, S.; Gil-Moreno, A.; et al. Genetic Analysis of Uterine Aspirates Improves the Diagnostic Value and Captures the Intra-Tumor Heterogeneity of Endometrial Cancers. Mod. Pathol. 2017, 30, 134–145. [Google Scholar] [CrossRef] [Green Version]
- Salk, J.J.; Schmitt, M.W.; Loeb, L.A. Enhancing the Accuracy of Next-Generation Sequencing for Detecting Rare and Subclonal Mutations. Nat. Rev. Genet. 2018, 19, 269–285. [Google Scholar] [CrossRef]
- Broad Institute Picard Toolkit; Broad Institute, GitHub Repository, 2019. Available online: https://broadinstitute.github.io/picard/ (accessed on 28 May 2021).
- Fulcrum Genomics Fgbio. 2020. Available online: http://fulcrumgenomics.github.io/fgbio/ (accessed on 29 May 2021).
- Li, H. Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Smith, T.; Heger, A.; Sudbery, I. UMI-Tools: Modeling Sequencing Errors in Unique Molecular Identifiers to Improve Quantification Accuracy. Genome Res. 2017, 27, 491–499. [Google Scholar] [CrossRef] [Green Version]
- Lai, Z.; Markovets, A.; Ahdesmaki, M.; Chapman, B.; Hofmann, O.; McEwen, R.; Johnson, J.; Dougherty, B.; Barrett, J.C.; Dry, J.R. VarDict: A Novel and Versatile Variant Caller for next-Generation Sequencing in Cancer Research. Nucleic Acids Res. 2016, 44, e108. [Google Scholar] [CrossRef] [PubMed]
- Benjamin, D.; Sato, T.; Cibulskis, K.; Getz, G.; Stewart, C.; Lichtenstein, L. Calling Somatic SNVs and Indels with Mutect2. Available online: https://www.biorxiv.org/content/10.1101/861054v1 (accessed on 22 June 2022).
- Robinson, J.T.; Thorvaldsdóttir, H.; Wenger, A.M.; Zehir, A.; Mesirov, J.P. Variant review with the Integrative Genomics Viewer. Cancer Res. 2017, 77, e31–e34. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional Annotation of Genetic Variants from High-Throughput Sequencing Data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wang, K. Genomic Variant Annotation and Prioritization with ANNOVAR and WANNOVAR. Nat. Protoc. 2015, 10, 1556–1566. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.; Leongamornlert, D.; Coorens, T.H.H.; Sanders, M.A.; Ellis, P.; Dentro, S.C.; Dawson, K.J.; Butler, T.; Rahbari, R.; Mitchell, T.J.; et al. The Mutational Landscape of Normal Human Endometrial Epithelium. Nature 2020, 580, 640–646. [Google Scholar] [CrossRef] [PubMed]
- Tamborero, D.; Rubio-Perez, C.; Deu-Pons, J.; Schroeder, M.P.; Vivancos, A.; Rovira, A.; Tusquets, I.; Albanell, J.; Rodon, J.; Tabernero, J.; et al. Cancer Genome Interpreter Annotates the Biological and Clinical Relevance of Tumor Alterations. Genome Med. 2018, 10, 25. [Google Scholar] [CrossRef]

| Participants | Epidemiologic Questionnaire | Blood | Vaginal Self-Samples | Cervical Pap-Brush Samples | Endometrial Aspirates | Tumour Samples | |
|---|---|---|---|---|---|---|---|
| Endometrial cancer 1 | 182 | 180 | 174 | 175 | 168 | 161 | 165 |
| Ovarian cancer | 69 | 69 | 67 | 66 | 62 | 56 | 52 |
| MMR pathogenic variant carriers 2 | 104 | 103 | 1 | 104 | 102 | 90 | NA |
| BRCA pathogenic variant carriers 3 | 98 | 98 | 0 | 98 | 98 | 0 | NA |
| Healthy women attending CC screening | 119 | 119 | 33 | 119 | 118 | 0 | NA |
| Controls with gynaecologic benign conditions | 190 | 146 | 117 | 176 | 151 | 105 | NA |
| Hospital controls (non-gynaecologic) | 76 | 72 | 76 | 48 | 0 | 0 | NA |
| TOTAL | 838 | 787 | 468 | 786 | 699 | 412 | 217 |
| High Risk Populations 1 | Controls 2 | EC 3 | OC | p-Values 4 | |
|---|---|---|---|---|---|
| n (%) | n (%) | n (%) | n (%) | ||
| Participants | 202 | 385 | 182 | 69 | |
| Epidemiologic questionnaire | 201 (99.5) | 337 (87.5) | 180 (98.9) | 69 (100.0) | |
| Age | 0.740/0.124 | ||||
| <60 | 188 (93.1) | 165 (42.9) | 51 (28.0) | 26 (37.7) | |
| 60–69 | 12 (5.9) | 108 (28.1) | 61 (33.5) | 22 (31.9) | |
| ≥70 | 2 (1.0) | 112 (29.1) | 70 (38.5) | 21 (30.4) | |
| Education 5 | 0.426/0.997 | ||||
| High School or below | 63 (31.3) | 240 (71.2) | 131 (72.8) | 52 (75.4) | |
| Some college/associate | 64 (31.8) | 70 (20.8) | 29 (16.1) | 12 (17.4) | |
| College or above | 74 (36.8) | 27 (8.0) | 20 (11.1) | 5 (7.2) | |
| BMI 5 | <0.001/0.500 | ||||
| <18.5 | 10 (5.0) | 4 (1.2) | 1 (0.6) | 3 (4.3) | |
| 18.5–24.99 | 110 (54.7) | 99 (29.4) | 26 (14.4) | 21 (30.4) | |
| 25–29.99 | 46 (22.9) | 131 (38.9) | 52 (28.9) | 24 (34.8) | |
| ≥30 | 28 (13.9) | 89 (26.4) | 95 (52.8) | 20 (29.0) | |
| Previous history of cancer (other than EC or OC) | 0.346/0.791 | ||||
| Yes | 56 (27.7) | 47 (12.2) | 29 (15.9) | 8 (11.6) | |
| No | 146 (57.2) | 338 (87.8) | 153 (84.1) | 61 (88.4) | |
| Menopausal status 5 | 0.001/0.633 | ||||
| Premenopausal | 151 (75.1) | 63 (18.7) | 7 (3.9) | 13 (18.8) | |
| Perimenopausal | 14 (7.0) | 29 (8.6) | 12 (6.7) | 2 (2.9) | |
| Postmenopausal | 36 (17.9) | 245 (72.7) | 161 (89.4) | 54 (78.3) | |
| Parity 5 | 0.792/0.122 | ||||
| Nulliparous | 77 (38.3) | 36 (10.7) | 26 (14.4) | 10 (14.5) | |
| 1 | 43 (21.4) | 70 (20.8) | 28 (15.6) | 20 (29.0) | |
| ≥2 | 81 (40.3) | 229 (68.0) | 126 (70.0) | 39 (56.5) | |
| Hormonal contraception5 | 0.002/0.483 | ||||
| Never | 53 (26.4) | 114 (33.8) | 107 (59.4) | 34 (49.3) | |
| Ever | 148 (73.6) | 221 (65.6) | 72 (40.0) | 35 (50.7) | |
| Tobacco consumption 5 | 0.651/0.794 | ||||
| Never | 81 (40.3) | 193 (57.3) | 126 (70.0) | 48 (69.6) | |
| Ever | 120 (59.7) | 144 (42.7) | 54 (30.0) | 21 (30.4) | |
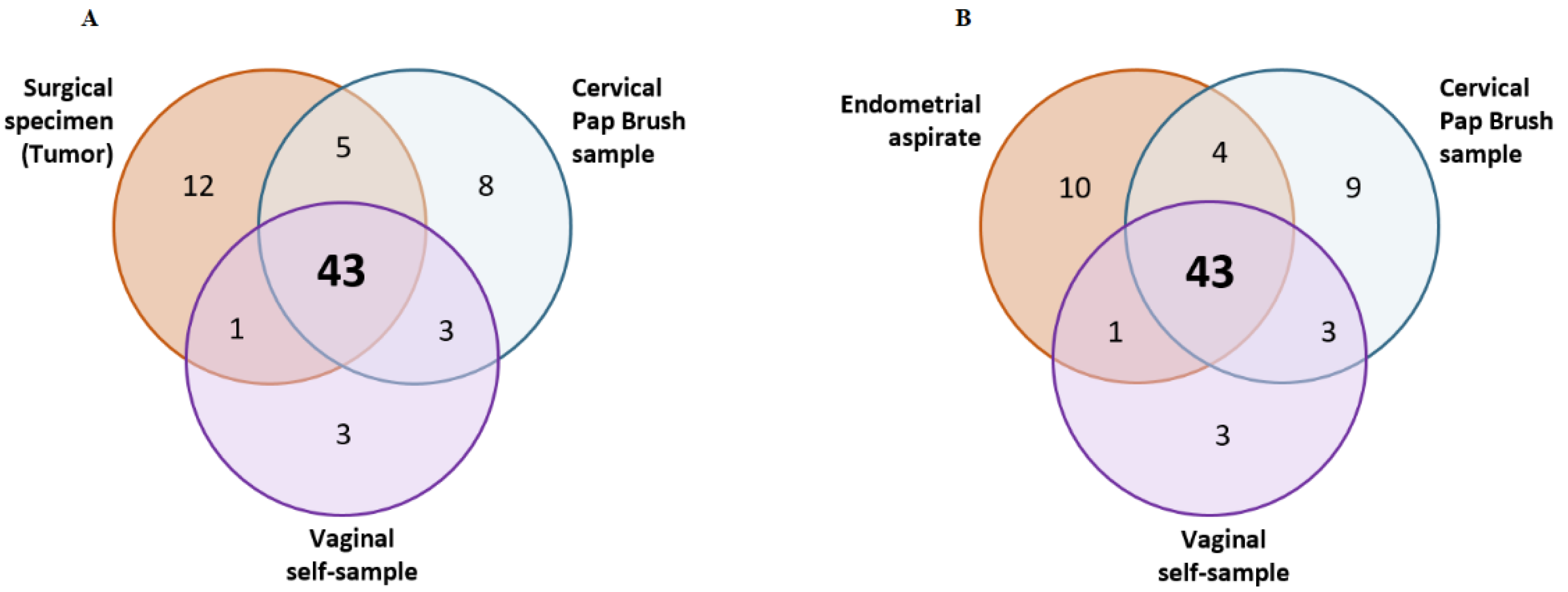
| Patient ID | Case-Control Status | Tumor 1 | Endometrial Aspirates 2 | Cervical Pap Brush Samples 3 | Vaginal Self-Samples 3 | |||
|---|---|---|---|---|---|---|---|---|
| Total Nº variants | Total Nº variants | Total Nº variants | Total Nº variants | |||||
| S110444 | Control | NA | 0 | 0 | 0 | |||
| S110449 | Control | NA | 0 | 1 | 0 | |||
| S110565 | Control | NA | 0 | 0 | 0 | |||
| S110574 | Control | NA | 0 | 0 | 0 | |||
| Total Nº variants | Total Nº variants | Nº variants also identified in tumour | Total Nº variants | Nº variants also identified in tumour | Total Nº variants | Nº variants also identified in tumour | ||
| S110036 | Case | 4 | 4 | 4 | 4 | 3 | 2 | 2 |
| S110081 | Case | 25 | 20 | 20 | 24 | 21 | 19 | 19 |
| S110124 | Case | 4 | 4 | 4 | 0 | 0 | 0 | 0 |
| S110424 | Case | 3 | 4 | 3 | 3 | 3 | 2 | 2 |
| S110435 | Case | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| S110451 | Case | 3 | 3 | 3 | 1 | 1 | 0 | 0 |
| S110474 | Case | 11 | 12 | 10 | 14 | 10 | 16 | 10 |
| S110475 | Case | 5 | 5 | 5 | 6 | 4 | 5 | 5 |
| S110501 | Case | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Total | 61 | 58 | 55 | 58 | 48 | 50 | 44 | |
| % (95% CI) | 95 (86–98) 5 | 90 (80–95) 4 | 84 (73–92) 5 | 79 (67–87) 4 | 90 (79–96) 5 | 72 (60–82) 4 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peremiquel-Trillas, P.; Paytubi, S.; Pelegrina, B.; Frias-Gomez, J.; Carmona, Á.; Martínez, J.M.; de Francisco, J.; Benavente, Y.; Barahona, M.; Briansó, F.; et al. An Integrated Approach for the Early Detection of Endometrial and Ovarian Cancers (Screenwide Study): Rationale, Study Design and Pilot Study. J. Pers. Med. 2022, 12, 1074. https://doi.org/10.3390/jpm12071074
Peremiquel-Trillas P, Paytubi S, Pelegrina B, Frias-Gomez J, Carmona Á, Martínez JM, de Francisco J, Benavente Y, Barahona M, Briansó F, et al. An Integrated Approach for the Early Detection of Endometrial and Ovarian Cancers (Screenwide Study): Rationale, Study Design and Pilot Study. Journal of Personalized Medicine. 2022; 12(7):1074. https://doi.org/10.3390/jpm12071074
Chicago/Turabian StylePeremiquel-Trillas, Paula, Sonia Paytubi, Beatriz Pelegrina, Jon Frias-Gomez, Álvaro Carmona, José Manuel Martínez, Javier de Francisco, Yolanda Benavente, Marc Barahona, Ferran Briansó, and et al. 2022. "An Integrated Approach for the Early Detection of Endometrial and Ovarian Cancers (Screenwide Study): Rationale, Study Design and Pilot Study" Journal of Personalized Medicine 12, no. 7: 1074. https://doi.org/10.3390/jpm12071074







