Abstract
The introduction of personalized medicine, through the increasing multi-omics characterization of disease, brings new challenges to disease modeling. The scope of this review was a broad evaluation of the relevance, validity, and predictive value of the current preclinical methodologies applied in stratified medicine approaches. Two case models were chosen: oncology and brain disorders. We conducted a scoping review, following the Joanna Briggs Institute guidelines, and searched PubMed, EMBASE, and relevant databases for reports describing preclinical models applied in personalized medicine approaches. A total of 1292 and 1516 records were identified from the oncology and brain disorders search, respectively. Quantitative and qualitative synthesis was performed on a final total of 63 oncology and 94 brain disorder studies. The complexity of personalized approaches highlights the need for more sophisticated biological systems to assess the integrated mechanisms of response. Despite the progress in developing innovative and complex preclinical model systems, the currently available methods need to be further developed and validated before their potential in personalized medicine endeavors can be realized. More importantly, we identified underlying gaps in preclinical research relating to the relevance of experimental models, quality assessment practices, reporting, regulation, and a gap between preclinical and clinical research. To achieve a broad implementation of predictive translational models in personalized medicine, these fundamental deficits must be addressed.
1. Introduction
The emergence of personalized medicine (PM) (refer to Box 1 for definition), through phenotype presentation and individual omics characterization, demands preclinical models which can recapitulate clinical features and provide predictive “personalized” data. This has meant a shift in translational research away from the “one size fits all” demonstration of efficacy, towards developing more sophisticated and targeted preclinical models. There are many definitions of translational research; in this review we define it as a scientific process to improve human health via a “bench-to-bedside” approach. Translational methods that integrate the increasing molecular categorization of diseases can inform a stratified medicine approach, and potentially predict treatment responses in subgroups of patients with certain biological signatures.
Box 1. Personalized medicine.
According to the European Council Conclusion on personalized medicine for patients personalized medicine is “a medical model using characterisation of individuals’ phenotypes and genotypes (e.g., molecular profiling, medical imaging, lifestyle data) for tailoring the right therapeutic strategy for the right person at the right time, and/or to determine the predisposition to disease and/or to deliver timely and targeted prevention [1]”. In the context of the PERMIT project, we applied the following common operational definition of personalized medicine research: a set of comprehensive methods, (methodological, statistical, validation, or technologies) to be applied in the different phases of the development of a personalized approach to treatment, diagnosis, prognosis, or risk prediction. Ideally, robust and reproducible methods should cover all the steps between the generation of the hypothesis (e.g., a given stratum of patients could better respond to a treatment), its validation and preclinical development, and up to the definition of its value in a clinical setting.
Rodent models are by far the most used in vivo models in translational research [2]. In fact, much of our current understanding of mechanisms of disease is based on research in these animals, but there is concern about the limited translational power [3,4,5,6,7]. In recent decades, animal models have been refined through precise genetic modifications, and there are well established genetically engineered mouse models (GEMMs) for many diseases [8,9]. The “mouse avatar” concept, where patient tissue is directly xenografted in vivo and studied, is now an established technique in the field of oncology [10,11]. This model has been applied in co-clinical trials, where mouse experiments are developed in parallel with human clinical trials in order to enable real-time transfer of information [12]. Correspondingly, the emergence of advanced in vitro techniques, such as the development of 3D cell cultures and organoids, and the integration of microfluidics to create organ-on-chip technologies, bring the promise of patient-derived personalized cellular models in the future [13,14,15,16,17,18]. Patient-derived cancer organoids are already being employed for drug-screening applications [19]. However, currently, one of the main challenges with both these patient-derived models is the absence of a functional immune system. In silico modeling, referring to mathematical and computational models of biological systems, is another approach which can complement the personalized approach, aiming to make predictions on drug targets, drug efficacy, and patient responses [20,21].
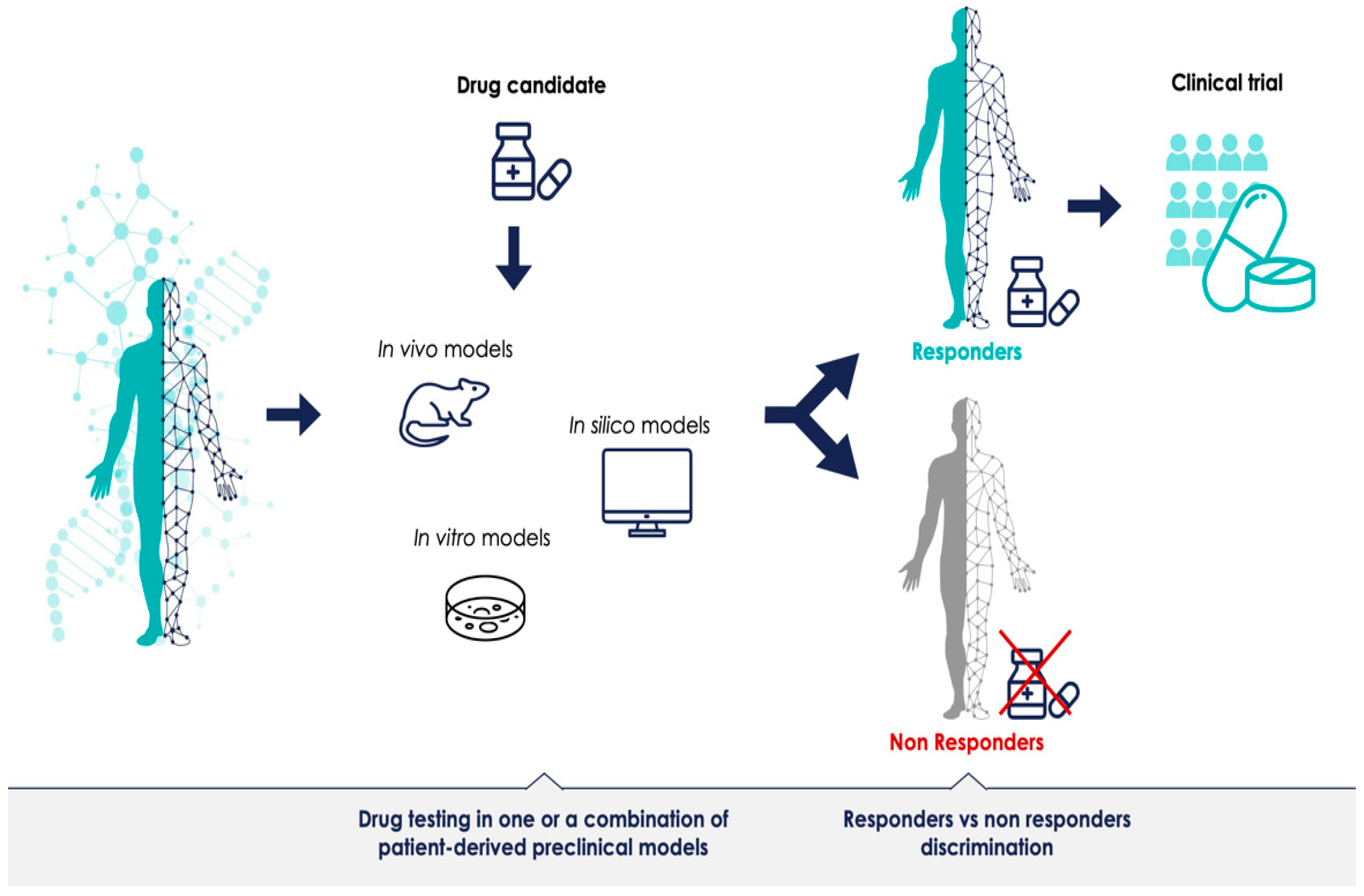
The promise of PM is that each patient’s treatment can be optimally tailored to their disease. Presumably, models—or a combination of models—that can successfully discriminate between responders and non-responders for a given treatment can provide predictive data prior to therapeutic clinical trials (Figure 1). Most preclinical models have been generated to understand disease mechanisms; for instance, defining a specific phenotype or a biological molecular mechanism. However, if preclinical model systems should have a predictive value as well, meaning that the results obtained with the model predict outcomes in humans and can support the decision to initiate and authorize clinical trials in PM, they must generate reliable data. The advancement of preclinical research in this regard is promising, but most of the methodologies are still in their infancy. There is a need for more robust preclinical resources which can validate biomarkers and demonstrate the clinical utility of the stratified approach. The identification of bottlenecks and challenges of preclinical methods is the first step towards defining a shared PM development strategy, and can lay the foundation for more successful clinical trials across the sector.
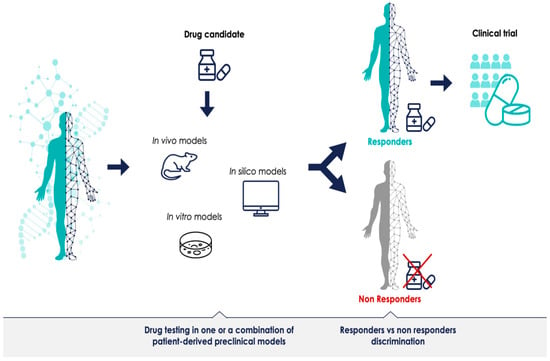
Figure 1.
Predictive patient-derived translational models for personalized medicine. Preclinical development in clinically relevant models with robust predictions could improve clinical trials for personalized medicine.
The scope of this review is a broad focus on the current preclinical methodologies used to support PM approaches. We aim to highlight the advantages and disadvantages of applying the existing preclinical model systems in the domain of PM and evaluate the status of emerging models. As translational research is a very broad topic, we decided to concentrate on two case models to make the search more manageable. We chose oncology and brain disorders, specifically neuropsychiatric, neurodegenerative, and neurodevelopmental disorders. We reviewed in vivo, in vitro, and in silico methods, and we assessed the models for relevance, validity, and predictive value in the context of PM, highlighting advantages and disadvantages.
This scoping review is part of the PERMIT project (PERsonalized MedIcine Trials), which aims to map the methods for personalized medicine research and build recommendations for robustness and reproducibility of different stages of the development programs. Although several categorization may be proposed, the PERMIT project considers four main building blocks of the personalized medicine research pipeline: (1) design, building and management of stratification and validation cohorts; (2) application of machine learning methods for patient stratification; (3) use of preclinical methods for translational development, including the use of preclinical models used to assign treatments to patient clusters; and (4) evaluation of treatments in randomized clinical trials [22,23,24]. This scoping review covers the third building block in this framework.
2. Materials and Methods
We conducted a scoping review, following the methodological framework suggested by the Joanna Briggs Institute [25,26,27]. The framework consists of six stages: (1) identifying the research questions; (2) identifying relevant studies; (3) study selection; (4) charting the data; (5) collating, summarizing, and reporting results; and (6) consultation.
The scoping review approach was considered by the PERMIT consortium to be the most suitable to respond to the broad scope of the field. Compared to systematic reviews that aim to answer specific questions, scoping reviews are used to determine the scope of available evidence in a given field and examine how research is conducted in that field, and they are useful to examine areas that are emerging, to clarify key concepts, and to identify gaps [28]. A study protocol was published in Zenodo before conducting the review (https://zenodo.org/record/3770937, accessed on 30 April 2022) [29]. Due to the iterative nature of scoping reviews, deviations from the protocol are expected and duly reported when occurred. We used the PRISMA-ScR (Preferred Reporting Items for Systematic reviews and Meta-Analyses extension for Scoping Reviews) checklist to report our results [30].
2.1. Research Questions
The main research questions addressed were:
- Which preclinical models are currently used to provide validity data prior to therapeutic clinical trials of PM in oncology and brain disorders and what are the pros and cons of the applied methods?
- Are the current preclinical models predictive for the outcome of PM trials?
2.2. Study Identification
Relevant studies and documents were identified, balancing feasibility with the breadth and comprehensiveness of searches. We searched PubMed, EMBASE, the Cochrane Library, Web of Science, and PsycInfo (search dates: March–June 2020) for research papers and (systematic) reviews in the fields of brain disorders and oncology to identify the most common methodological approaches. Online Supplementary File S1 reports the search strategies applied. We limited our search from 2005 to April 2020 and restricted inclusion to English, French, German, Italian, and Spanish languages. We also searched for gray literature [31] on relevant websites and by consulting partners of the PERMIT project.
2.3. Study Selection and Eligibility Criteria
The title and abstracts of records identified by the literature search were screened by two independent reviewers (EO, VF) using the Rayyan online tool [32]. The full-text publications of the relevant articles related to oncology were retrieved and examined by VF, whereas EO retrieved and assessed the articles related to brain disorders to confirm eligibility. Discrepancies were solved by discussion among the review team and the methods group (CG, RB, MF).
Research papers and reviews describing preclinical methods (i.e., cellular, organoid, animal, and in silico models) were considered in the broad context of PM development. We focused on reports assessing the validity, reliability, and predictive value of these methodologies applied to the two case models used in this study, oncology and brain disorders. No restrictions in terms of types of publications were included. References with a focus on disease specific issues, or which did not focus on a personalized approach of the methodology, were excluded. We also excluded congress reports and abstracts.
2.4. Charting the Data
We designed a data extraction form using an Excel file (Online Supplementary File S2). The general study characteristics extracted were as follows: first author name, title of article, year of publication, and type of publication. In addition, for each preclinical model referred to in the paper, we collected information on its definition, the preclinical model type, methodology, advantages, disadvantages, internal and external validity—as previously defined—and applications in PM. During the data extraction phase, the aspects covered by one or more research questions were summarized in tables by one reviewer for each search (VF, EO). Since many narrative reviews have been published about preclinical models, we decided to extract data first from reviews, adding relevant missing information from the remaining research papers. This aspect was not specified in the protocol, but was agreed among the authors before starting the extraction phase process. Assessment of the methodological quality of individual studies included in the analysis was not within the remit of this scoping review.
2.5. Consultation Exercise
The members of the PERMIT consortium, associated partners, and the PERMIT project Scientific Advisory Board discussed the preliminary findings of the scoping review in an two-hour long online workshop, held on 2 December 2020.
2.6. Patient and Public Involvement
The European Patients’ Forum is a member of the PERMIT project. Although not directly involved in the conduction of the scoping review, they received the draft review protocol to collect comments and feedback.
3. Results
3.1. Study Selection and General Characteristics of Reports
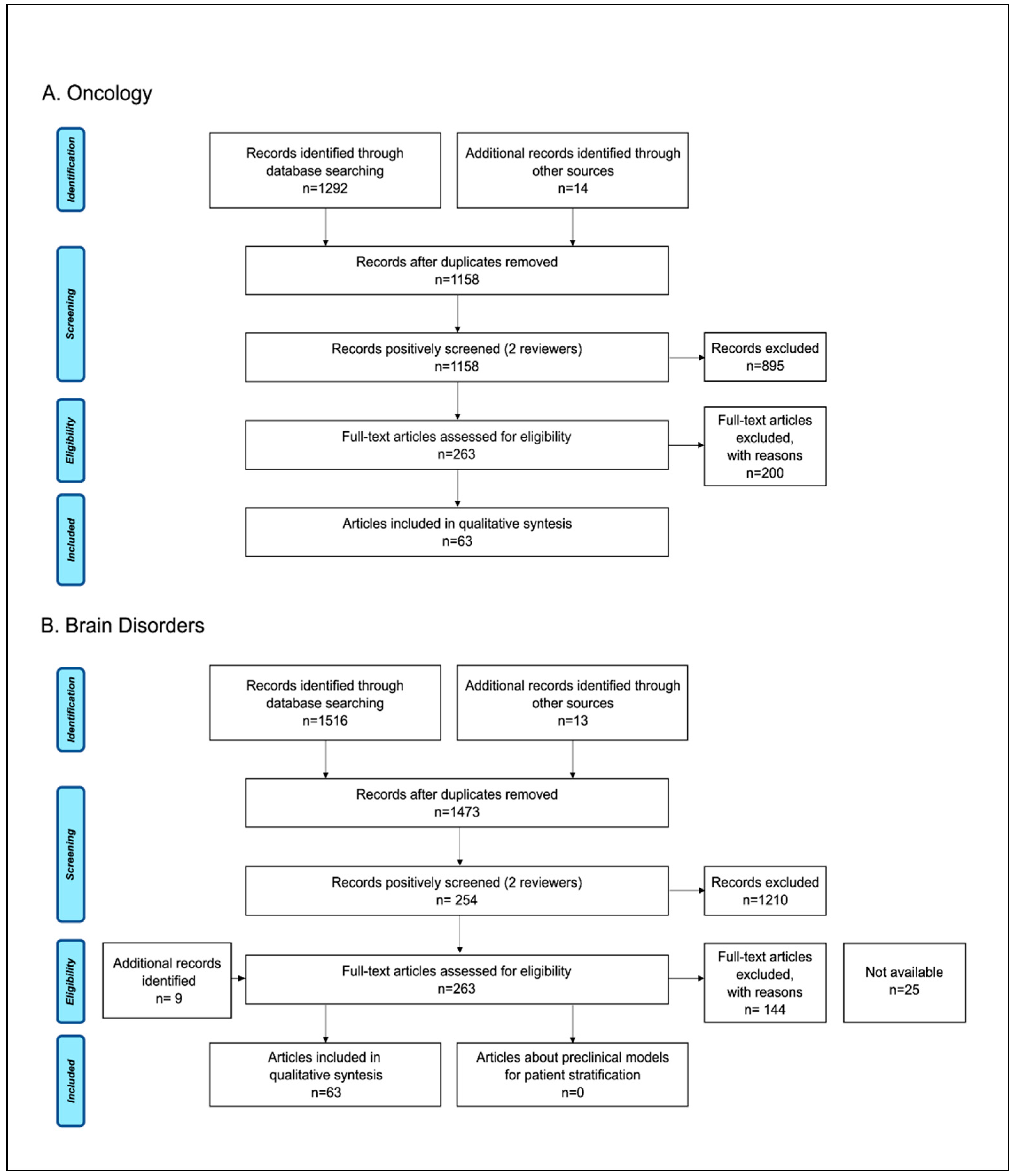
A total of 1292 records were identified from the oncology search, with an additional 14 records identified through manual search. After removal of duplication, 1158 records remained, and initial screening left 263 articles for full-text evaluation. A final total of 63 studies (3 systematic reviews, 54 narrative reviews, and 6 research papers) met the inclusion criteria and were reviewed for the analysis. For the brain disorders use case, we identified 1516 articles through the literature search and 22 additional records through manual search. After screening of the 1473 unique articles, a full-text review was performed on 263 articles. A total of 94 studies (54 reviews and 40 research papers) met the inclusion criteria and were included in the qualitative synthesis. Figure 2 reports the process for article selection. A list of the included studies can be found in the open access database Zenodo (https://zenodo.org/record/6087847, accessed on 30 April 2022).
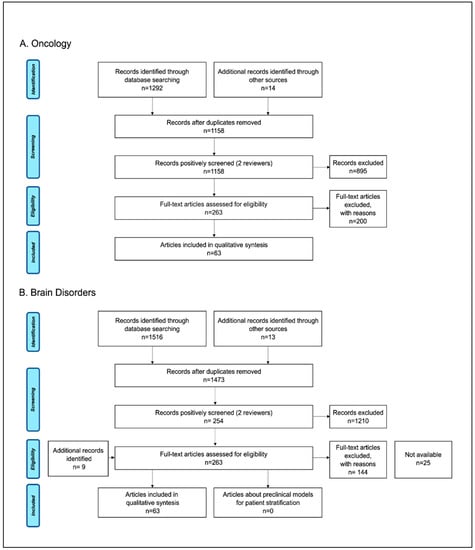
Figure 2.
Study selection flow diagram: PRISMA flow-chart of data collection process for the (A) oncology and (B) brain disorders literature searches.
The year of publication ranged from 2005 to 2020. The median year was 2017 for oncology, and for brain disorders, the range was 2008 to 2020, and the median was 2016. The types of papers reviewed were mainly narrative reviews (86% in the oncology search and 87% in the brain disorder search); only 5% were systematic reviews in the oncology search, no systematic reviews were identified in the brain disorders search. The remaining papers were research articles.
The literature search, including searching gray literature, did not yield any results in either use case (brain disorders and oncology) where the specific preclinical data that provided the support for approving a clinical trial in PM were reported. Therefore, the included papers were mainly narrative reviews and research articles describing preclinical models and their potential application of PM development. Below, we provide a brief synthesis of the main application towards personalized medicine for the different model systems in two selected use cases. Detailed analysis about specific methods was beyond the scope of this study.
3.2. In Vivo Models for PM
Animal models were the most described model for PM approaches, comprising 47% of references in the oncology use case, of which 2/3 related to mouse models. The remainder described various, less commonly used animal-derived models such as chicken chorioallantoic membrane, zebrafish, and companion animal comparative oncology (the study of naturally developing cancers in pets as models for human disease). In the brain disorders use case, 62% of papers referring to animal models were included in the qualitative synthesis. Among them, more than half described rodent models and the rest regarded less commonly used animal models, such as zebrafish, tapeworm, and fruit fly, among others.
Animal models can provide an important instrument for understanding pathogenic mechanisms, identifying drug targets, and developing new therapeutic approaches, but challenges remain for recapitulating the human phenotypes of diseases and to discriminate between successful and unsuccessful treatments [33]. In oncology, the field has progressed some way towards meeting these requirements through the development of patient-derived xenografts (PDXs), where patient tumors are implanted directly into immunodeficient mice [34]. These mouse models largely encapsulate the inter- and intra-tumor heterogeneity observed in cancer, and patient stratification modeling has been attempted in these models; nevertheless, the approach is limited by variable engraftment rates and issues relating to the validity of disease representation in the model [35]. One of the main limitations of PDXs is their immunodeficient status, a prerequisite to facilitate xenotransplantation, but which limits the evaluation of immunological effects. To overcome this, various methodologies are employed to generate a competent human immune system in these models, which are called humanized PDX, leading to different degrees of immune reconstitution. Currently, the major limitation of this approach is the durability and quality of engraftment of the human immune system [11,36,37,38]. Another approach for preclinical modeling of patient stratification in oncology is the co-clinical trials approach, which has been achieved using both PDXs and GEMMs, but the time and cost involved limit the applicability [39].
None of the articles about brain disorders described animal models which can recapitulate the heterogeneity of the human disease phenotypes. Very recent studies in synucleinopathies, diseases characterized by pathological accumulation of aggregated asyn protein, describe animal models that incorporate the two aspects of α-synuclein pathobiology, attempting to reproduce the phenotypic differences seen in the clinical setting. The predictive value of these models remains to be defined, but it can be a first step for the development of preclinical personalized approaches in the field [40,41]. The opportunity to develop chimeric humanized animal models using patient-derived CNS stem cells in vivo has been explored [15], but this technology needs to be further developed and validated before the potential in PM endeavors can be realized.
Advantages and disadvantages of the application of rodent models in PM are summarized in Table 1.

Table 1.
Rodent models for personalized medicine.
3.3. In Vitro Models for PM
Cell culture techniques have been used by researchers for more than 100 years. For much of that time, two-dimensional (2D) monolayer cultures were the gold standard in determining the in vitro efficacy and safety of drug candidates. In the last few years, the emergence of in vitro techniques that can more accurately recapitulate the physiologic features observed in patients, such as three-dimensional (3D) cell culture and organoids, have brought the promise of personalized cellular approaches. Cellular models for PM were included in 18% of the oncology, and 28% of the brain disorders references. In oncology, patient-derived cellular models and tumor explants have been used successfully in drug screening for the most effective therapy [42,43,44]. In brain disorders, the same has been attempted with human lymphoblastoid cell lines (LCL) and induced pluripotent stem cells (iPSC) [16,45,46,47,48].
Organoids can be established to form healthy organs through stem cell initiation, or from tissues directly derived from the patient, and they have the potential to provide disease modeling for infectious disease, genetic disease, PM, drug discovery (screening and toxicology), and regenerative medicine [49,50]. The organoid model is increasingly used for PM in oncology and is described in 23% of the references. Patient-derived organoids have been used as a tool to predict chemotherapy response in individual patients [51,52,53,54]; however, the main disadvantage of this model for the personalized approach is the inconsistency in the organoid growth rate, and the possibility of overgrowth of non-tumor cell populations. In addition, the organoid model does not provide any information about toxicology. Organoid development represents a breakthrough for the study of brain function, evolution, and disorders [55]; however, in our search, only 3% of references referred to brain organoids in PM, and the availability of tissue is a limiting factor [56,57,58].
All these cellular models are limited by the lack of perfusion and biochemical and physical interaction with the surrounding microenvironment. The development of organ-on-chip technologies attempts to overcome some of the limitations of cellular models, and more accurately model personalized drug therapy [13]. Microphysiological systems (MPS), where engineered organ-on-chip technologies are combined with organoids, have the potential to facilitate assessment of pharmacological and toxicological effect [59,60,61]. In oncology, MPS tumor models can replicate the tumor microenvironment in a physiologically relevant manner by incorporating a vascular system, co-culturing with relevant cell types, mimicking elevated interstitial fluid pressure and shear stresses [62]. In brain disorders, the ability to model disease features, microenvironmental parameters, and the complexity of the human central nervous system is highly dependent on the chip [63]. MPS models hold great promise for the future; however, there are still technical, regulatory, and ethical challenges to overcome before patient-derived organ chips are available for clinical evaluation of PM strategies [64]. For a summary of in vitro methods for PM, refer to Table 2.

Table 2.
In vitro methods for personalized medicine.
3.4. In Silico Models
In silico modeling, a process of integrating machine learning approaches to biological analysis and simulation, aims to make predictions on drug targets, drug efficacy, and patient responses [20]. This is an emerging field for PM, comprising 12% of the records in oncology, and 8% in brain disorders describing this approach. In silico models in PM aim to couple clinical data with mathematical methods to create subject-specific organ models and design new, personalized protocols, as well as to determine patient stratification. Departing from different patient-specific parameters, they can capture inter- and intra-patient variability [21]. Despite the enormous opportunities offered by these models, a full-scale adaption of patient-specific implementation is still far from reality; the main limitation relates to the prediction accuracy, which depends on the quality and quantity of the input data, and the lack of standards and model validation [65]. The current pros and cons are summarized in Table 3.

Table 3.
In silico models for personalized medicine.
3.5. Are the Current Preclinical Models Predictive for PM Trials?
In this scoping review, we were not able to identify any articles which directly report on the success rate of PM trials based on accrued preclinical evidence. However, we found several reviews referring to the issues facing preclinical research when applying PM in both oncology (n = 6) and brain disorders (n = 21). In summary, advancing drug development and biomarker research in the era of PM is highly dependent on choosing the right preclinical model for the right molecular pathway to be explored [66]. Some authors expressed the opinion that retrospective analysis of the preclinical data used to support a failed clinical program should be published to help advance the field [67,68], and others raised the question of whether the more advanced models fit within the established drug-development paradigm, calling for a rethink of the existing anticancer drug discovery pipeline [69].
3.6. Main Gaps Identified
To allow for safe development and implementation of PM, appropriate preclinical models generating reliable and predictive data need to be available. Despite the progress in evolving numerous sophisticated model systems, to date, there are fundamental deficits that prevent their broad implementation in PM. As part of the consultation phase of the scoping review, a Gap Analysis Workshop was organized. This workshop took place on 1 December and 2 December 2020 online, and was attended by representatives of all project beneficiaries, as well as by three associated partners, the European Medicines Agency (EMA), the European Network for Health Technology Assessment (EUnetHTA), and the Clinical Trials Coordination Group (CTCG-HMA). From this process, we have identified five main gaps in translational methods, which we believe must be addressed to further develop robust models for PM.
- The first gap is a lack of clinically relevant experimental models for personalized medicine. Despite technical advances and more sophisticated preclinical models, to date, there are knowledge gaps in biology and an inability to recapitulate human phenotypes for many diseases, which is a challenge for translation and prediction of preclinical data to human PM clinical trials. There is also an apparent deficit in validating preclinical methods for clinical relevance; in other words, how well the model represents the phenotype of disease or clustering of patients.
- The second gap is the lack of standards for methods, validation procedures, and the lack of quality assessment systems. The fact is that preclinical models are often not robust enough for translation. Some of the hurdles for model validation are that this type of work is not academically rewarded, it is time consuming, and it is expensive.
- The third gap is the lack of accurate reporting and the lack of reporting negative results, which then further leads to a lack of systematic reviews and meta-analyses on methods, and these are important tools for evidence-based medicine. Access to preclinical data supporting clinical trials is challenging. There is a publication bias toward positive experiments, and methods are often not reported in sufficient detail to attempt reproducibility of results.
- The fourth gap relates to regulation, and the lack of harmonized guidelines for evaluating the relevance and robustness of preclinical evidence.
- The last gap we identified is the lack of involvement between preclinical and clinical research, and the need for a better definition for patient engagement.
4. Discussion
4.1. Principal Findings
Traditionally, preclinical models have been used as simplified models of human conditions, managed with a high degree of standardization, kept in pathogen-free environments, and treated identically to remove the influences of known variables. The increasing multi-omics characterization of disease, generated by advances in molecular characterization and bioengineering, brings new challenges to disease modeling, which becomes even more evident when associated with personalized treatment decisions. The complexity of PM highlights the need for more sophisticated biological systems to assess the integrated mechanisms of response. This scoping review investigated how current preclinical methods can support decision makers in approving clinical trials in PM. In the field of oncology, where the personalized approach is the most advanced, preclinical models which can recapitulate the patient tumor heterogeneity exist; nevertheless, the approach of modeling patient clustering through this approach is not yet widely used for various reasons. In brain disorders, there is no availability of models which can fully recapitulate patient phenotypes, and there is a dearth in the understanding of the disease mechanisms occurring at an individual level. Emerging models, such as organ-on-chip and in silico models, have been proposed to close the translational gap in the future. However, this is reliant on technologies which are still in their infancy, and additional fundamental issues in preclinical research remain unsolved.
4.2. Limitations of the Scope
As the preclinical research topic is broad, we decided to concentrate on two case studies to make the search manageable. These were oncology and brain disorders. It might have been informative to also include other disease areas, but we decided to narrow our focus to the two use cases, which possibly represent the two extremes in relation to availability of preclinical models in PM. The main limitation of our scope is that we were unable to find information about the specific preclinical evidence supporting the decision to approve clinical trials, and therefore we could not directly assess the translatability of the model used. One of the reasons for this is probably that most clinical trials are sponsored by industry, and preclinical data generated by the pharmaceutical industry are often not published. Through our gray literature search, we could identify two registries for preclinical trial protocols (www.preclinicaltrials.eu, accessed on 6 July 2022; www.animalstudyregistry.org, accessed on 6 July 2022), but there is no requirement to use such registries. The number of registered preclinical studies is 118 and 113, respectively, at the time of publication, and they are not linked to subsequent clinical data. In contrast, there is a legal requirement to register clinical trials; a search on clinicaltrials.gov at the same timepoint showed 420,268 entries. This scoping review was primarily aimed to inform the development of the subsequent recommendations, and even if the search might be perceived as out of date, we have continued to monitor the literature in the field during these two years and added relevant papers.
4.3. Challenges of Preclinical Research in PM
The probable gaps identified in this review are not novel issues in preclinical research [70,71]. The low rate of translation is evident when looking at the high attrition rates in drug and medical device development, and it has been suggested that this could be explained by faults in the design, conduct, and reporting of preclinical studies [7,72,73,74,75]. Attempts at addressing the issues relating to preclinical study reporting have been made through the development of reporting checklists, such as the ARRIVE guidelines for in vivo experiments [76,77], but despite being endorsed by over 1000 scientific journals, non-compliance with standard reporting checklists was the major finding in three systematic reviews of PDX models, where only one study was found to fully comply with the guidelines [36,78,79].
Furthermore, the systematic validation of the model systems often fails, in terms of internal, external, construct, and predictive validity [80]. The internal validity in preclinical methods, i.e., the risk of bias, can be addressed by systematic reviews, which can improve the success and reproducibility of subsequent translational clinical studies in this era of PM. However, only three systematic reviews were identified in the oncology search and none in the brain disorders search, and this deficit in preclinical research has already been addressed by the SYRCLE initiative [81]. The absence of standardized protocols and guidelines, because of the large variation in methodology across preclinical studies, makes quantitative analysis of bias challenging across studies, and researchers have made calls for more harmonized approaches [82]. Another relevant point to be addressed is the low availability of negative data. Negative results are not appealing for publication, meaning that the results of thousands of experiments that fail to confirm the reliability of preclinical models do not see the light of day. However, this is not necessarily a result of publication bias with the journals, but rather that scientists do not submit negative studies [83,84,85]. It results in a waste of time and resources from other scientists in repeating negative findings and, consequently, the deceleration of the translational pipeline. This is even more true in an industry setting, where in-house data are not generally published for reasons of competitiveness [86,87]. Therefore, the scientific community should address this issue, showing awareness of the richness of negative results in research.
The predictive validity of preclinical research is challenging when modeling complex disease processes. One approach could be to test the hypotheses in several different models, which could capture various aspects of the heterogeneity of the human pathophysiological processes. Another is to make sure that the model adequately represents the human disease condition for the question being asked. Several tools and guidelines for assessing the clinical relevance preclinical research have been published [88,89,90,91], but so far, they have not been broadly implemented. In the end, it is important to remember that a model system can never be a complete reflection of a human. However, by choosing the most appropriate model for the question asked, striving to make sure that the model is fully validated, and using complementary models to fill in the gaps, enough evidence can be gathered to move through the translational phase. To an extent, it is the drive to develop animal-free methods which is fueling the development of advanced in vitro and in silico models. Animal research is strictly regulated from an ethical point of view, but not from a qualitative perspective, and this deficit is becoming more evident when in vivo models are applied for PM [92].
The disappointing results of PM clinical trials cannot entirely be attributed to the lack of preclinical models that recapitulate human disease phenotypes. Continued efforts should be directed towards deep phenotyping of patients, and identification of reliable biomarkers to identify patient subgroups [70], as well as better definition of criteria for patient selection in clinical trials [93].
Based on the findings of this scoping review, it is our opinion that all preclinical research will benefit from better guidance regarding the clinical relevance and validity of models. When preclinical methods are applied to personalized medicine development, such as modeling molecular analyses of patient samples and treatment outcomes, the gaps become even more evident. These challenges need to be addressed at global, national, regional, and local levels.
5. Conclusions
When adequately designed and conducted, preclinical experiments may contribute invaluable information to our knowledge of medicine, including the discovery and development of new drugs, and can be essential tools to bridge the translational gap between preclinical and clinical research. In fact, appropriate preclinical models should be an integral contributor to interventional clinical trial success rates. We are at a key moment for the era of PM, and research in this domain is constantly evolving and generating new knowledge. However, PM development has proven to be an extremely ambitious enterprise at the preclinical level. The gaps identified in this scoping review are the first step towards building recommendations for more robust translational research in PM and corresponding to different scientific domains. The challenges can only be faced with concomitant action on all levels, with the involvement of all relevant stakeholders involved in this field, from researchers to policy makers and regulators, and should be viewed as aspects to work on, rather than obstacles, as they form the foundation of personalized preclinical research.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jpm12071177/s1, File S1: Search strategy; File S2: data extraction form.
Author Contributions
Conceptualization, V.F., E.O., F.B., A.L.A. and E.M.; methodology, C.G., M.F. and R.B.; validation, V.F., E.O.; formal analysis, V.F., E.O.; data curation, V.F., E.O., F.B.; writing—original draft preparation, V.F., E.O., R.B., C.G.; writing—review and editing, V.F., F.B., A.U., A.L.A., M.F. and E.M.; visualization, E.O., V.F. The members of the PERMIT group were involved in the preparation or revision of the joint protocol of the four scoping reviews of the PERMIT series, attended the joint workshop (consultation exercise), or contributed to one of the other scoping reviews of the PERMIT series. Project administration, P.G., J.D.; funding acquisition, J.D. The PERMIT group is Florie Brion Bouvier, Montserrat Carmona Rodriguez, Maria del Mar Polo-de Santos, Jacques Demotes-Mainard, Paula Garcia, Enrico Glaab, Rainer Girgenrath, Alexander Grundmann, Josep Maria Haro, Frank Hulstaert, Iñaki Imaz-Iglesia, Pascale Jonckheer, Setefilla Luengo Matos, Albert Sanchez Niubo, Raphael Porcher, Armin Rauschenberger, Luis M. Sánchez-Gómez, Lorena San Miguel, Cecilia Superchi, Teresa Torres, Anna Monistrol Mula. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the European Union’s Horizon 2020 research and innovation programme under grant agreement No. 874825.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Copies of searches and data extraction sheets are publicly available on the online platform Zenodo (https://zenodo.org/record/6087847, published on 15 February 2022), as part of the database collection for all scoping reviews conducted in the PERMIT project.
Acknowledgments
The authors thank Vanna Pistotti for her assistance with search strategy development and conduct, and Jake Fairnie, EATRIS ERIC, for assisting with the figure design.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- EU. 2015/C 421/03 Council conclusions on personalised medicine for patients. Off. J. Eur. Union 2015, 421, 2–5. [Google Scholar]
- European Commission. 2019 Report on the Statistics on the Use of Animals for Scientific Purposes in the Member States of the European Union in 2015–2017; CELEX: Philadelphia, PA, USA, 2020. [Google Scholar]
- Nestler, E.J.; Hyman, S.E. Animal models of neuropsychiatric disorders. Nat. Neurosci 2010, 13, 1161–1169. [Google Scholar] [CrossRef] [PubMed]
- Day, C.P.; Merlino, G.; Van Dyke, T. Preclinical mouse cancer models: A maze of opportunities and challenges. Cell 2015, 163, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Van Dam, D.; De Deyn, P.P. Animal models in the drug discovery pipeline for Alzheimer’s disease. Br. J. Pharmacol. 2011, 164, 1285–1300. [Google Scholar] [CrossRef] [PubMed]
- Crawley, J.N. Translational animal models of autism and neurodevelopmental disorders. Dialogues Clin. Neurosci. 2012, 14, 293–305. [Google Scholar] [CrossRef]
- Mak, I.W.; Evaniew, N.; Ghert, M. Lost in translation: Animal models and clinical trials in cancer treatment. Am. J. Transl. Res. 2014, 6, 114–118. [Google Scholar]
- Aartsma-Rus, A.; van Putten, M. The use of genetically humanized animal models for personalized medicine approaches. Dis. Model. Mech. 2019, 13, dmm041673. [Google Scholar] [CrossRef]
- Li, H.; Auwerx, J. Mouse Systems Genetics as a Prelude to Precision Medicine. Trends Genet. 2020, 36, 259–272. [Google Scholar] [CrossRef]
- Bhimani, J.; Ball, K.; Stebbing, J. Patient-derived xenograft models-the future of personalised cancer treatment. Br. J. Cancer 2020, 122, 601–602. [Google Scholar] [CrossRef]
- Byrne, A.T.; Alferez, D.G.; Amant, F.; Annibali, D.; Arribas, J.; Biankin, A.V.; Bruna, A.; Budinska, E.; Caldas, C.; Chang, D.K.; et al. Interrogating open issues in cancer precision medicine with patient-derived xenografts. Nat. Rev. Cancer 2017, 17, 254–268. [Google Scholar] [CrossRef]
- Clohessy, J.G.; Pandolfi, P.P. Mouse hospital and co-clinical trial project--from bench to bedside. Nat. Rev. Clin. Oncol. 2015, 12, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Cavero, I.; Guillon, J.M.; Holzgrefe, H.H. Human organotypic bioconstructs from organ-on-chip devices for human-predictive biological insights on drug candidates. Expert Opin. Drug. Saf. 2019, 18, 651–677. [Google Scholar] [CrossRef] [PubMed]
- Fong, E.L.S.; Toh, T.B.; Yu, H.; Chow, E.K.-H. 3D Culture as a Clinically Relevant Model for Personalized Medicine. SLAS Technol. Transl. Life Sci. Innov. 2017, 22, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Nair, R.R.; Corrochano, S.; Gasco, S.; Tibbit, C.; Thompson, D.; Maduro, C.; Ali, Z.; Fratta, P.; Arozena, A.A.; Cunningham, T.J.; et al. Uses for humanised mouse models in precision medicine for neurodegenerative disease. Mamm. Genome 2019, 30, 173–191. [Google Scholar] [CrossRef] [PubMed]
- Haggarty, S.J.; Silva, M.C.; Cross, A.; Brandon, N.J.; Perlis, R.H. Advancing drug discovery for neuropsychiatric disorders using patient-specific stem cell models. Mol. Cell Neurosci. 2016, 73, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Song, L.; Bejoy, J.; Zhao, J.; Kanekiyo, T.; Bu, G.; Zhou, Y.; Li, Y. Modeling Neurodegenerative Microenvironment Using Cortical Organoids Derived from Human Stem Cells. Tissue Eng. Part A 2018, 24, 1125–1137. [Google Scholar] [CrossRef] [PubMed]
- McCammon, J.M.; Sive, H. Addressing the Genetics of Human Mental Health Disorders in Model Organisms. Annu. Rev. Genom. Hum. Genet. 2015, 16, 173–197. [Google Scholar] [CrossRef]
- Driehuis, E.; Kretzschmar, K.; Clevers, H. Establishment of patient-derived cancer organoids for drug-screening applications. Nat. Protoc. 2020, 15, 3380–3409. [Google Scholar] [CrossRef]
- Chakravarty, K.; Antontsev, V.; Bundey, Y.; Varshney, J. Driving success in personalized medicine through AI-enabled computational modeling. Drug. Discov. Today 2021, 26, 1459–1465. [Google Scholar] [CrossRef]
- Chase, J.G.; Preiser, J.C.; Dickson, J.L.; Pironet, A.; Chiew, Y.S.; Pretty, C.G.; Shaw, G.M.; Benyo, B.; Moeller, K.; Safaei, S.; et al. Next-generation, personalised, model-based critical care medicine: A state-of-the art review of in silico virtual patient models, methods, and cohorts, and how to validation them. Biomed. Eng. Online 2018, 17, 24. [Google Scholar] [CrossRef]
- Torres Moral, T.; Sanchez-Niubo, A.; Monistrol-Mula, A.; Gerardi, C.; Banzi, R.; Garcia, P.; Demotes-Mainard, J.; Haro, J.M.; the PERMIT Group. Methods for Stratification and Validation Cohorts: A Scoping Review. J. Pers. Med. 2022, 12, 688. [Google Scholar] [CrossRef] [PubMed]
- Glaab, E.; Rauschenberger, A.; Banzi, R.; Gerardi, C.; Garcia, P.; Demotes, J. Biomarker discovery studies for patient stratification using machine learning analysis of omics data: A scoping review. BMJ Open 2021, 11, e053674. [Google Scholar] [CrossRef]
- Superchi, C.; Brion Bouvier, F.; Gerardi, C.; Carmona, M.; San Miguel, L.; Sánchez-Gómez, L.M.; Imaz-Iglesia, I.; Garcia, P.; Demotes, J.; Banzi, R.; et al. Study designs for clinical trials applied to personalised medicine: A scoping review. BMJ Open 2022, 12, e052926. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef]
- Tricco, A.C.; Lillie, E.; Zarin, W.; O’Brien, K.; Colquhoun, H.; Kastner, M.; Levac, D.; Ng, C.; Sharpe, J.P.; Wilson, K.; et al. A scoping review on the conduct and reporting of scoping reviews. BMC Med. Res. Methodol. 2016, 16, 15. [Google Scholar] [CrossRef]
- Peters, M.D.; Godfrey, C.M.; Khalil, H.; McInerney, P.; Parker, D.; Soares, C.B. Guidance for conducting systematic scoping reviews. Int J. Evid. Based Healthc. 2015, 13, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Munn, Z.; Peters, M.D.J.; Stern, C.; Tufanaru, C.; McArthur, A.; Aromataris, E. Systematic review or scoping review? Guidance for authors when choosing between a systematic or scoping review approach. BMC Med. Res. Methodol. 2018, 18, 143. [Google Scholar] [CrossRef]
- Banzi, R.; Gerardi, C.; Fratelli, M.; Garcia, P.; Torres, T.; Abad, J.M.H.; Niubo, A.S.; Glaab, E.; Oldoni, E.; Bietrix, F.; et al. Methodological approaches for personalised medicine: Protocol for a series of scoping reviews. Zenodo 2020. [Google Scholar] [CrossRef]
- Tricco, A.C.; Lillie, E.; Zarin, W.; O’Brien, K.K.; Colquhoun, H.; Levac, D.; Moher, D.; Peters, M.D.J.; Horsley, T.; Weeks, L.; et al. PRISMA Extension for Scoping Reviews (PRISMA-ScR): Checklist and Explanation. Ann. Intern. Med. 2018, 169, 467–473. [Google Scholar] [CrossRef]
- Paez, A. Gray literature: An important resource in systematic reviews. J. Evid. Based Med. 2017, 10, 233–240. [Google Scholar] [CrossRef]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan—a web and mobile app for systematic reviews. Syst. Rev. 2016, 5, 210. [Google Scholar] [CrossRef] [PubMed]
- Leenaars, C.H.C.; Kouwenaar, C.; Stafleu, F.R.; Bleich, A.; Ritskes-Hoitinga, M.; De Vries, R.B.M.; Meijboom, F.L.B. Animal to human translation: A systematic scoping review of reported concordance rates. J. Transl. Med. 2019, 17, 223. [Google Scholar] [CrossRef]
- Malaney, P.; Nicosia, S.V.; Davé, V. One mouse, one patient paradigm: New avatars of personalized cancer therapy. Cancer Lett. 2014, 344, 1–12. [Google Scholar] [CrossRef]
- Shi, J.; Li, Y.; Jia, R.; Fan, X. The fidelity of cancer cells in PDX models: Characteristics, mechanism and clinical significance. Int. J. Cancer 2020, 146, 2078–2088. [Google Scholar] [CrossRef] [PubMed]
- Collins, A.T.; Lang, S.H. A systematic review of the validity of patient derived xenograft (PDX) models: The implications for translational research and personalised medicine. PeerJ 2018, 6, e5981. [Google Scholar] [CrossRef]
- Tentler, J.J.; Tan, A.C.; Weekes, C.D.; Jimeno, A.; Leong, S.; Pitts, T.M.; Arcaroli, J.J.; Messersmith, W.A.; Eckhardt, S.G. Patient-derived tumour xenografts as models for oncology drug development. Nat. Rev. Clin. Oncol. 2012, 9, 338–350. [Google Scholar] [CrossRef] [PubMed]
- Saito, R.; Kobayashi, T.; Kashima, S.; Matsumoto, K.; Ogawa, O. Faithful preclinical mouse models for better translation to bedside in the field of immuno-oncology. Int. J. Clin. Oncol. 2020, 25, 831–841. [Google Scholar] [CrossRef]
- Kucherlapati, R. Genetically modified mouse models for biomarker discovery and preclinical drug testing. Clin. Cancer Res. 2012, 18, 625–630. [Google Scholar] [CrossRef][Green Version]
- Just, M.K.; Gram, H.; Theologidis, V.; Jensen, P.H.; Nilsson, K.P.R.; Lindgren, M.; Knudsen, K.; Borghammer, P.; Van Den Berge, N. Alpha-Synuclein Strain Variability in Body-First and Brain-First Synucleinopathies. Front. Aging Neurosci 2022, 14, 907293. [Google Scholar] [CrossRef]
- Van Den Berge, N.; Ulusoy, A. Animal models of brain-first and body-first Parkinson’s disease. Neurobiol. Dis. 2022, 163, 105599. [Google Scholar] [CrossRef]
- Yang, C.; Xia, B.R.; Jin, W.L.; Lou, G. Circulating tumor cells in precision oncology: Clinical applications in liquid biopsy and 3D organoid model. Cancer Cell Int. 2019, 19, 341. [Google Scholar] [CrossRef] [PubMed]
- Powley, I.R.; Patel, M.; Miles, G.; Pringle, H.; Howells, L.; Thomas, A.; Kettleborough, C.; Bryans, J.; Hammonds, T.; MacFarlane, M.; et al. Patient-derived explants (PDEs) as a powerful preclinical platform for anti-cancer drug and biomarker discovery. Br. J. Cancer 2020, 122, 735–744. [Google Scholar] [CrossRef] [PubMed]
- Braunschweig, T.; Chung, J.Y.; Hewitt, S.M. Tissue microarrays: Bridging the gap between research and the clinic. Expert Rev. Proteom. 2005, 2, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Christian, K.M.; Song, H.; Ming, G.L. Modeling psychiatric disorders with patient-derived iPSCs. Curr. Opin. Neurobiol. 2016, 36, 118–127. [Google Scholar] [CrossRef]
- Nam, H.Y.; Shim, S.M.; Han, B.G.; Jeon, J.P. Human lymphoblastoid cell lines: A goldmine for the biobankomics era. Pharmacogenomics 2011, 12, 907–917. [Google Scholar] [CrossRef] [PubMed]
- Penney, J.; Ralvenius, W.T.; Tsai, L.H. Modeling Alzheimer’s disease with iPSC-derived brain cells. Mol. Psychiatry 2020, 25, 148–167. [Google Scholar] [CrossRef]
- Silva, M.C.; Haggarty, S.J. Human pluripotent stem cell-derived models and drug screening in CNS precision medicine. Ann. N. Y. Acad. Sci. 2020, 1471, 18–56. [Google Scholar] [CrossRef]
- Aboulkheyr Es, H.; Montazeri, L.; Aref, A.R.; Vosough, M.; Baharvand, H. Personalized Cancer Medicine: An Organoid Approach. Trends Biotechnol. 2018, 36, 358–371. [Google Scholar] [CrossRef]
- Bartfeld, S.; Clevers, H. Stem cell-derived organoids and their application for medical research and patient treatment. J. Mol. Med. 2017, 95, 729–738. [Google Scholar] [CrossRef]
- Phan, N.; Hong, J.J.; Tofig, B.; Mapua, M.; Elashoff, D.; Moatamed, N.A.; Huang, J.; Memarzadeh, S.; Damoiseaux, R.; Soragni, A. A simple high-throughput approach identifies actionable drug sensitivities in patient-derived tumor organoids. Commun. Biol. 2019, 2, 78. [Google Scholar] [CrossRef]
- Granat, L.M.; Kambhampati, O.; Klosek, S.; Niedzwecki, B.; Parsa, K.; Zhang, D. The promises and challenges of patient-derived tumor organoids in drug development and precision oncology. Animal. Model. Exp. Med. 2019, 2, 150–161. [Google Scholar] [CrossRef] [PubMed]
- Kondo, J.; Inoue, M. Application of Cancer Organoid Model for Drug Screening and Personalized Therapy. Cells 2019, 8, 470. [Google Scholar] [CrossRef] [PubMed]
- Weeber, F.; Ooft, S.N.; Dijkstra, K.K.; Voest, E.E. Tumor Organoids as a Pre-clinical Cancer Model for Drug Discovery. Cell Chem. Biol. 2017, 24, 1092–1100. [Google Scholar] [CrossRef] [PubMed]
- Sidhaye, J.; Knoblich, J.A. Brain organoids: An ensemble of bioassays to investigate human neurodevelopment and disease. Cell Death Differ. 2021, 28, 52–67. [Google Scholar] [CrossRef] [PubMed]
- Mason, J.O.; Price, D.J. Building brains in a dish: Prospects for growing cerebral organoids from stem cells. Neuroscience 2016, 334, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Meng, X.; Liu, Y.; Song, D.; Jiang, C.; Cai, J. Applications of brain organoids in neurodevelopment and neurological diseases. J. Biomed. Sci. 2021, 28, 30. [Google Scholar] [CrossRef]
- Rossetti, A.C.; Koch, P.; Ladewig, J. Drug discovery in psychopharmacology: From 2D models to cerebral organoids. Dialogues Clin. Neurosci 2019, 21, 203–224. [Google Scholar] [CrossRef]
- Sontheimer-Phelps, A.; Hassell, B.A.; Ingber, D.E. Modelling cancer in microfluidic human organs-on-chips. Nat. Rev. Cancer 2019, 19, 65–81. [Google Scholar] [CrossRef]
- Kankala, R.K.; Wang, S.B.; Chen, A.Z. Microengineered Organ-on-a-chip Platforms towards Personalized Medicine. Curr. Pharm. Des. 2018, 24, 5354–5366. [Google Scholar] [CrossRef]
- Skardal, A.; Shupe, T.; Atala, A. Organoid-on-a-chip and body-on-a-chip systems for drug screening and disease modeling. Drug Discov. Today 2016, 21, 1399–1411. [Google Scholar] [CrossRef]
- Shang, M.; Soon, R.H.; Lim, C.T.; Khoo, B.L.; Han, J. Microfluidic modelling of the tumor microenvironment for anti-cancer drug development. Lab. Chip. 2019, 19, 369–386. [Google Scholar] [CrossRef] [PubMed]
- Haring, A.P.; Sontheimer, H.; Johnson, B.N. Microphysiological Human Brain and Neural Systems-on-a-Chip: Potential Alternatives to Small Animal Models and Emerging Platforms for Drug Discovery and Personalized Medicine. Stem. Cell Rev. Rep. 2017, 13, 381–406. [Google Scholar] [CrossRef] [PubMed]
- Marx, U.; Akabane, T.; Andersson, T.B.; Baker, E.; Beilmann, M.; Beken, S.; Brendler-Schwaab, S.; Cirit, M.; David, R.; Dehne, E.M.; et al. Biology-inspired microphysiological systems to advance patient benefit and animal welfare in drug development. Altex 2020, 37, 365–394. [Google Scholar] [CrossRef] [PubMed]
- Kamel Boulos, M.N.; Zhang, P. Digital Twins: From Personalised Medicine to Precision Public Health. J. Pers. Med. 2021, 11, 745. [Google Scholar] [CrossRef]
- Klinghammer, K.; Walther, W.; Hoffmann, J. Choosing wisely-Preclinical test models in the era of precision medicine. Cancer Treat. Rev. 2017, 55, 36–45. [Google Scholar] [CrossRef]
- Gould, S.E.; Junttila, M.R.; de Sauvage, F.J. Translational value of mouse models in oncology drug development. Nat. Med. 2015, 21, 431–439. [Google Scholar] [CrossRef]
- Lieu, C.H.; Tan, A.C.; Leong, S.; Diamond, J.R.; Eckhardt, S.G. From bench to bedside: Lessons learned in translating preclinical studies in cancer drug development. J. Natl. Cancer Inst. 2013, 105, 1441–1456. [Google Scholar] [CrossRef]
- Liu, Z.; Delavan, B.; Roberts, R.; Tong, W. Lessons Learned from Two Decades of Anticancer Drugs. Trends Pharmacol. Sci. 2017, 38, 852–872. [Google Scholar] [CrossRef]
- Seyhan, A.A. Lost in translation: The valley of death across preclinical and clinical divide–identification of problems and overcoming obstacles. Transl. Med. Commun. 2019, 4, 18. [Google Scholar] [CrossRef]
- Godlee, F. We need better animal research, better reported. BMJ 2018, 360, k124. [Google Scholar] [CrossRef]
- Begley, C.G.; Ioannidis, J.P. Reproducibility in science: Improving the standard for basic and preclinical research. Circ. Res. 2015, 116, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Collins, F.S.; Tabak, L.A. Policy: NIH plans to enhance reproducibility. Nature 2014, 505, 612–613. [Google Scholar] [CrossRef] [PubMed]
- Perrin, S. Preclinical research: Make mouse studies work. Nature 2014, 507, 423–425. [Google Scholar] [CrossRef] [PubMed]
- Ioannidis, J.P. Acknowledging and Overcoming Nonreproducibility in Basic and Preclinical Research. JAMA 2017, 317, 1019–1020. [Google Scholar] [CrossRef]
- Kilkenny, C.; Browne, W.J.; Cuthill, I.C.; Emerson, M.; Altman, D.G. Improving bioscience research reporting: The ARRIVE guidelines for reporting animal research. PLoS Biol. 2010, 8, e1000412. [Google Scholar] [CrossRef]
- Du Sert, N.P.; Ahluwalia, A.; Alam, S.; Avey, M.T.; Baker, M.; Browne, W.J.; Clark, A.; Cuthill, I.C.; Dirnagl, U.; Emerson, M.; et al. Reporting animal research: Explanation and elaboration for the ARRIVE guidelines 2.0. PLoS Biol. 2020, 18, e3000411. [Google Scholar] [CrossRef]
- Brown, K.M.; Xue, A.; Mittal, A.; Samra, J.S.; Smith, R.; Hugh, T.J. Patient-derived xenograft models of colorectal cancer in pre-clinical research: A systematic review. Oncotarget 2016, 7, 66212–66225. [Google Scholar] [CrossRef]
- Koga, Y.; Ochiai, A. Systematic Review of Patient-Derived Xenograft Models for Preclinical Studies of Anti-Cancer Drugs in Solid Tumors. Cells 2019, 8, 418. [Google Scholar] [CrossRef]
- 46th ESAO Congress 3–7 September 2019 Hannover, Germany Abstracts. Int. J. Artif. Organs 2019, 42, 386–474. [CrossRef]
- Hooijmans, C.R.; Rovers, M.M.; de Vries, R.B.; Leenaars, M.; Ritskes-Hoitinga, M.; Langendam, M.W. SYRCLE’s risk of bias tool for animal studies. BMC Med. Res. Methodol. 2014, 14, 43. [Google Scholar] [CrossRef]
- Landis, S.C.; Amara, S.G.; Asadullah, K.; Austin, C.P.; Blumenstein, R.; Bradley, E.W.; Crystal, R.G.; Darnell, R.B.; Ferrante, R.J.; Fillit, H.; et al. A call for transparent reporting to optimize the predictive value of preclinical research. Nature 2012, 490, 187–191. [Google Scholar] [CrossRef] [PubMed]
- Olson, C.M.; Rennie, D.; Cook, D.; Dickersin, K.; Flanagin, A.; Hogan, J.W.; Zhu, Q.; Reiling, J.; Pace, B. Publication Bias in Editorial Decision Making. JAMA 2002, 287, 2825–2828. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.P.; Boyd, E.A.; Holroyd-Leduc, J.M.; Bacchetti, P.; Bero, L.A. Predictors of publication: Characteristics of submitted manuscripts associated with acceptance at major biomedical journals. Med. J. Aust. 2006, 184, 621–626. [Google Scholar] [CrossRef]
- Okike, K.; Kocher, M.S.; Mehlman, C.T.; Heckman, J.D.; Bhandari, M. Publication bias in orthopaedic research: An analysis of scientific factors associated with publication in the Journal of Bone and Joint Surgery (American Volume). J. Bone Joint Surg Am. 2008, 90, 595–601. [Google Scholar] [CrossRef] [PubMed]
- Pratte, M.; Ganeshamoorthy, S.; Carlisle, B.; Kimmelman, J. How well are Phase 2 cancer trial publications supported by preclinical efficacy evidence? Int. J. Cancer 2019, 145, 3370–3375. [Google Scholar] [CrossRef]
- Federico, C.A.; Carlisle, B.; Kimmelman, J.; Fergusson, D.A. Late, never or non-existent: The inaccessibility of preclinical evidence for new drugs. Br. J. Pharmacol. 2014, 171, 4247–4254. [Google Scholar] [CrossRef]
- Gurusamy, K.S.; Moher, D.; Loizidou, M.; Ahmed, I.; Avey, M.T.; Barron, C.C.; Davidson, B.; Dwek, M.; Gluud, C.; Jell, G.; et al. Clinical relevance assessment of animal preclinical research (RAA) tool: Development and explanation. PeerJ 2021, 9, e10673. [Google Scholar] [CrossRef]
- Ferreira, G.S.; Veening-Griffioen, D.H.; Boon, W.P.C.; Moors, E.H.M.; van Meer, P.J.K. Levelling the Translational Gap for Animal to Human Efficacy Data. Animals 2020, 10, 1199. [Google Scholar] [CrossRef]
- Bahr-Hosseini, M.; Bikson, M.; Iacoboni, M.; Liebeskind, D.S.; Hinman, J.D.; Carmichael, S.T.; Saver, J.L. PRIMED2 Preclinical Evidence Scoring Tool to Assess Readiness for Translation of Neuroprotection Therapies. Transl. Stroke Res. 2022, 13, 222–227. [Google Scholar] [CrossRef]
- Schäfer-Korting, M.; Zoschke, C. How Qualification of 3D Disease Models Cuts the Gordian Knot in Preclinical Drug Development. Handb. Exp. Pharmacol. 2021, 265, 29–56. [Google Scholar] [CrossRef]
- National Academies of Sciences, Engineering, and Medicine; Division on Earth and Life Studies; Institute for Laboratory Animal Research; Roundtable on Science and Welfare in Laboratory Animal Use. The National Academies Collection: Reports funded by National Institutes of Health. In Advancing Disease Modeling in Animal-Based Research in Support of Precision Medicine: Proceedings of a Workshop; National Academies Press: Washington, DC, USA, 2018. [Google Scholar] [CrossRef]
- Folke, J.; Ferreira, N.; Brudek, T.; Borghammer, P.; Van Den Berge, N. Passive Immunization in Alpha-Synuclein Preclinical Animal Models. Biomolecules 2022, 12, 168. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).