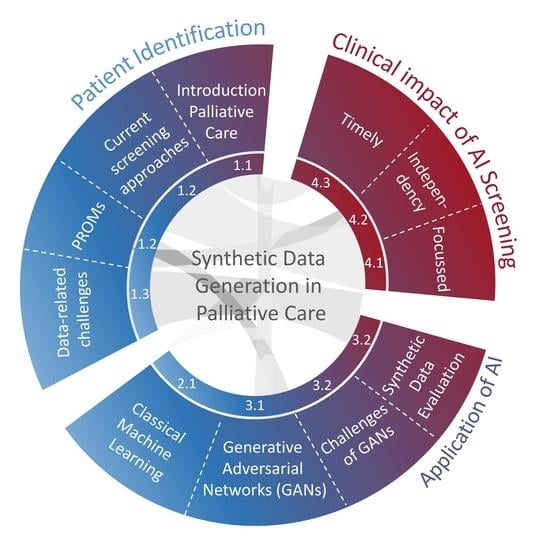
Contribution of Synthetic Data Generation towards an Improved Patient Stratification in Palliative Care
Abstract
1. Introduction and Definition of Palliative Care
1.1. Literature Screening Methodology
1.2. Current Screening for Patients in Need for Palliative Care
1.3. Data-Related Challenges That Limit A General Use of AI in Palliative Care
- Palliative care is a transient process and highly case specific. There is an ongoing controversial debate on the most important parameters that are used to define and effectively screen the need for palliative care
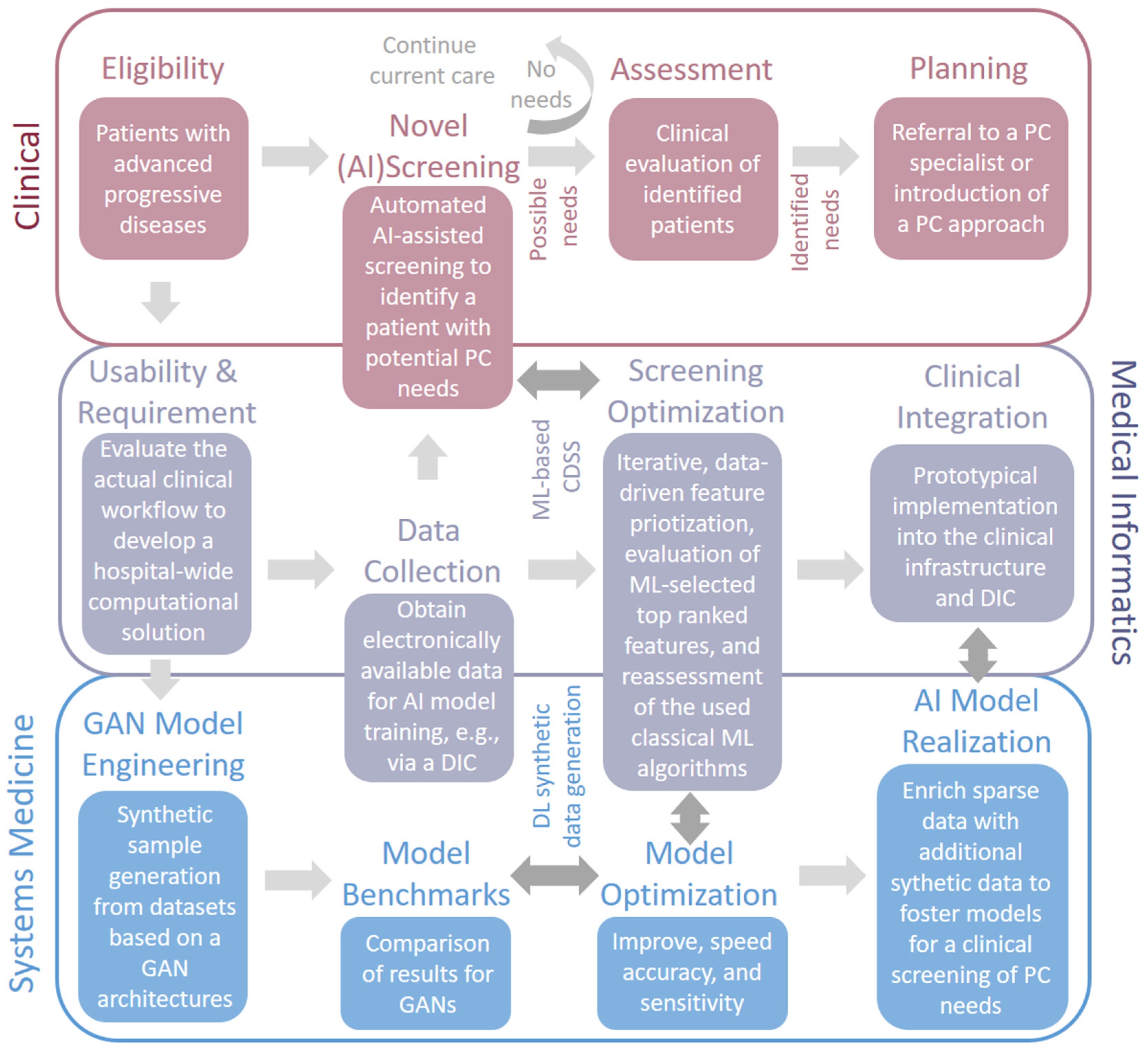
2. Existing and Prospective Applications of AI for Palliative Care
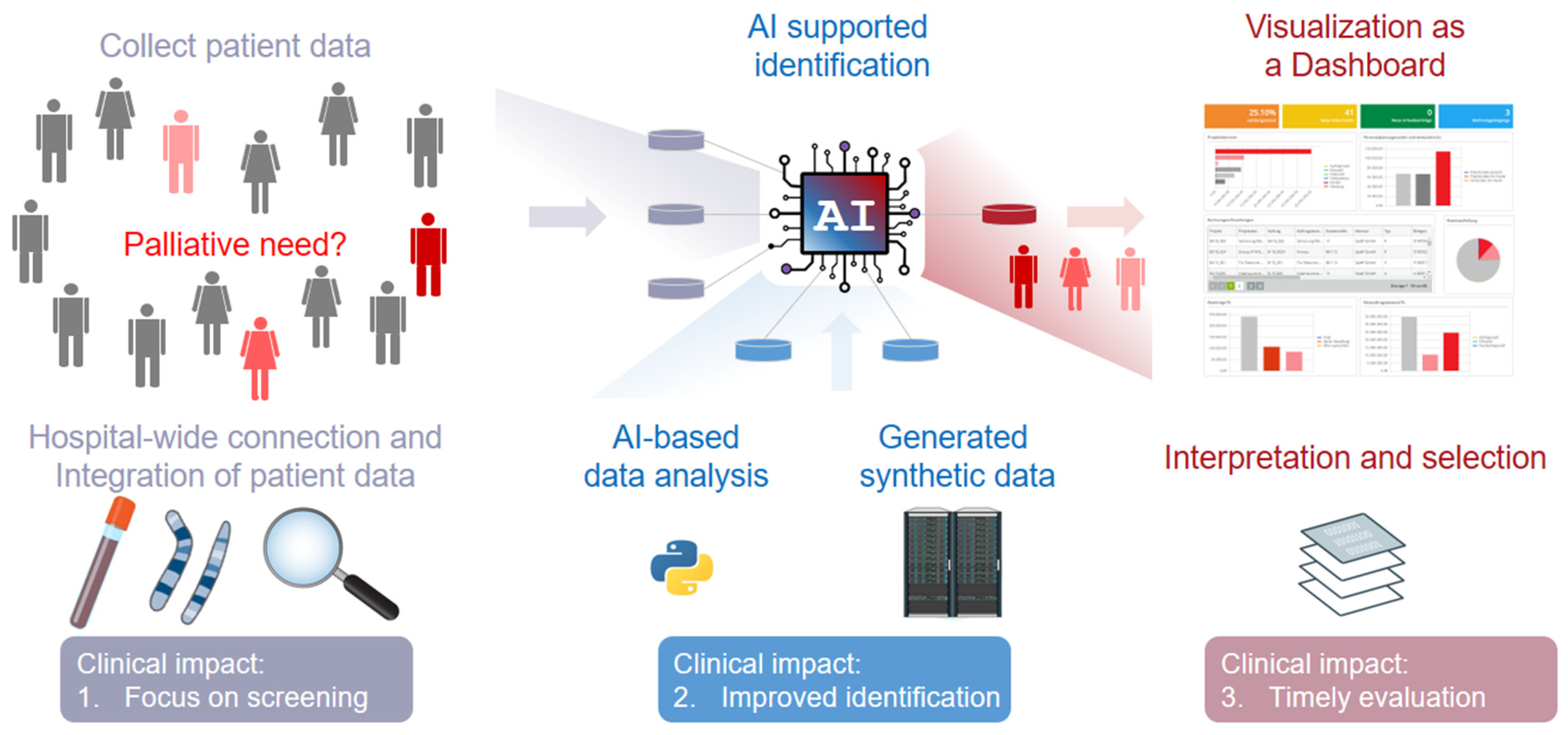
3. Potential Impact of Synthetic Data Generation Towards an Improved Identification of Patients in Need of Palliative Care
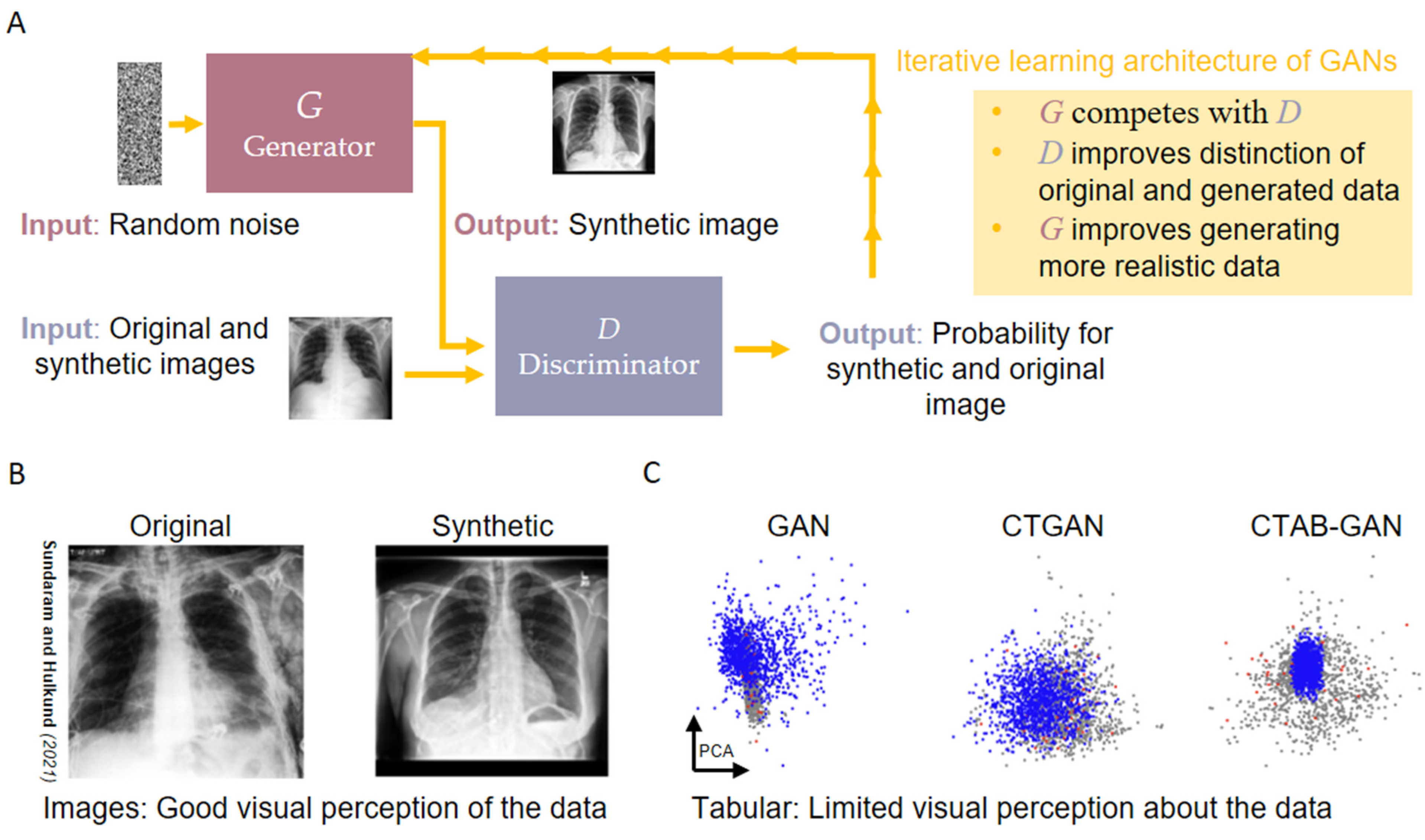
3.1. Synthetic Data Generation via Generative Adversarial Networks
3.2. Domain Level Challenges Concerning the Use of GANs for Clinical Problems
4. Clinical Impact of AI and GAN-Based Screening Solutions in Palliative Care
4.1. Clinical Impact 1: Set A Focus on Screening Rather Than Prognosis
4.2. Clinical Impact 2: Identification of Patients with Palliative Care Needs and Its Barriers
4.3. Clinical Impact 3: Evaluation of the Correct Timing to Specialized Palliative Care
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Radbruch, L.; De Lima, L.; Knaul, F.; Wenk, R.; Ali, Z.; Bhatnaghar, S.; Blanchard, C.; Bruera, E.; Buitrago, R.; Burla, C.; et al. Redefining Palliative Care-A New Consensus-Based Definition. J. Pain Symptom Manag. 2020, 60, 754–764. [Google Scholar] [CrossRef] [PubMed]
- Lopes, I.M.; Guarda, T.; Oliveira, P. General Data Protection Regulation in Health Clinics. J. Med. Syst. 2020, 44, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kamal, A.H.; Bausewein, C.; Casarett, D.J.; Currow, D.C.; Dudgeon, D.J.; Higginson, I.J. Standards, Guidelines, and Quality Measures for Successful Specialty Palliative Care Integration into Oncology: Current Approaches and Future Directions. J. Clin. Oncol. 2020, 38, 987–994. [Google Scholar] [CrossRef] [PubMed]
- Hui, D.; Bruera, E. Integrating palliative care into the trajectory of cancer care. Nat. Rev. Clin. Oncol. 2016, 13, 159. [Google Scholar] [CrossRef] [PubMed]
- Rangachari, D.; Smith, T.J.; Kimmel, S. Integrating Palliative Care in Oncology: The Oncologist as a Primary Palliative Care Provider. Cancer J. 2013, 19, 373. [Google Scholar] [CrossRef]
- Schenker, Y.; Arnold, R. The Next Era of Palliative Care. JAMA 2015, 314, 1565. [Google Scholar] [CrossRef]
- Schenker, Y.; Crowley-Matoka, M.; Dohan, D.; Rabow, M.W.; Smith, C.B.; White, D.B.; Chu, E.; Tiver, G.A.; Einhorn, S.; Arnold, R.M. Oncologist Factors That Influence Referrals to Subspecialty Palliative Care Clinics. J. Oncol. Pract. 2014, 10, e37. [Google Scholar] [CrossRef]
- Buurman, B.M.; Van Munster, B.C.; Korevaar, J.C.; Abu-Hanna, A.; Levi, M.; De Rooij, S.E. Prognostication in acutely admitted older patients by nurses and physicians. J. Gen. Intern. Med. 2008, 23, 1883–1889. [Google Scholar] [CrossRef][Green Version]
- Glare, P.; Plakovic, K.; Schloms, A.; Egan, B.; Epstein, A.S.; Kelsen, D.; Saltz, L. Study using the NCCN guidelines for palliative care to screen patients for palliative care needs and referral to palliative care specialists. J. Natl. Compr. Cancer Netw. 2013, 11, 1087–1096. [Google Scholar] [CrossRef]
- Weissman, D.E.; Meier, D.E. Identifying patients in need of a palliative care assessment in the hospital setting: A consensus report from the Center to Advance Palliative Care. J. Palliat. Med. 2011, 14, 17–23. [Google Scholar] [CrossRef]
- Trout, A.; Kirsh, K.L.; Peppin, J.F. Development and implementation of a palliative care consultation tool. Palliat. Support. Care 2012, 10, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Stiel, S.; Matthes, M.E.; Bertram, L.; Ostgathe, C.; Elsner, F.; Radbruch, L. Validierung der neuen Fassung des Minimalen Dokumentationssystems (MIDOS2) für Patienten in der Palliativmedizin: Deutsche Version der Edmonton Symptom Assessment Scale (ESAS). Schmerz 2010, 24, 596–604. [Google Scholar] [CrossRef] [PubMed]
- Bruera, E.; Kuehn, N.; Miller, M.J.; Selmser, P.; Macmillan, K. The Edmonton Symptom Assessment System (ESAS): A simple method for the assessment of palliative care patients. J. Palliat. Care 1991, 7, 6–9. [Google Scholar] [CrossRef]
- Murtagh, F.E.M.; Ramsenthaler, C.; Firth, A.; Groeneveld, E.I.; Lovell, N.; Simon, S.T.; Denzel, J.; Guo, P.; Bernhardt, F.; Schildmann, E.; et al. A brief, patient- and proxy-reported outcome measure in advanced illness: Validity, reliability and responsiveness of the Integrated Palliative care Outcome Scale (IPOS). Palliat. Med. 2019, 33, 1045–1057. [Google Scholar] [CrossRef] [PubMed]
- Mehnert, A.; Lehmann, C.; Cao, P.; Koch, U. Die erfassung psychosozialer belastungen und ressourcen in der onkologie—Ein literaturüberblick zu screeningmethoden und entwicklungstrends. PPmP Psychother. Psychosom. Med. Psychol. 2006, 56, 462–479. [Google Scholar] [CrossRef] [PubMed]
- Sandham, M.H.; Hedgecock, E.A.; Siegert, R.J.; Narayanan, A.; Hocaoglu, M.B.; Higginson, I.J. Intelligent Palliative Care Based on Patient-Reported Outcome Measures. J. Pain Symptom Manag. 2022, 63, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Alt-Epping, B.; Solar, S.; Lordick, F.; Mehnert-Theuerkauf, A.; Oechsle, K.; van Oorschot, B.; Thomas, M.; Asendorf, T. Niederschwelliges Screening versus multidimensionales Assessment von Symptomen und psychosozialen Belastungen bei Krebspatienten ab dem Zeitpunkt der Inkurabilität (SCREBEL). Forum Fam. Plan. West. Hemisph. 2020, 35, 143–144. [Google Scholar] [CrossRef]
- Moss, A.H.; Lunney, J.R.; Culp, S.; Auber, M.; Kurian, S.; Rogers, J.; Dower, J.; Abraham, J. Prognostic significance of the “surprise” question in cancer patients. J. Palliat. Med. 2010, 13, 837–840. [Google Scholar] [CrossRef]
- Bausewein, C.; Fegg, M.; Radbruch, L.; Nauck, F.; Von Mackensen, S.; Borasio, G.D.; Higginson, I.J. Validation and clinical application of the german version of the palliative care outcome scale. J. Pain Symptom Manag. 2005, 30, 51–62. [Google Scholar] [CrossRef]
- Roch, C.; Heckel, M.; van Oorschot, B.; Alt-Epping, B.; Tewes, M. Screening for Palliative Care Needs: Pilot Data From German Comprehensive Cancer Centers. JCO Oncol. Pract. 2021, 17, e1584–e1591. [Google Scholar] [CrossRef]
- Cava, W.L.; Bauer, C.; Moore, J.H.; Pendergrass, S.A. Interpretation of machine learning predictions for patient outcomes in electronic health records. AMIA Annu. Symp. Proc. 2019, 2019, 572. [Google Scholar] [PubMed]
- Simon, S.T.; Pralong, A.; Radbruch, L.; Bausewein, C.; Voltz, R. The Palliative Care of Patients with Incurable Cancer. Dtsch. Arztebl. Int. 2020, 117, 108. [Google Scholar] [CrossRef] [PubMed]
- Levy, M.H.; Adolph, M.D.; Back, A.; Block, S.; Codada, S.N.; Dalal, S.; Deshields, T.L.; Dexter, E.; Dy, S.M.; Knight, S.J.; et al. Palliative care. J. Natl. Compr. Cancer Netw. 2012, 10, 1284–1309. [Google Scholar] [CrossRef] [PubMed]
- Hui, D.; Cherny, N.I.; Wu, J.; Liu, D.; Latino, N.J.; Strasser, F. Indicators of integration at ESMO Designated Centres of Integrated Oncology and Palliative Care. ESMO Open 2018, 3, e000372. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.J.; Temin, S.; Alesi, E.R.; Abernethy, A.P.; Balboni, T.A.; Basch, E.M.; Ferrell, B.R.; Loscalzo, M.; Meier, D.E.; Paice, J.A.; et al. American Society of Clinical Oncology provisional clinical opinion: The integration of palliative care into standard oncology care. J. Clin. Oncol. 2012, 30, 880–887. [Google Scholar] [CrossRef]
- Coppen, R.; Van Veen, E.B.; Groenewegen, P.P.; Hazes, J.M.W.; De Jong, J.D.; Kievit, J.; De Neeling, J.N.D.; Reijneveld, S.A.; Verheij, R.A.; Vroom, E. Will the trilogue on the EU Data Protection Regulation recognise the importance of health research? Eur. J. Public Health 2015, 25, 757. [Google Scholar] [CrossRef][Green Version]
- Murdoch, B. Privacy and artificial intelligence: Challenges for protecting health information in a new era. BMC Med. Ethics 2021, 22, 1–5. [Google Scholar] [CrossRef]
- Liaw, S.T.; Liyanage, H.; Kuziemsky, C.; Terry, A.L.; Schreiber, R.; Jonnagaddala, J.; de Lusignan, S. Ethical Use of Electronic Health Record Data and Artificial Intelligence: Recommendations of the Primary Care Informatics Working Group of the International Medical Informatics Association. Yearb. Med. Inform. 2020, 29, 51. [Google Scholar] [CrossRef]
- Olatunji, I.E.; Rauch, J.; Katzensteiner, M.; Khosla, M. A Review of Anonymization for Healthcare Data. Big Data, 2022; online ahead of print. [Google Scholar] [CrossRef]
- Csányi, G.M.; Nagy, D.; Vági, R.; Vadász, J.P.; Orosz, T. Challenges and Open Problems of Legal Document Anonymization. Symmetry 2021, 13, 1490. [Google Scholar] [CrossRef]
- Zuo, Z.; Watson, M.; Budgen, D.; Hall, R.; Kennelly, C.; Al Moubayed, N. Data Anonymization for Pervasive Health Care: Systematic Literature Mapping Study. JMIR Med. Inf. 2021, 9, e29871. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, A.; Shmatikov, V. Robust de-anonymization of large sparse datasets. Proc. IEEE Symp. Secur. Priv. 2008, 111–125. [Google Scholar] [CrossRef]
- Douriez, M.; Doraiswamy, H.; Freire, J.; Silva, C.T. Anonymizing NYC taxi data: Does it matter? In Proceedings of the 2016 IEEE International Conference on Data Science and Advanced Analytics (DSAA), Montreal, QC, Canada, 17–19 October 2016; pp. 140–148. [Google Scholar] [CrossRef]
- Chen, R.J.; Lu, M.Y.; Chen, T.Y.; Williamson, D.F.K.; Mahmood, F. Synthetic data in machine learning for medicine and healthcare. Nat. Biomed. Eng. 2021, 5, 493–497. [Google Scholar] [CrossRef] [PubMed]
- Vayena, E.; Blasimme, A. Health Research with Big Data: Time for Systemic Oversight. J. Law. Med. Ethics 2018, 46, 119. [Google Scholar] [CrossRef] [PubMed]
- McLennan, S.; Fiske, A.; Tigard, D.; Müller, R.; Haddadin, S.; Buyx, A. Embedded ethics: A proposal for integrating ethics into the development of medical AI. BMC Med. Ethics 2022, 23, 1–10. [Google Scholar] [CrossRef]
- Bohr, A.; Memarzadeh, K. The rise of artificial intelligence in healthcare applications. Artif. Intell. Healthc. 2020, 25, 25–60. [Google Scholar] [CrossRef]
- Meskó, B.; Görög, M. A short guide for medical professionals in the era of artificial intelligence. NPJ Digit. Med. 2020, 3, 1–8. [Google Scholar] [CrossRef]
- Maragatham, G.; Devi, S. LSTM Model for Prediction of Heart Failure in Big Data. J. Med. Syst. 2019, 43, 1–13. [Google Scholar] [CrossRef]
- Storick, V.; O’Herlihy, A.; Abdelhafeez, S.; Ahmed, R.; May, P. Improving palliative care with machine learning and routine data: A rapid review. HRB Open Res. 2019, 2, 13. [Google Scholar] [CrossRef] [PubMed]
- Mather, H.; Guo, P.; Firth, A.; Davies, J.M.; Sykes, N.; Landon, A.; Murtagh, F.E.M. Phase of Illness in palliative care: Cross-sectional analysis of clinical data from community, hospital and hospice patients. Palliat. Med. 2018, 32, 404. [Google Scholar] [CrossRef] [PubMed]
- Lind, S.; Wallin, L.; Fürst, C.J.; Beck, I. The integrated palliative care outcome scale for patients with palliative care needs: Factors related to and experiences of the use in acute care settings. Palliat. Support. Care 2019, 17, 561–568. [Google Scholar] [CrossRef]
- Avati, A.; Jung, K.; Harman, S.; Downing, L.; Ng, A.; Shah, N.H. Improving palliative care with deep learning. BMC Med. Inform. Decis. Mak. 2018, 18, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Mashima, Y.; Tamura, T.; Kunikata, J.; Tada, S.; Yamada, A.; Tanigawa, M.; Hayakawa, A.; Tanabe, H.; Yokoi, H. Using Natural Language Processing Techniques to Detect Adverse Events From Progress Notes Due to Chemotherapy. Cancer Inform. 2022, 21, 11769351221085064. [Google Scholar] [CrossRef] [PubMed]
- Swan, A.; Azhar, A.; Anderson, A.E.; Williams, J.L.; Liu, D.; Bruera, E. Empowering the Health and Well-Being of the Palliative Care Workforce: Evaluation of a Weekly Self-Care Checklist. J. Pain Symptom Manag. 2021, 61, 817–823. [Google Scholar] [CrossRef] [PubMed]
- Kuosmanen, L.; Hupli, M.; Ahtiluoto, S.; Haavisto, E. Patient participation in shared decision-making in palliative care—An integrative review. J. Clin. Nurs. 2021, 30, 3415–3428. [Google Scholar] [CrossRef]
- Forbat, L.; Chapman, M.; Lovell, C.; Liu, W.M.; Johnston, N. Improving specialist palliative care in residential care for older people: A checklist to guide practice. BMJ Support. Palliat. Care 2018, 8, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Tai, S.Y.; Lee, C.Y.; Wu, C.Y.; Hsieh, H.Y.; Huang, J.J.; Huang, C.T.; Chien, C.Y. Symptom severity of patients with advanced cancer in palliative care unit: Longitudinal assessments of symptoms improvement. BMC Palliat. Care 2016, 15, 1–7. [Google Scholar] [CrossRef]
- Glare, P.A.; Chow, K. Validation of a Simple Screening Tool for Identifying Unmet Palliative Care Needs in Patients with Cancer. J. Oncol. Pract. 2015, 11, e81–e86. [Google Scholar] [CrossRef]
- Kingma, D.P.; Welling, M. Auto-Encoding Variational Bayes. arXiv 2013. [Google Scholar] [CrossRef]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Networks. Commun. ACM 2014, 63, 139–144. [Google Scholar] [CrossRef]
- Baowaly, M.K.; Lin, C.C.; Liu, C.L.; Chen, K.T. Synthesizing electronic health records using improved generative adversarial networks. J. Am. Med. Inform. Assoc. 2019, 26, 228–241. [Google Scholar] [CrossRef]
- Elbattah, M.; Loughnane, C.; Guérin, J.L.; Carette, R.; Cilia, F.; Dequen, G. Variational Autoencoder for Image-Based Augmentation of Eye-Tracking Data. J. Imaging 2021, 7, 83. [Google Scholar] [CrossRef] [PubMed]
- García-Ordás, M.T.; Benítez-Andrades, J.A.; García-Rodríguez, I.; Benavides, C.; Alaiz-Moretón, H. Detecting Respiratory Pathologies Using Convolutional Neural Networks and Variational Autoencoders for Unbalancing Data. Sensors 2020, 20, 1214. [Google Scholar] [CrossRef] [PubMed]
- Simidjievski, N.; Bodnar, C.; Tariq, I.; Scherer, P.; Andres Terre, H.; Shams, Z.; Jamnik, M.; Liò, P. Variational Autoencoders for Cancer Data Integration: Design Principles and Computational Practice. Front. Genet. 2019, 10, 1205. [Google Scholar] [CrossRef] [PubMed]
- Akrami, H.; Aydore, S.; Leahy, R.M.; Joshi, A.A. Robust Variational Autoencoder for Tabular Data with Beta Divergence. arXiv 2020. [Google Scholar] [CrossRef]
- Xu, L.; Skoularidou, M.; Cuesta-Infante, A.; Veeramachaneni, K. Modeling Tabular data using Conditional GAN. Adv. Neural Inf. Process. Syst. 2019, 32, 7335–7345. [Google Scholar] [CrossRef]
- Hernandez, M.; Epelde, G.; Alberdi, A.; Cilla, R.; Rankin, D. Synthetic data generation for tabular health records: A systematic review. Neurocomputing 2022, 493, 28–45. [Google Scholar] [CrossRef]
- Georges-Filteau, J.; Cirillo, E. Synthetic Observational Health Data with GANs: From slow adoption to a boom in medical research and ultimately digital twins? arXiv 2020. [Google Scholar] [CrossRef]
- Goncalves, A.; Ray, P.; Soper, B.; Stevens, J.; Coyle, L.; Sales, A.P. Generation and evaluation of synthetic patient data. BMC Med. Res. Methodol. 2020, 20, 1–40. [Google Scholar] [CrossRef]
- Bej, S.; Davtyan, N.; Wolfien, M.; Nassar, M.; Wolkenhauer, O. LoRAS: An oversampling approach for imbalanced datasets. Mach. Learn. 2021, 110, 279–301. [Google Scholar] [CrossRef]
- Bej, S.; Schulz, K.; Srivastava, P.; Wolfien, M.; Wolkenhauer, O. A Multi-Schematic Classifier-Independent Oversampling Approach for Imbalanced Datasets. IEEE Access 2021, 9, 123358–123374. [Google Scholar] [CrossRef]
- Han, H.; Wang, W.Y.; Mao, B.H. Borderline-SMOTE: A New Over-Sampling Method in Imbalanced Data Sets Learning. In Proceedings of the International Conference on Intelligent Computing, Hefei, China, 23–26 August 2005; pp. 878–887. [Google Scholar] [CrossRef]
- KammounAmina; SlamaRim; TabiaHedi; OuniTarek; AbidMohmed Generative Adversarial Networks for face generation: A survey. ACM Comput. Surv. 2021. [CrossRef]
- Schultz, K.; Bej, S.; Hahn, W.; Wolfien, M.; Srivastava, P.; Wolkenhauer, O. Convex space learning improves deep-generative oversampling for tabular imbalanced classification on smaller datasets. arXiv 2022. [Google Scholar] [CrossRef]
- Choi, E.; Biswal, S.; Malin, B.; Duke, J.; Stewart, W.F.; Org, S.; Sun, J. Generating Multi-label Discrete Patient Records using Generative Adversarial Networks. In Proceedings of the 2nd Machine Learning for Healthcare Conference, Boston, MA, USA, 18–19 August 2017. [Google Scholar] [CrossRef]
- Park, N.; Mohammadi, M.; Gorde, K.; Jajodia, S.; Park, H.; Kim, Y. Data Synthesis based on Generative Adversarial Networks. Proc. VLDB Endow. 2018, 11, 1071–1083. [Google Scholar] [CrossRef]
- Camino, R.D.; Hammerschmidt, C.A.; State, R. Generating Multi-Categorical Samples with Generative Adversarial Networks. arXiv 2018. [Google Scholar] [CrossRef]
- Xu, L.; Veeramachaneni, K. Synthesizing Tabular Data using Generative Adversarial Networks. arXiv 2018. [Google Scholar] [CrossRef]
- Coutinho-Almeida, J.; Rodrigues, P.P.; Cruz-Correia, R.J. GANs for Tabular Healthcare Data Generation: A Review on Utility and Privacy. Lect. Notes Comput. Sci. 2021, 12986, 282–291. [Google Scholar] [CrossRef]
- Wen, B.; Colon, L.O.; Subbalakshmi, K.P.; Chandramouli, R. Causal-TGAN: Generating Tabular Data Using Causal Generative Adversarial Networks. arXiv 2021. [Google Scholar] [CrossRef]
- Kim, J.; Jeon, J.; Lee, J.; Hyeong, J.; Park, N. OCT-GAN: Neural ODE-based Conditional Tabular GANs. arXiv 2021, 10. [Google Scholar] [CrossRef]
- Kunar, A.; Birke, R.; Zhao, Z.; Chen, L. DTGAN: Differential Private Training for Tabular GANs. arXiv 2021. [Google Scholar] [CrossRef]
- Zhao, Z.; Kunar, A.; Van der Scheer, H.; Birke, R.; Chen, L.Y. CTAB-GAN: Effective Table Data Synthesizing. arXiv 2021. [Google Scholar] [CrossRef]
- Tantipongpipat, U.T.; Waites, C.; Boob, D.; Siva, A.A.; Cummings, R. Differentially Private Synthetic Mixed-Type Data Generation For Unsupervised Learning. Intell. Decis. Technol. 2019, 15, 779–807. [Google Scholar] [CrossRef]
- Bej, S.; Sarkar, J.; Biswas, S.; Mitra, P.; Chakrabarti, P.; Wolkenhauer, O. Identification and epidemiological characterization of Type-2 diabetes sub-population using an unsupervised machine learning approach. Nutr. Diabetes 2022, 12, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Marcy, T.W.; Kaplan, B.; Connolly, S.W.; Michel, G.; Shiffman, R.N.; Flynn, B.S. Developing a Decision Support System for Tobacco Use Counseling Using Primary Care Physicians. Inform. Prim. Care 2008, 16, 101. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fraccaro, P.; Casteleiro, M.A.; Ainsworth, J.; Buchan, I. Adoption of clinical decision support in multimorbidity: A systematic review. JMIR Med. Inform. 2015, 3, e3503. [Google Scholar] [CrossRef] [PubMed]
- Brunner, J.; Chuang, E.; Goldzweig, C.; Cain, C.L.; Sugar, C.; Yano, E.M. User-centered design to improve clinical decision support in primary care. Int. J. Med. Inform. 2017, 104, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Jokela, T. Assessments of Usability Engineering Processes: Experiences from Experiments. In Proceedings of the 36th Annual Hawaii International Conference on System Sciences, Big Island, HI, USA, 6–9 January 2003. [Google Scholar]
- Hui, D.; Meng, Y.-C.; Bruera, S.; Geng, Y.; Hutchins, R.; Mori, M.; Strasser, F.; Bruera, E. Referral Criteria for Outpatient Palliative Cancer Care: A Systematic Review. Oncologist 2016, 21, 895–901. [Google Scholar] [CrossRef]
- ElMokhallalati, Y.; Bradley, S.H.; Chapman, E.; Ziegler, L.; Murtagh, F.E.M.; Johnson, M.J.; Bennett, M.I. Identification of patients with potential palliative care needs: A systematic review of screening tools in primary care. Palliat. Med. 2020, 34, 989–1005. [Google Scholar] [CrossRef]
- Gaertner, J.; Wolf, J.; Hallek, M.; Glossmann, J.P.; Voltz, R. Standardizing integration of palliative care into comprehensive cancer therapy--a disease specific approach. Support. Care Cancer 2011, 19, 1037–1043. [Google Scholar] [CrossRef]
- Erweiterte S3-Leitlinie Palliativmedizin für Patienten mit Einer Nicht-Heilbaren Krebserkrankung; Kohlhammer: Stuttgart, Germany, 2020; ISBN 978-3-17-038390-6.
- Adelson, K.; Paris, J.; Horton, J.R.; Hernandez-Tellez, L.; Ricks, D.; Morrison, R.S.; Smith, C.B. Standardized Criteria for Palliative Care Consultation on a Solid Tumor Oncology Service Reduces Downstream Health Care Use. J. Oncol. Pract. 2017, 13, e431–e438. [Google Scholar] [CrossRef]
- Ostgathe, C.; Wendt, K.N.; Heckel, M.; Kurkowski, S.; Klein, C.; Krause, S.W.; Fuchs, F.S.; Bayer, C.M.; Stiel, S. Identifying the need for specialized palliative care in adult cancer patients-development and validation of a screening procedure based on proxy assessment by physicians and filter questions. BMC Cancer 2019, 19, 646. [Google Scholar] [CrossRef] [PubMed]
- Bentler, S.E.; Liu, L.; Obrizan, M.; Cook, E.A.; Wright, K.B.; Geweke, J.F.; Chrischilles, E.A.; Pavlik, C.E.; Wallace, R.B.; Ohsfeldt, R.L.; et al. The aftermath of hip fracture: Discharge placement, functional status change, and mortality. Am. J. Epidemiol. 2009, 170, 1290–1299. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.P.; Temel, J.S.; Balboni, T.; Glare, P. A review of the trials which examine early integration of outpatient and home palliative care for patients with serious illnesses. Ann. Palliat. Med. 2015, 4, 99–121. [Google Scholar] [CrossRef] [PubMed]
- Haun, M.W.; Estel, S.; Rücker, G.; Friederich, H.C.; Villalobos, M.; Thomas, M.; Hartmann, M. Early palliative care for adults with advanced cancer. Cochrane Database Syst. Rev. 2017, 6, 6. [Google Scholar] [CrossRef] [PubMed]
- Berendt, J.; Stiel, S.; Simon, S.T.; Schmitz, A.; van Oorschot, B.; Stachura, P.; Ostgathe, C. Integrating Palliative Care into Comprehensive Cancer Centers: Consensus-Based Development of Best Practice Recommendations. Oncologist 2016, 21, 1241–1249. [Google Scholar] [CrossRef]
- Tewes, M.; Rettler, T.; Wolf, N.; Hense, J.; Schuler, M.; Teufel, M.; Beckmann, M. Predictors of outpatients’ request for palliative care service at a medical oncology clinic of a German comprehensive cancer center. Support. Care Cancer 2018, 26, 3641–3647. [Google Scholar] [CrossRef]
- Zimmermann, C.; Swami, N.; Krzyzanowska, M.; Leighl, N.; Rydall, A.; Rodin, G.; Tannock, I.; Hannon, B. Perceptions of palliative care among patients with advanced cancer and their caregivers. CMAJ 2016, 188, E217–E227. [Google Scholar] [CrossRef]
- Kavalieratos, D.; Corbelli, J.; Zhang, D.; Dionne-Odom, J.N.; Ernecoff, N.C.; Hanmer, J.; Hoydich, Z.P.; Ikejiani, D.Z.; Klein-Fedyshin, M.; Zimmermann, C.; et al. Association Between Palliative Care and Patient and Caregiver Outcomes: A Systematic Review and Meta-analysis. JAMA 2016, 316, 2104–2114. [Google Scholar] [CrossRef]
- Bennardi, M.; Diviani, N.; Gamondi, C.; Stüssi, G.; Saletti, P.; Cinesi, I.; Rubinelli, S. Palliative care utilization in oncology and hemato-oncology: A systematic review of cognitive barriers and facilitators from the perspective of healthcare professionals, adult patients, and their families. BMC Palliat. Care 2020, 19, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Okuyama, T.; Akechi, T.; Yamashita, H.; Toyama, T.; Nakaguchi, T.; Uchida, M.; Furukawa, T.A. Oncologists’ recognition of supportive care needs and symptoms of their patients in a breast cancer outpatient consultation. Jpn. J. Clin. Oncol. 2011, 41, 1251–1258. [Google Scholar] [CrossRef][Green Version]
- Gaertner, J.; Siemens, W.; Meerpohl, J.J.; Antes, G.; Meffert, C.; Xander, C.; Stock, S.; Mueller, D.; Schwarzer, G.; Becker, G. Effect of specialist palliative care services on quality of life in adults with advanced incurable illness in hospital, hospice, or community settings: Systematic review and meta-analysis. BMJ 2017, 357, 2925. [Google Scholar] [CrossRef]
- Hui, D.; De La Cruz, M.; Mori, M.; Parsons, H.A.; Kwon, J.H.; Torres-Vigil, I.; Kim, S.H.; Dev, R.; Hutchins, R.; Liem, C.; et al. Concepts and definitions for “supportive care”, “best supportive care”, “palliative care”, and “hospice care” in the published literature, dictionaries, and textbooks. Support. Care Cancer 2013, 21, 659–685. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hahn, W.; Schütte, K.; Schultz, K.; Wolkenhauer, O.; Sedlmayr, M.; Schuler, U.; Eichler, M.; Bej, S.; Wolfien, M. Contribution of Synthetic Data Generation towards an Improved Patient Stratification in Palliative Care. J. Pers. Med. 2022, 12, 1278. https://doi.org/10.3390/jpm12081278
Hahn W, Schütte K, Schultz K, Wolkenhauer O, Sedlmayr M, Schuler U, Eichler M, Bej S, Wolfien M. Contribution of Synthetic Data Generation towards an Improved Patient Stratification in Palliative Care. Journal of Personalized Medicine. 2022; 12(8):1278. https://doi.org/10.3390/jpm12081278
Chicago/Turabian StyleHahn, Waldemar, Katharina Schütte, Kristian Schultz, Olaf Wolkenhauer, Martin Sedlmayr, Ulrich Schuler, Martin Eichler, Saptarshi Bej, and Markus Wolfien. 2022. "Contribution of Synthetic Data Generation towards an Improved Patient Stratification in Palliative Care" Journal of Personalized Medicine 12, no. 8: 1278. https://doi.org/10.3390/jpm12081278
APA StyleHahn, W., Schütte, K., Schultz, K., Wolkenhauer, O., Sedlmayr, M., Schuler, U., Eichler, M., Bej, S., & Wolfien, M. (2022). Contribution of Synthetic Data Generation towards an Improved Patient Stratification in Palliative Care. Journal of Personalized Medicine, 12(8), 1278. https://doi.org/10.3390/jpm12081278