Artificial Intelligence-Based Medical Data Mining
Abstract
1. Introduction
1.1. Medical vs. Non-Medical Literature Text Mining
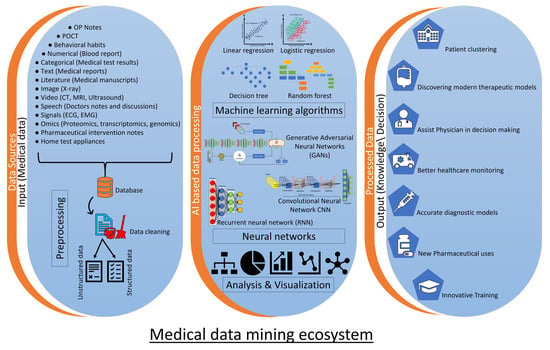
1.2. Use of Artificial Intelligence and Machine Learning in Medical Literature Data Mining
Natural Language Processing
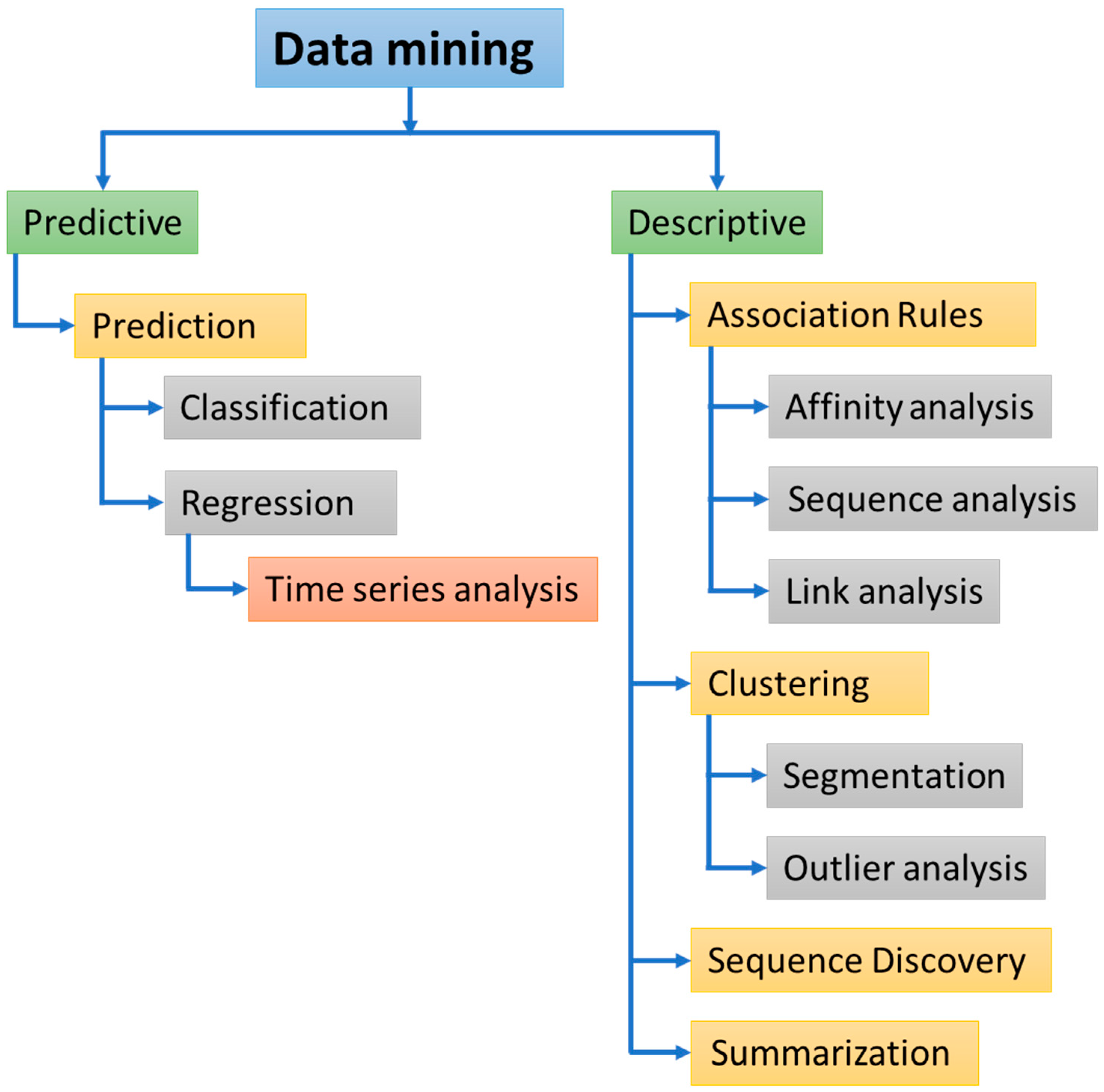
2. Standard Process for Data Mining
2.1. Business Understanding
2.2. Data Understanding
2.2.1. Literature Extraction/Data Gathering
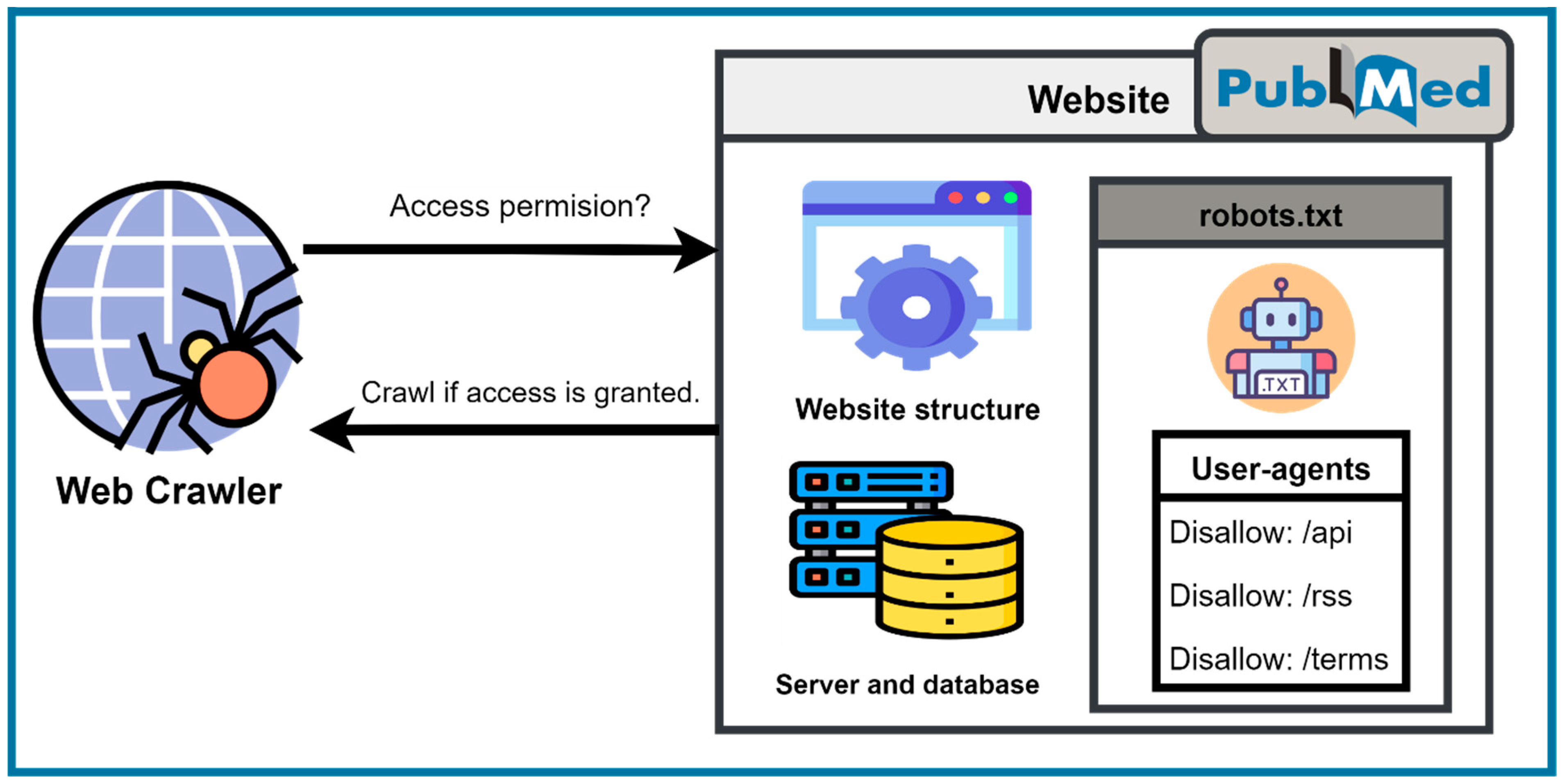
Access Restriction
Data Collection from Different Sources
- Hospital management software (Patient data/Clinical narratives).
- Clinical trials.
- Research data in Medicine.
- Publication platforms for Medicine (PubMed, for instance).
- Pharmaceuticals and regulatory data.
2.3. Data Preparation
2.3.1. Data Cleaning/Data Transformation
- NumPy is a quick and easy-to-use open-source Python library for data processing. Because many of the most well-known Python libraries, including Pandas and Matplotlib, are based on NumPy, it is a fundamentally crucial library for the data science environment. The primary purpose of the NumPy library is the straightforward manipulation of large multidimensional arrays, vectors, and matrices. For numerical calculations, NumPy also offers effectively implemented functions [48].
- Data processing tasks such as data cleaning, data manipulation, and data analysis are performed using the well-known Python library Pandas. The Python Data Analysis Library is referred to as “Pandas”. Multiple modules for reading, processing, and writing CSV, JSON, and Excel files are available in the library. Although there are many data cleaning tools available, managing and exploring data with the Pandas library is incredibly quick and effective [49].
- An open-source Python library for automating data cleaning procedures is called DataCleaner. Pandas Dataframe and scikit-learn data preprocessing features comprise its two separate modules [50].
- Generation of Bibliographic Data is known as GROBID. It is a machine-learning library that has developed into a state-of-the-art open-source library for removing metadata from PDF-formatted technical and scientific documents. The library plans to reconstruct the logical structure of its original document in addition to simple bibliographic extraction in order to support large-scale advanced digital library processes and text analysis.
- 2.
- BioC is a straightforward and straightforward format for exchanging text data and annotations, as well as for simple text processing. Its primary goal is to provide an abundance of research data and articles for text mining and information retrieval. They are available in a variety of file formats, including BioC XML, BioC JSON, Unicode, and ASCII. These formats are available through a Web API or FTP [53].
2.3.2. Feature Engineering
2.3.3. Searching for Keywords
2.4. Modeling
2.5. Data Model Validation and Testing
2.6. Evaluation
2.7. Deployment
3. Conclusions and Future Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sumathy, K.L.; Chidambaram, M. Text Mining: Concepts, Applications, Tools and Issues—An Overview. Int. J. Comput. Appl. 2013, 80, 29–32. [Google Scholar] [CrossRef]
- Cios, K.J.; Moore, G.W. Uniqueness of medical data mining. Artif. Intell. Med. 2002, 26, 1–24. [Google Scholar] [CrossRef]
- Yang, Y.; Li, R.; Xiang, Y.; Lin, D.; Yan, A.; Chen, W.; Li, Z.; Lai, W.; Wu, X.; Wan, C.; et al. Standardization of Collection, Storage, Annotation, and Management of Data Related to Medical Artificial Intelligence. Intell. Med. 2021. [Google Scholar] [CrossRef]
- Thorpe, J.H.; Gray, E.A. Big data and public health: Navigating privacy laws to maximize potential. Public Health Rep. 2015, 130, 171–175. [Google Scholar] [CrossRef]
- McGuire, A.L.; Beskow, L.M. Informed consent in genomics and genetic research. Annu. Rev. Genom. Hum. Genet. 2010, 11, 361–381. [Google Scholar] [CrossRef]
- Tayefi, M.; Ngo, P.; Chomutare, T.; Dalianis, H.; Salvi, E.; Budrionis, A.; Godtliebsen, F. Challenges and opportunities beyond structured data in analysis of electronic health records. WIREs Comp. Stat. 2021, 13, 1–19. [Google Scholar] [CrossRef]
- Han, J.; Kamber, M.; Pei, J. Data Mining: Concepts and Techniques, 3rd ed.; Morgan Kaufmann Publisher: Waltham, MA, USA, 2011; ISBN 978-0-12-381479-1. [Google Scholar]
- Locke, S.; Bashall, A.; Al-Adely, S.; Moore, J.; Wilson, A.; Kitchen, G.B. Natural language processing in medicine: A review. Trends Anaesth. Crit. Care 2021, 38, 4–9. [Google Scholar] [CrossRef]
- Vyas, A. Top 14 Use Cases of Natural Language Processing in Healthcare. 6 July 2019. Available online: https://marutitech.com/use-cases-of-natural-language-processing-in-healthcare/ (accessed on 29 July 2022).
- Liu, Z.; Yang, M.; Wang, X.; Chen, Q.; Tang, B.; Wang, Z.; Xu, H. Entity recognition from clinical texts via recurrent neural network. BMC Med. Inform. Decis. Mak. 2017, 17, 67. [Google Scholar] [CrossRef]
- Deng, Y.; Faulstich, L.; Denecke, K. Concept Embedding for Relevance Detection of Search Queries Regarding CHOP. Stud. Health Technol. Inform. 2017, 245, 1260. [Google Scholar]
- Afzal, M.; Hussain, M.; Malik, K.M.; Lee, S. Impact of Automatic Query Generation and Quality Recognition Using Deep Learning to Curate Evidence from Biomedical Literature: Empirical Study. JMIR Med. Inform. 2019, 7, e13430. [Google Scholar] [CrossRef]
- Pandey, B.; Kumar Pandey, D.; Pratap Mishra, B.; Rhmann, W. A comprehensive survey of deep learning in the field of medical imaging and medical natural language processing: Challenges and research directions. J. King Saud Univ. Comput. Inf. Sci. 2021, 34, 5083–5099. [Google Scholar] [CrossRef]
- Shearer, C. The CRISP-DM Model: The New Blueprint for Data Mining. Int. J. Data Warehous. 2000, 5, 13–22. [Google Scholar]
- Costa, C.J.; Aparicio, J.T. POST-DS: A Methodology to Boost Data Science. In Proceedings of the 15th Iberian Conference on Information Systems and Technologies (CISTI), Sevilla, Spain, 24–27 June 2020; pp. 1–6, ISBN 978-989-54659-0-3. [Google Scholar]
- Catley, C.; Smith, K.; McGregor, C.; Tracy, M. Extending CRISP-DM to incorporate temporal data mining of multidimensional medical data streams: A neonatal intensive care unit case study. In Proceedings of the 22nd IEEE International Symposium on Computer-Based Medical Systems, Albuquerque, NM, USA, 2–5 August 2009; pp. 1–5, ISBN 978-1-4244-4878-4. [Google Scholar]
- Data Science Process Alliance. What Is CRISP DM? Available online: https://www.datascience-pm.com/crisp-dm-2/ (accessed on 16 April 2022).
- Martins, B.; Ferreira, D.; Neto, C.; Abelha, A.; Machado, J. Data Mining for Cardiovascular Disease Prediction. J. Med. Syst. 2021, 45, 6. [Google Scholar] [CrossRef]
- NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2013, 41, D8–D20. [Google Scholar] [CrossRef]
- Guo, C.X.; He, L.; Yin, J.Y.; Meng, X.G.; Tan, W.; Yang, G.P.; Bo, T.; Liu, J.P.; Lin, X.J.; Chen, X. Epidemiological and clinical features of pediatric COVID-19. BMC Med. 2020, 18, 250. [Google Scholar] [CrossRef] [PubMed]
- Miuțescu, A. Web Scraping vs. Web Crawling: Understand the Difference. WebScrapingAPI [Online]. 7 January 2021. Available online: https://www.webscrapingapi.com/web-scraping-vs-web-crawling/ (accessed on 19 April 2022).
- Octoparse. What Is Web Scraping—Basics & Practical Uses—DataDrivenInvestor. DataDrivenInvestor [Online]. 25 January 2022. Available online: https://medium.datadriveninvestor.com/what-is-web-scraping-basics-practical-uses-66e1063cfa74 (accessed on 19 April 2022).
- Batsakis, S.; Petrakis, E.G.; Milios, E. Improving the performance of focused web crawlers. Data Knowl. Eng. 2009, 68, 1001–1013. [Google Scholar] [CrossRef]
- Yuan, X.; MacGregor, M.H.; Harms, J. An efficient scheme to remove crawler traffic from the Internet. In Proceedings of the Eleventh International Conference on Computer Communications and Networks. Eleventh International Conference on Computer Communications and Networks, Miami, FL, USA, 14–16 October 2002; pp. 90–95, ISBN 0-7803-7553-X. [Google Scholar]
- DeVito, N.J.; Richards, G.C.; Inglesby, P. How we learnt to stop worrying and love web scraping. Nature 2020, 585, 621–622. [Google Scholar] [CrossRef]
- Kaur, A.; Chopra, D. Comparison of text mining tools. In Proceedings of the 5th International Conference on Reliability, Infocom Technologies and Optimization (ICRITO) (Trends and Future Directions), Noida, India, 9 July–9 September 2016; pp. 186–192, ISBN 978-1-5090-1489-7. [Google Scholar]
- Chandra, R.V.; Varanasi, B.S. Python Requests Essentials: Learn How to Integrate Your Applications Seamlessly with Web Services Using Python Requests; Packt: Birmingham, UK; Mumbai, India, 2015; ISBN 9781784395414. [Google Scholar]
- Kouzis-Loukas, D. Learning Scrapy: Learn the Art of Efficient Web Scraping and Crawling with Python; Packt: Birmingham, UK, 2016; ISBN 9781784390914. [Google Scholar]
- Richardson, L. Beautiful Soup Documentation. Available online: https://www.crummy.com/software/BeautifulSoup/bs4/doc/ (accessed on 16 April 2022).
- Sharma, P.R. Selenium with Python: A Beginner’s Guide; BPB: Delhi, India, 2019; ISBN 9789389328820. [Google Scholar]
- Gu, D.; Li, J.; Li, X.; Liang, C. Visualizing the knowledge structure and evolution of big data research in healthcare informatics. Int. J. Med. Inform. 2017, 98, 22–32. [Google Scholar] [CrossRef]
- Ristevski, B.; Chen, M. Big Data Analytics in Medicine and Healthcare. J. Integr. Bioinform. 2018, 15, 1–5. [Google Scholar] [CrossRef]
- Giffen, C.A.; Carroll, L.E.; Adams, J.T.; Brennan, S.P.; Coady, S.A.; Wagner, E.L. Providing Contemporary Access to Historical Biospecimen Collections: Development of the NHLBI Biologic Specimen and Data Repository Information Coordinating Center (BioLINCC). Biopreserv. Biobank. 2015, 13, 271–279. [Google Scholar] [CrossRef]
- Cimino, J.J.; Ayres, E.J.; Remennik, L.; Rath, S.; Freedman, R.; Beri, A.; Chen, Y.; Huser, V. The National Institutes of Health’s Biomedical Translational Research Information System (BTRIS): Design, contents, functionality and experience to date. J. Biomed. Inform. 2014, 52, 11–27. [Google Scholar] [CrossRef] [PubMed]
- Mayo-Wilson, E.; Doshi, P.; Dickersin, K. Are manufacturers sharing data as promised? BMJ 2015, 351, h4169. [Google Scholar] [CrossRef] [PubMed]
- Doll, K.M.; Rademaker, A.; Sosa, J.A. Practical Guide to Surgical Data Sets: Surveillance, Epidemiology, and End Results (SEER) Database. JAMA Surg. 2018, 153, 588–589. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.E.W.; Pollard, T.J.; Shen, L.; Lehman, L.W.H.; Feng, M.; Ghassemi, M.; Moody, B.; Szolovits, P.; Celi, L.A.; Mark, R.G. MIMIC-III, a freely accessible critical care database. Sci. Data 2016, 3, 160035. [Google Scholar] [CrossRef]
- Johnson, A.; Bulgarelli, L.; Pollard, T.; Horng, S.; Celi, L.A.; Mark, R. MIMIC-IV; Version 1.0; PhysioNet: Cambridge, MA, USA, 2021. [Google Scholar] [CrossRef]
- Johnson, A.E.W.; Pollard, T.J.; Berkowitz, S.J.; Greenbaum, N.R.; Lungren, M.P.; Deng, C.Y.; Mark, R.G.; Horng, S. MIMIC-CXR, a de-identified publicly available database of chest radiographs with free-text reports. Sci. Data 2019, 6, 317. [Google Scholar] [CrossRef]
- Ahluwalia, N.; Dwyer, J.; Terry, A.; Moshfegh, A.; Johnson, C. Update on NHANES Dietary Data: Focus on Collection, Release, Analytical Considerations, and Uses to Inform Public Policy. Adv. Nutr. 2016, 7, 121–134. [Google Scholar] [CrossRef]
- Vos, T.; Lim, S.S.; Abbafati, C.; Abbas, K.M.; Abbasi, M.; Abbasifard, M.; Abbasi-Kangevari, M.; Abbastabar, H.; Abd-Allah, F.; Abdelalim, A.; et al. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2020, 396, 1204–1222. [Google Scholar] [CrossRef]
- Palmer, L.J. UK Biobank: Bank on it. Lancet 2007, 369, 1980–1982. [Google Scholar] [CrossRef]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Davis, S.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef]
- Woolley, C.S.C.; Handel, I.G.; Bronsvoort, B.M.; Schoenebeck, J.J.; Clements, D.N. Is it time to stop sweeping data cleaning under the carpet? A novel algorithm for outlier management in growth data. PLoS ONE 2020, 15, e0228154. [Google Scholar] [CrossRef] [PubMed]
- Coupler.io Blog. Data Cleansing vs. Data Transformation|Coupler.io Blog. Available online: https://blog.coupler.io/data-cleansing-vs-data-transformation/#What_is_data_transformation (accessed on 24 June 2022).
- Elgabry, O. The Ultimate Guide to Data Cleaning—Towards Data Science. 28 February 2019. Available online: https://towardsdatascience.com/the-ultimate-guide-to-data-cleaning-3969843991d4 (accessed on 24 June 2022).
- Harris, C.R.; Millman, K.J.; van der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef] [PubMed]
- McKinney, W. Data structures for statistical computing in python. In Proceedings of the Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010; pp. 51–56. [Google Scholar]
- Gordon, B.; Fennessy, C.; Varma, S.; Barrett, J.; McCondochie, E.; Heritage, T.; Duroe, O.; Jeffery, R.; Rajamani, V.; Earlam, K.; et al. Evaluation of freely available data profiling tools for health data research application: A functional evaluation review. BMJ Open 2022, 12, e054186. [Google Scholar] [CrossRef] [PubMed]
- Lopez, P. GROBID: Combining Automatic Bibliographic Data Recognition and Term Extraction for Scholarship Publications. In Research and Advanced Technology for Digital Libraries; Agosti, M., Borbinha, J., Kapidakis, S., Papatheodorou, C., Tsakonas, G., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 473–474. ISBN 978-3-642-04345-1. [Google Scholar]
- Lo, K.; Wang, L.L.; Neumann, M.; Kinney, R.; Weld, D. S2ORC: The Semantic Scholar Open Research Corpus. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, Online, 5–10 July 2020; Jurafsky, D., Chai, J., Schluter, N., Tetreault, J., Eds.; Association for Computational Linguistics: Stroudsburg, PA, USA, 2020; pp. 4969–4983. [Google Scholar]
- Comeau, D.C.; Wei, C.H.; Doğan, R.I.; Lu, Z. PMC text mining subset in BioC: 2.3 million full text articles and growing. arXiv 2018, arXiv:1804.05957. [Google Scholar] [CrossRef]
- Rawat, T.; Khemchandani, V. Feature engineering (FE) tools and techniques for better classification performance. Int. J. Innov. Eng. Technol. 2017, 8, 169–179. [Google Scholar] [CrossRef]
- Heaton, J. An empirical analysis of feature engineering for predictive modeling. In Proceedings of the SoutheastCon 2016, Norfolk, VA, USA, 30 March–3 April 2016; IEEE: Manhattan, NY, USA, 2016; pp. 1–6, ISBN 978-1-5090-2246-5. [Google Scholar]
- Vijithananda, S.M.; Jayatilake, M.L.; Hewavithana, B.; Gonçalves, T.; Rato, L.M.; Weerakoon, B.S.; Kalupahana, T.D.; Silva, A.D.; Dissanayake, K.D. Feature extraction from MRI ADC images for brain tumor classification using machine learning techniques. BioMed Eng. OnLine 2022, 21, 52. [Google Scholar] [CrossRef]
- Rus, A. Keyword-Recherche: Die richtigen Keywords Finden Leicht Gemacht. Evergreen Media AR GmbH. 7 September 2021. Available online: https://www.evergreenmedia.at/ratgeber/keyword-recherche/ (accessed on 20 April 2022).
- Singh, V. Replace or Retrieve Keywords in Documents at Scale. 2017. Available online: https://arxiv.org/pdf/1711.00046 (accessed on 19 April 2022).
- Bird, S.; Klein, E.; Loper, E. Natural Language Processing with Python; O’Reilly: Beijing, China; Farnham, UK, 2009; ISBN 9780596516499. [Google Scholar]
- Honnibal, M. spaCy 2: Natural Language Understanding with Bloom Embeddings, Convolutional Neural Networks and Incremental Parsing. Sentometrics Research. 1 January 2017. Available online: https://sentometrics-research.com/publication/72/ (accessed on 19 April 2022).
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Müller, A.; Nothman, J.; Louppe, G.; et al. Scikit-learn: Machine Learning in Python. JMLR 2011, 12, 2825–2830. [Google Scholar]
- Pinto, A.; Oliveira, H.G.; Alves, A.O. Comparing the Performance of Different NLP Toolkits in Formal and Social Media Text. In Proceedings of the 5th Symposium on Languages, Applications and Technologies (SLATE’16), Maribor, Slovenia, 20–21 June 2016; Mernik, M., Leal, J.P., Oliveira, H.G., Eds.; Schloss Dagstuhl—Leibniz-Zentrum fuer Informatik GmbH: Wadern/Saarbruecken, Germany, 2016. ISBN 978-3-95977-006-4. [Google Scholar]
- Rehurek, R.; Sojka, P. Gensim-python framework for vector space modelling. NLP Cent. Fac. Inform. Masaryk. Univ. Brno Czech Repub. 2011, 3, 2. [Google Scholar]
- Nadif, M.; Role, F. Unsupervised and self-supervised deep learning approaches for biomedical text mining. Brief. Bioinform. 2021, 22, 1592–1603. [Google Scholar] [CrossRef]
- Wu, W.T.; Li, Y.J.; Feng, A.Z.; Li, L.; Huang, T.; Xu, A.D.; Lyu, J. Data mining in clinical big data: The frequently used databases, steps, and methodological models. Mil. Med. Res. 2021, 8, 44. [Google Scholar] [CrossRef]
- Berry, M.W. Supervised and Unsupervised Learning for Data Science; Springer: Berlin/Heidelberg, Germany, 2020; ISBN 978-3-030-22474-5. [Google Scholar]
- Zowalla, R.; Wetter, T.; Pfeifer, D. Crawling the German Health Web: Exploratory Study and Graph Analysis. J. Med. Internet Res. 2020, 22, e17853. [Google Scholar] [CrossRef] [PubMed]
- Tsioptsias, N.; Tako, A.; Robinson, S. (Eds.) Model Validation and Testing in Simulation: A Literature Review; Schloss Dagstuhl—Leibniz-Zentrum fuer Informatik GmbH: Wadern/Saarbruecken, Germany, 2016; p. 11. [Google Scholar]
- Dong, H.; Liu, Y.; Zeng, W.-F.; Shu, K.; Zhu, Y.; Chang, C. A Deep Learning-Based Tumor Classifier Directly Using MS Raw Data. Proteomics 2020, 20, e1900344. [Google Scholar] [CrossRef] [PubMed]
- OWOX. What Is Data Visualization: Definition, Examples, Principles, Tools. Available online: https://www.owox.com/blog/articles/data-visualization/ (accessed on 12 April 2022).
- Berger, M.; McDonough, K.; Seversky, L.M. cite2vec: Citation-Driven Document Exploration via Word Embeddings. IEEE Trans. Vis. Comput. Graph. 2017, 23, 691–700. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Kang, K.; Park, D.; Choo, J.; Elmqvist, N. TopicLens: Efficient Multi-Level Visual Topic Exploration of Large-Scale Document Collections. IEEE Trans. Vis. Comput. Graph. 2017, 23, 151–160. [Google Scholar] [CrossRef]
- Beck, F.; Koch, S.; Weiskopf, D. Visual Analysis and Dissemination of Scientific Literature Collections with SurVis. IEEE Trans. Vis. Comput. Graph. 2016, 22, 180–189. [Google Scholar] [CrossRef]
- McCurdy, N.; Lein, J.; Coles, K.; Meyer, M. Poemage: Visualizing the Sonic Topology of a Poem. IEEE Trans. Vis. Comput. Graph. 2016, 22, 439–448. [Google Scholar] [CrossRef]
- Brehmer, M.; Ingram, S.; Stray, J.; Munzner, T. Overview: The Design, Adoption, and Analysis of a Visual Document Mining Tool for Investigative Journalists. IEEE Trans. Vis. Comput. Graph. 2014, 20, 2271–2280. [Google Scholar] [CrossRef]
- Hu, M.; Wongsuphasawat, K.; Stasko, J. Visualizing Social Media Content with SentenTree. IEEE Trans. Vis. Comput. Graph. 2017, 23, 621–630. [Google Scholar] [CrossRef]
- Hinrichs, U.; Forlini, S.; Moynihan, B. Speculative Practices: Utilizing InfoVis to Explore Untapped Literary Collections. IEEE Trans. Vis. Comput. Graph. 2016, 22, 429–438. [Google Scholar] [CrossRef]
- Kwon, B.C.; Kim, S.-H.; Lee, S.; Choo, J.; Huh, J.; Yi, J.S. VisOHC: Designing Visual Analytics for Online Health Communities. IEEE Trans. Vis. Comput. Graph. 2016, 22, 71–80. [Google Scholar] [CrossRef]
- Liu, C.-Y.; Chen, M.-S.; Tseng, C.-Y. IncreSTS: Towards Real-Time Incremental Short Text Summarization on Comment Streams from Social Network Services. IEEE Trans. Knowl. Data Eng. 2015, 27, 2986–3000. [Google Scholar] [CrossRef]
- Castellà, Q.; Sutton, C. Word storms: Multiples of word clouds for visual comparison of documents. In Proceedings of the 23rd International Conference on World Wide Web—WWW ‘14, Seoul, Korea, 7–11 April 2014; Chung, C.-W., Broder, A., Shim, K., Suel, T., Eds.; ACM Press: New York, NY, USA, 2014; pp. 665–676, ISBN 9781450327442. [Google Scholar]
- Felix, C.; Pandey, A.V.; Bertini, E. TextTile: An Interactive Visualization Tool for Seamless Exploratory Analysis of Structured Data and Unstructured Text. IEEE Trans. Vis. Comput. Graph. 2017, 23, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.Y.; Sallaberry, A.; Klein, K.; Takatsuka, M.; Roche, M. SentiCompass: Interactive visualization for exploring and comparing the sentiments of time-varying twitter data. In Proceedings of the 2015 IEEE Pacific Visualization Symposium (PacificVis), Hangzhou, China, 14–17 April 2015; pp. 129–133, ISBN 978-1-4673-6879-7. [Google Scholar]
- Gao, T.; Hullman, J.R.; Adar, E.; Hecht, B.; Diakopoulos, N. NewsViews: An automated pipeline for creating custom geovisualizations for news. In Proceedings of the SIGCHI Conference on Human Factors in Computing Systems, Toronto, ON, Canada, 26 April–1 May 2014; Jones, M., Palanque, P., Schmidt, A., Grossman, T., Eds.; ACM: New York, NY, USA, 2014; pp. 3005–3014, ISBN 9781450324731. [Google Scholar]
- Ren, D.; Zhang, X.; Wang, Z.; Li, J.; Yuan, X. WeiboEvents: A Crowd Sourcing Weibo Visual Analytic System. In Proceedings of the 2014 IEEE Pacific Visualization Symposium (PacificVis), Yokohama, Japan, 4–7 March 2014; pp. 330–334, ISBN 978-1-4799-2874-3. [Google Scholar]
- Garcia Esparza, S.; O’Mahony, M.P.; Smyth, B. CatStream: Categorising tweets for user profiling and stream filtering. In Proceedings of the 2013 International Conference on Intelligent User Interfaces—IUI ’13, Santa Monica, CL, USA, 19–22 March 2013; Kim, J., Nichols, J., Szekely, P., Eds.; ACM Press: New York, NY, USA, 2013; p. 25, ISBN 9781450319652. [Google Scholar]
- Glueck, M.; Naeini, M.P.; Doshi-Velez, F.; Chevalier, F.; Khan, A.; Wigdor, D.; Brudno, M. PhenoLines: Phenotype Comparison Visualizations for Disease Subtyping via Topic Models. IEEE Trans. Vis. Comput. Graph. 2018, 24, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Xu, A.; Gou, L.; Liu, H.; Akkiraju, R.; Shen, H.-W. SocialBrands: Visual analysis of public perceptions of brands on social media. In Proceedings of the 2016 IEEE Conference on Visual Analytics Science and Technology (VAST), Baltimore, MD, USA, 23–28 October 2016; pp. 71–80, ISBN 978-1-5090-5661-3. [Google Scholar]
- Wang, X.; Liu, S.; Liu, J.; Chen, J.; Zhu, J.; Guo, B. TopicPanorama: A Full Picture of Relevant Topics. IEEE Trans. Vis. Comput. Graph. 2016, 22, 2508–2521. [Google Scholar] [CrossRef]
- Zhao, J.; Cao, N.; Wen, Z.; Song, Y.; Lin, Y.-R.; Collins, C. #FluxFlow: Visual Analysis of Anomalous Information Spreading on Social Media. IEEE Trans. Vis. Comput. Graph. 2014, 20, 1773–1782. [Google Scholar] [CrossRef]
- Zhao, J.; Gou, L.; Wang, F.; Zhou, M. PEARL: An interactive visual analytic tool for understanding personal emotion style derived from social media. In Proceedings of the 2014 IEEE Conference on Visual Analytics Science and Technology (VAST), Paris, France, 25–31 October 2014; pp. 203–212, ISBN 978-1-4799-6227-3. [Google Scholar]
- Isenberg, P.; Heimerl, F.; Koch, S.; Isenberg, T.; Xu, P.; Stolper, C.D.; Sedlmair, M.; Chen, J.; Moller, T.; Stasko, J. Vispubdata.org: A Metadata Collection About IEEE Visualization (VIS) Publications. IEEE Trans. Vis. Comput. Graph. 2017, 23, 2199–2206. [Google Scholar] [CrossRef]
- Chen, W.; Lao, T.; Xia, J.; Huang, X.; Zhu, B.; Hu, W.; Guan, H. GameFlow: Narrative Visualization of NBA Basketball Games. IEEE Trans. Multimed. 2016, 18, 2247–2256. [Google Scholar] [CrossRef]
- Hoque, E.; Carenini, G. MultiConVis: A Visual Text Analytics System for Exploring a Collection of Online Conversations. In Proceedings of the 21st International Conference on Intelligent User Interfaces, Sonoma, CL, USA, 7–10 March 2016; Nichols, J., Mahmud, J., O’Donovan, J., Conati, C., Zancanaro, M., Eds.; ACM: New York, NY, USA, 2016; pp. 96–107, ISBN 9781450341370. [Google Scholar]
- Hullman, J.; Diakopoulos, N.; Adar, E. Contextifier: Automatic Generation of Annotated Stock Visualizations. In Proceedings of the SIGCHI Conference on Human Factors in Computing Systems, Paris, France, 27 April–2 May 2013; Mackay, W.E., Brewster, S., Bødker, S., Eds.; ACM: New York, NY, USA, 2013; pp. 2707–2716, ISBN 9781450318990. [Google Scholar]
- Viégas, F.; Wattenberg, M.; Hebert, J.; Borggaard, G.; Cichowlas, A.; Feinberg, J.; Orwant, J.; Wren, C. Google + Ripples: A Native Visualization of Information Flow. In Proceedings of the 22nd International Conference on World Wide Web—WWW ’13, Rio de Janeiro, Brazil, 13–17 May 2013; Schwabe, D., Almeida, V., Glaser, H., Baeza-Yates, R., Moon, S., Eds.; ACM: New York, NY, USA, 2013; pp. 1389–1398, ISBN 9781450320351. [Google Scholar]
- El-Assady, M.; Sevastjanova, R.; Gipp, B.; Keim, D.; Collins, C. NEREx: Named-Entity Relationship Exploration in Multi-Party Conversations. Comput. Graph. Forum 2017, 36, 213–225. [Google Scholar] [CrossRef]
- Fu, S.; Zhao, J.; Cui, W.; Qu, H. Visual Analysis of MOOC Forums with iForum. IEEE Trans. Vis. Comput. Graph. 2017, 23, 201–210. [Google Scholar] [CrossRef]
- Shen, Q.; Wu, T.; Yang, H.; Wu, Y.; Qu, H.; Cui, W. NameClarifier: A Visual Analytics System for Author Name Disambiguation. IEEE Trans. Vis. Comput. Graph. 2017, 23, 141–150. [Google Scholar] [CrossRef]
- Madhavan, K.; Elmqvist, N.; Vorvoreanu, M.; Chen, X.; Wong, Y.; Xian, H.; Dong, Z.; Johri, A. DIA2: Web-based Cyberinfrastructure for Visual Analysis of Funding Portfolios. IEEE Trans. Vis. Comput. Graph. 2014, 20, 1823–1832. [Google Scholar] [CrossRef] [PubMed]
- Shahaf, D.; Yang, J.; Suen, C.; Jacobs, J.; Wang, H.; Leskovec, J. Information cartography: Creating Zoomable, Large-Scale Maps of Information. In Proceedings of the 19th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Chicago, IL, USA, 11–14 August 2013; Ghani, R., Senator, T.E., Bradley, P., Parekh, R., He, J., Grossman, R.L., Uthurusamy, R., Dhillon, I.S., Koren, Y., Eds.; ACM: New York, NY, USA, 2013; pp. 1097–1105, ISBN 9781450321747. [Google Scholar]
- Wu, Y.; Liu, S.; Yan, K.; Liu, M.; Wu, F. OpinionFlow: Visual Analysis of Opinion Diffusion on Social Media. IEEE Trans. Vis. Comput. Graph. 2014, 20, 1763–1772. [Google Scholar] [CrossRef] [PubMed]
- Abdul-Rahman, A.; Lein, J.; Coles, K.; Maguire, E.; Meyer, M.; Wynne, M.; Johnson, C.R.; Trefethen, A.; Chen, M. Rule-based Visual Mappings—with a Case Study on Poetry Visualization. Comput. Graph. Forum 2013, 32, 381–390. [Google Scholar] [CrossRef]
- Dou, W.; Yu, L.; Wang, X.; Ma, Z.; Ribarsky, W. HierarchicalTopics: Visually exploring large text collections using topic hierarchies. IEEE Trans. Vis. Comput. Graph. 2013, 19, 2002–2011. [Google Scholar] [CrossRef]
- Cao, N.; Lin, Y.R.; Sun, X.; Lazer, D.; Liu, S.; Qu, H. Whisper: Tracing the Spatiotemporal Process of Information Diffusion in Real Time. IEEE Trans. Vis. Comput. Graph. 2012, 18, 2649–2658. [Google Scholar] [CrossRef]
- Rohrdantz, C.; Hund, M.; Mayer, T.; Wälchli, B.; Keim, D.A. The World’s Languages Explorer: Visual Analysis of Language Features in Genealogical and Areal Contexts. Comput. Graph. Forum 2012, 31, 935–944. [Google Scholar] [CrossRef]
- Stoffel, F.; Jentner, W.; Behrisch, M.; Fuchs, J.; Keim, D. Interactive Ambiguity Resolution of Named Entities in Fictional Literature. Comput. Graph. Forum 2017, 36, 189–200. [Google Scholar] [CrossRef]
- Oelke, D.; Kokkinakis, D.; Keim, D.A. Fingerprint Matrices: Uncovering the dynamics of social networks in prose literature. Comput. Graph. Forum 2013, 32, 371–380. [Google Scholar] [CrossRef][Green Version]
- Angus, D.; Smith, A.; Wiles, J. Conceptual recurrence plots: Revealing patterns in human discourse. IEEE Trans. Vis. Comput. Graph. 2012, 18, 988–997. [Google Scholar] [CrossRef]
- Butler, P.; Chakraborty, P.; Ramakrishan, N. The Deshredder: A visual analytic approach to reconstructing shredded documents. In Proceedings of the 2012 IEEE Conference on Visual Analytics Science and Technology (VAST), Seattle, WA, USA, 14–19 October 2012; pp. 113–122, ISBN 978-1-4673-4753-2. [Google Scholar]
- Chuang, J.; Manning, C.D.; Heer, J. Termite: Visualization techniques for assessing textual topic models. In Proceedings of the International Working Conference on Advanced Visual Interfaces—AVI ‘12, Capri Island, Naples, Italy, 21–25 May 2012; Tortora, G., Levialdi, S., Tucci, M., Eds.; ACM: New York, NY, USA, 2012; p. 74, ISBN 9781450312875. [Google Scholar]
- Cho, I.; Dou, W.; Wang, D.X.; Sauda, E.; Ribarsky, W. VAiRoma: A Visual Analytics System for Making Sense of Places, Times, and Events in Roman History. IEEE Trans. Vis. Comput. Graph. 2016, 22, 210–219. [Google Scholar] [CrossRef]
- Heimerl, F.; Han, Q.; Koch, S.; Ertl, T. CiteRivers: Visual Analytics of Citation Patterns. IEEE Trans. Vis. Comput. Graph. 2016, 22, 190–199. [Google Scholar] [CrossRef] [PubMed]
- Gad, S.; Javed, W.; Ghani, S.; Elmqvist, N.; Ewing, T.; Hampton, K.N.; Ramakrishnan, N. ThemeDelta: Dynamic Segmentations over Temporal Topic Models. IEEE Trans. Vis. Comput. Graph. 2015, 21, 672–685. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Wu, Y.; Liu, S.; Peng, T.-Q.; Zhu, J.J.H.; Liang, R. EvoRiver: Visual Analysis of Topic Coopetition on Social Media. IEEE Trans. Vis. Comput. Graph. 2014, 20, 1753–1762. [Google Scholar] [CrossRef] [PubMed]
- Dou, W.; Wang, X.; Skau, D.; Ribarsky, W.; Zhou, M.X. LeadLine: Interactive visual analysis of text data through event identification and exploration. In Proceedings of the 2012 IEEE Conference on Visual Analytics Science and Technology (VAST), Seattle, WA, USA, 14–19 October 2012; pp. 93–102, ISBN 978-1-4673-4753-2. [Google Scholar]
- Fulda, J.; Brehmel, M.; Munzner, T. TimeLineCurator: Interactive Authoring of Visual Timelines from Unstructured Text. IEEE Trans. Vis. Comput. Graph. 2016, 22, 300–309. [Google Scholar] [CrossRef]
- Janicke, S.; Focht, J.; Scheuermann, G. Interactive Visual Profiling of Musicians. IEEE Trans. Vis. Comput. Graph. 2016, 22, 200–209. [Google Scholar] [CrossRef]
- El-Assady, M.; Gold, V.; Acevedo, C.; Collins, C.; Keim, D. ConToVi: Multi-Party Conversation Exploration using Topic-Space Views. Comput. Graph. Forum 2016, 35, 431–440. [Google Scholar] [CrossRef]
- Hoque, E.; Carenini, G. ConVis: A Visual Text Analytic System for Exploring Blog Conversations. Comput. Graph. Forum 2014, 33, 221–230. [Google Scholar] [CrossRef]
- Oesterling, P.; Scheuermann, G.; Teresniak, S.; Heyer, G.; Koch, S.; Ertl, T.; Weber, G.H. Two-stage framework for a topology-based projection and visualization of classified document collections. In Proceedings of the 2010 IEEE Symposium on Visual Analytics Science and Technology (VAST), Salt Lake City, UT, USA, 25–26 October 2010; pp. 91–98, ISBN 978-1-4244-9488-0. [Google Scholar]
- Thom, D.; Kruger, R.; Ertl, T. Can Twitter Save Lives? A Broad-Scale Study on Visual Social Media Analytics for Public Safety. IEEE Trans. Vis. Comput. Graph. 2016, 22, 1816–1829. [Google Scholar] [CrossRef]
- Mashima, D.; Kobourov, S.G.; Hu, Y. Visualizing Dynamic Data with Maps. IEEE Trans. Vis. Comput. Graph. 2012, 18, 1424–1437. [Google Scholar] [CrossRef] [PubMed]
- Thom, D.; Bosch, H.; Koch, S.; Worner, M.; Ertl, T. Spatiotemporal anomaly detection through visual analysis of geolocated Twitter messages. In Proceedings of the 2012 IEEE Pacific Visualization Symposium (PacificVis), Songdo, Korea, 28 February–2 March 2012; pp. 41–48, ISBN 978-1-4673-0866-3. [Google Scholar]
- Siddiqui, A.T. Data Visualization: A Study of Tools and Challenges. Asian J. Technol. Manag. Res. 2021, 11, 18–23. [Google Scholar]



| Products/Research Prototypes | Treatment/Field of Study | Company/Institution | Reference |
|---|---|---|---|
| MergePACS™ | Clinical Radiology Imaging | IBM Watson | Merge PACS—Overview|IBM |
| BiometryAssist™ | Diagnostic Ultrasound | Samsung Medison | https://www.intel.com/content/www/us/en/developer/tools/oneapi/application-catalog/full-catalog/diagnostic-ultrasound.html (accessed on 17 February 2022) |
| LaborAssist™ | Diagnostic Ultrasound | Samsung Medison | |
| Breast Cancer Detection Solution | Ultrasound, mammography, MRI | Huiying’s solution | https://builders.intel.com/ai/solutionscatalog/breast-cancer-detection-solution-657 (accessed on 17 February 2022) |
| CT solution | Early detection of COVID-19 | Huiying’s solution | https://builders.intel.com/ai/solutionscatalog/ct-solution-for-early-detection-of-covid-19-704 (accessed on 17 February 2022) |
| Dr. Pecker CT Pneumonia CAD System | Classification and quantification of COVID-19 | Jianpei Technology | https://www.intel.com/content/www/us/en/developer/tools/oneapi/application-catalog/full-catalog/dr-pecker-ct-pneumonia-cad-system.html (accessed on 17 February 2022) |
| Requests | Scrapy | Beautiful Soup | Selenium | |
|---|---|---|---|---|
| What is it? | HTTP library for Python | Open-source web framework written in Python | python library | Open-source application framework tool and python library |
| Goal | Sending HTTP/1.1 requests using Python |
|
|
|
| Ideal usage | Used for simple and low-level complex web scraping tasks |
|
|
|
| Advantage |
|
|
|
|
| Selectors | None | JCSS and XPath | CSS | CSS and Xpath |
| Documentation | Detailed and simple to understand | Detailed and simple to understand | Detailed and simple to understand | Detailed and very complex |
| GitHub stars | 46.8 k | 42.7 k | - | 22.7 k |
| Reference | Chandra and Varanasi [27] | Kouzis-Loukas [28] | Richardson [29] | Sharma [30] |
| Databases/Registries | Trial Numbers | Provided by | Location | Founded Year | URL |
|---|---|---|---|---|---|
| ClinicalTrials.gov | 405,612 | U.S. National Library of Medicine | Bethesda, MD, USA | 1997 | https://clinicaltrials.gov/ (accessed on 11 April 2022) |
| Cochrane Central Register of Controlled Trials (CENTRAL) | 1,854,672 | a component of Cochrane Library | London, UK | 1996 | https://www.cochranelibrary.com/central (accessed on 11 April 2022) |
| WHO International Clinical Trials Registry Platform (ICTRP) | 353,502 | World Health Organization | Geneva, Switzerland | - | https://trialsearch.who.int/ (accessed on 11 April 2022) |
| The European Union Clinical Trials Database | 60,321 | European Medicines Agency | Amsterdam, The Netherlands | 2004 | https://www.clinicaltrialsregister.eu/ctr-search/search (accessed on 11 April 2022) |
| CenterWatch | 50,112 | - | Boston, MA, USA | 1994 | http://www.centerwatch.com/clinical-trials/listings/ (accessed on 11 April 2022) |
| German Clinical Trials Register (Deutsches Register Klinischer Studien—DRKS) | >13,000 | Federal Institute for Drugs and Medical Devices | Cologne, Germany | https://www.bfarm.de/EN/BfArM/Tasks/German-Clinical-Trials-Register/_node.html (accessed on 11 April 2022) |
| Databases | No. of Datasets | Owned by | Domains | Available Resources | URL | Ref |
|---|---|---|---|---|---|---|
| Biologic Specimen and Data Repository Information Coordinating Center (BioLINCC) | 262 | National Institute of Health, Calverton, MD, USA | Cardiovascular, pulmonary, and hematological | Specimens and Study Datasets | https://biolincc.nhlbi.nih.gov/studies/ (accessed on 4 April 2022) | [33] |
| Biomedical Translational Research Information System (BTRIS) | Five billion rows of data | Bethesda, MD, USA | Multiple subjects | Study Datasets | https://btris.nih.gov/ (accessed on 4 April 2022) | [34] |
| Clinical Data Study Request | 3135 | The consortium of clinical study Sponsors | Multiple subjects | Study Datasets | https://www.clinicalstudydatarequest.com/ (accessed on 4 April 2022) | [35] |
| Surveillance, Epidemiology, and End Results (SEER) | - | National Cancer Institute, Bethesda, MD, USA | Cancer (All types)—Stage and histological details | Study Datasets | https://seer.cancer.gov/ (accessed on 4 April 2022) | [36] |
| Medical Information Mart for Intensive Care (MIMIC) MIMIC-III | 53,423 patients | MIT Laboratory for Computational Physiology, Cambridge, MA, USA | Intensive Care | Patient data (vital signs, medications, laboratory measurements, observations and notes charted by care providers, survival data, hospital length of stay, imaging reports, diagnostic codes, procedure codes, and fluid balance) | https://mimic.mit.edu/ (accessed on 4 April 2022) | [37,38] |
| MIMIC-CXR | 65,379 patients (377,110 images of chest radiographs) | [39] | ||||
| National Health and Nutrition Examination Survey (NHANES) | - | Centers for disease control and prevention, Hyattsville, MD, USA | Dietary assessment and other nutrition surveillance | data nutritional status, dietary intake, anthropometric measurements, laboratory tests, biospecimens, and clinical findings. | https://www.cdc.gov/nchs/nhanes/index.htm (accessed on 4 April 2022) | [40] |
| Global Burden of Disease (GBDx) | - | Institute for Health Metrics and Evaluation, Seattle, WA, USA | Epidemic patterns and disease burden | Surveys, censuses, vital statistics, and other health-related data | https://ghdx.healthdata.org/ (accessed on 4 April 2022) | [41] |
| UK Biobank (UKB) | 0.5 million | Stockport, UK | In-depth genetic and health information | Genetic, biospecimens, and health data | https://www.ukbiobank.ac.uk/ (accessed on 4 April 2022) | [42] |
| The Cancer Genome Atlas (TCGA) | molecularly characterized over 20,000 cancer samples spanning 33 cancer types | National Cancer Institute, NIH, Bethesda, MD, USA | Cancer genomics | over 2.5 petabytes of epigenomic, proteomic, transcriptomic, and genomic data | https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga (accessed on 4 April 2022) | [43] |
| Gene Expression Omnibus (GEO) | 4,981,280 samples | National Center for Bioinformatics (NCBI), NIH, Bethesda, MD, USA | Sequencing and gene expression | 4348 datasets available | https://www.ncbi.nlm.nih.gov/geo/ (accessed on 4 April 2022) | [44] |
| Source | Articles (Million) | Launched by | Publication Type | Topic | Online | Link |
|---|---|---|---|---|---|---|
| PubMed | 33 | National Center for Biotechnology Information (NCBI) | Abstracts | Biomedical and life sciences | 1996 | https://www.ncbi.nlm.nih.gov/pubmed/ (accessed on 4 April 2022) |
| PubMed Central (PMC) | 7.6 | National Center for Biotechnology Information (NCBI) | Full text | Biomedical and life sciences | 2000 | https://www.ncbi.nlm.nih.gov/pmc/ (accessed on 4 April 2022) |
| Cochrane Library | - | Cochrane | Abstracts and full text | Healthcare | - | https://www.cochranelibrary.com/search (accessed on 4 April 2022) |
| bioRxiv | - | Cold Spring Harbor Laboratory (CSHL) | Unpublished preprints | Biological sciences | 2013 | https://www.biorxiv.org/ (accessed on 4 April 2022) |
| medRxiv | - | Cold Spring Harbor Laboratory (CSHL) | Unpublished manuscripts | Health sciences | 2019 | https://www.medrxiv.org/ (accessed on 4 April 2022) |
| arXiv | 2.05 | Cornell Tech | Non-peer-reviewed | Multidisciplinary | 1991 | https://arxiv.org/ (accessed on 4 April 2022) |
| Google Scholar | 100 (in 2014) | full text or metadata | Multidisciplinary | 2004 | https://scholar.google.com/ (accessed on 4 April 2022) | |
| Semantic Scholar | 205.25 | Allen Institute for Artificial Intelligence | Abstracts and full text | Multidisciplinary | 2015 | https://www.semanticscholar.org/ (accessed on 4 April 2022) |
| Elsevier | 17 (as of 2018) | Elsevier | Abstracts and full text | Multidisciplinary | 1880 | https://www.elsevier.com/ (accessed on 4 April 2022) |
| Springer Nature | - | Springer Nature Group | Abstracts and full text | Multidisciplinary | 2015 | https://www.springernature.com/ (accessed on 4 April 2022) |
| Springer | - | Springer Nature | Abstracts and full text | Multidisciplinary | 1842 | https://link.springer.com/ (accessed on 4 April 2022) |
| Natural Language Toolkit | SpaCy | Scikit-Learn NLP Toolkit | Gensim | |
|---|---|---|---|---|
| What is it? | open-source python platform for handling human language data | open-source python library for advanced natural language processing | machine learning software library for the Python programming language | fastest python library for the training of vector embedding |
| Features |
| |||
| Advantage |
|
|
|
|
| NLP Tasks |
|
|
|
|
| GitHub stars | 10.4 k | 22.4 k | 49 k | 12.9 k |
| Website | nltk.org (accessed on 16 March 2022) | spacy.io (accessed on 16 March 2022) | scikit-learn.org (accessed on 16 March 2022) | radimrehurek.com/gensim/ (accessed on 16 March 2022) |
| Reference | Bird et al. [59] | Honnibal [60] | Pedregosa et al. [61], Pinto et al. [62] | Rehurek and Sojka [63] |
| Visualization Style | Tool [Reference] |
|---|---|
| Text marking/highlighting | cite2vec [71], TopicLens [72], SurVis [73], Poemage [74], Overview [75] |
| Tags or word cloud | SentenTree [76], InfoVis [77], VisOHC [78], IncreSTS [79], Word storms [80] |
| Bar charts | TextTile [81], SentiCompass [82], NewsViews [83], WeiboEvents [84], CatStream [85] |
| Scatterplot | PhenoLines [86], SocialBrands [87], TopicPanorama [88], #FluxFlow [89], PEARL [90] |
| Line chart | Vispubdata.org [91], GameFlow [92], MultiConVis [93], Contextifier [94], Google+Ripples [95] |
| Node-link | NEREx [96], iForum [97], NameClarifier [98], DIA2 [99], Information Cartography [100] |
| Tree | OpinionFlow [101], Rule-based Visual Mappings [102], HierarchicalTopics [103], Whisper [104], The World’s Languages Explorer [105] |
| Matrix | Interactive Ambiguity Resolution [106], Fingerprint Matrices [107], Conceptual recurrence plots [108], The Deshredder [109], Termite [110] |
| Stream graph timeline | VAiRoma [111], CiteRivers [112], ThemeDelta [113], EvoRiver [114], LeadLine [115] |
| Flow timeline | TimeLineCurator [116], Interactive visual profiling [117] |
| Radial visualization | ConToVi [118], ConVis [119] |
| 3D visualization | Two-stage Framework [120] |
| Maps/Geo chart | Can Twitter save lives? [121], Visualizing Dynamic Data with Maps [122], Spatiotemporal Anomaly Detection [123] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zia, A.; Aziz, M.; Popa, I.; Khan, S.A.; Hamedani, A.F.; Asif, A.R. Artificial Intelligence-Based Medical Data Mining. J. Pers. Med. 2022, 12, 1359. https://doi.org/10.3390/jpm12091359
Zia A, Aziz M, Popa I, Khan SA, Hamedani AF, Asif AR. Artificial Intelligence-Based Medical Data Mining. Journal of Personalized Medicine. 2022; 12(9):1359. https://doi.org/10.3390/jpm12091359
Chicago/Turabian StyleZia, Amjad, Muzzamil Aziz, Ioana Popa, Sabih Ahmed Khan, Amirreza Fazely Hamedani, and Abdul R. Asif. 2022. "Artificial Intelligence-Based Medical Data Mining" Journal of Personalized Medicine 12, no. 9: 1359. https://doi.org/10.3390/jpm12091359
APA StyleZia, A., Aziz, M., Popa, I., Khan, S. A., Hamedani, A. F., & Asif, A. R. (2022). Artificial Intelligence-Based Medical Data Mining. Journal of Personalized Medicine, 12(9), 1359. https://doi.org/10.3390/jpm12091359