Abstract
To date, multiple efforts have been made to use genome-wide association studies (GWAS) to untangle the genetic basis for SARS-CoV-2 infection susceptibility and severe COVID-19. However, data on the genetic-related effects of SARS-CoV-2 infection on the presence of accompanying and long-term post-COVID-19 neurological symptoms in younger individuals remain absent. We aimed to examine the possible association between SNPs found in a GWAS of COVID-19 outcomes and three phenotypes: SARS-CoV-2 infection, neurological complications during disease progression, and long-term neurological complications in young adults with a mild-to-moderate disease course. University students (N = 336, age 18–25 years, European ancestry) with or without COVID-19 and neurological symptoms in anamnesis comprised the study sample. Logistic regression was performed with COVID-19-related phenotypes as outcomes, and the top 25 SNPs from GWAS meta-analyses and an MR study linking COVID-19 and cognitive deficits were found. We replicated previously reported associations of the FURIN and SLC6A20 gene variants (OR = 2.36, 95% CI 1.31–4.24) and OR = 1.94, 95% CI 1.08–3.49, respectively) and remaining neurological complications (OR = 2.12, 95% CI 1.10–4.35 for SLC6A20), while NR1H2 (OR = 2.99, 95% CI 1.39–6.69) and TMPRSS2 (OR = 2.03, 95% CI 1.19–3.50) SNPs were associated with neurological symptoms accompanying COVID-19. Our findings indicate that genetic variants related to a severe COVID-19 course in adults may contribute to the occurrence of neurological repercussions in individuals at a young age.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which causes coronavirus disease 2019 (COVID-19), has provoked a tremendous burden on society worldwide, including considerable economic costs and a high mortality and morbidity rate. In addition, the long-term consequences of the disease and its effect on various systems of the body can negatively affect many aspects of an individual’s life.
The effect of the SARS-CoV-2 virus on the respiratory system, cardiovascular system, and gastrointestinal tract has been repeatedly demonstrated [1,2]. Together with epidemiological data on disease severity related to anthropometric parameters and concomitant chronic pathology [1], the role of genetic factors in COVID-19 illness and disease severity has been reported (for review, see [2,3,4]). Twin studies have elucidated a substantial genetic influence for predicted COVID-19, with a concordance rate of 80% in MZ twins [4]. The largest study was conducted by the COVID-19 Host Genetics Initiative (COVID19 HGI) (released on 15 June 2021) [5], which established a link between host genes and three COVID-19 outcomes in a genome-wide association study (GWAS) meta-analysis. A list of genetic variants associated with overall disease susceptibility (112,612 cases, 2,474,079 controls, mainly Europeans), hospitalization due to COVID-19 (24,274 cases, 2,061,529 controls, mainly Europeans), and critical COVID-19 (8779 cases, 1,001,875 controls, mainly Europeans) was reported [5]. In summary, the following genetic loci were shown to have genome-wide statistical significance in several COVID-19 GWAS: the blood type ABO locus; a cluster of immune system genes (SLC6A20, SMRR1, and the immunoglobulin lambda locus) [6,7]; a gene cluster encoding antiviral restriction enzyme activators neighboring the TYK2, DPP9, and IFNAR2 genes [8]; the IVNS1ABP/SWT1 gene cluster [7]; and the MYH14, SETX, ATXN1, and SCN11A genes [9]. A significant association of exonic variants in the ACE2, TMPRSS2, and FURIN genes with COVID-19 incidence was detected by another research group using whole-exome sequencing [10].
Together with the well-established negative effects of SARS-CoV-2 on the pulmonary system, gastrointestinal tract, and cardiovascular system, the virus can also penetrate the blood–brain barrier, affect both neurons and glial cells, and cause neurological complications [3]. Different pathomechanisms underlie the development of neurological impairments, including post-COVID-19 syndrome, such as chronic fatigue, cognitive dysfunction, mood and sleep disorders, pain syndromes, and smell and taste impairments [2]. Previous research has indicated a causal relationship between genetically mediated hospitalized and critical COVID-19 and later memory problems, such as Alzheimer’s disease (AD) and other types of dementia [11]. Interestingly, the ApoE ε4 allele, which was linked to cognitive impairment and functioning [12], was also reported as being associated with an increased risk of severe COVID-19 [13] and infection incidence [14].
An exaggerated susceptibility rate for SARS-CoV-2 infection, more severe forms of the disease [15], and the hyperactivity of immune responses [16] have been detected in adults and the elderly. However, pronounced neurological problems accompany the disease course and manifest as long-term repercussions even in young adults and children [17,18]. Nonetheless, data on the genetic-related effects of SARS-CoV-2 infection on the presence of accompanying neurological symptoms and long-term neurological consequences in younger individuals are lacking. Although several specific findings of gene variants implicated in disease susceptibility [19] and severe COVID-19 [20] in children have been reported, it was also suggested that coronaviral infection may result in an enhanced rate of mental health problems later in life [2]. Therefore, the identification of plausible host genetic variants associated with an elevated risk of developing neurological repercussions during SARS-CoV-2 infection and over a long-term period in younger individuals is of relevance. It will help to develop practical recommendations that can be given to genetically predisposed individuals to reduce the probable mental problems caused by SARS-CoV-2 infection.
As previously reported, neurological impairments are more frequently associated with a severe COVID-19 course, including critical illness and hospitalization [2,21,22]. Therefore, we hypothesized that young adults with long-term neurological symptoms could be the carriers of risky SNP alleles, which have previously been associated with mental problems and severe disease courses in adults.
Herein, we aimed to examine a possible association between SNPs identified in a GWAS of COVID-19 outcomes and three phenotypes, i.e., SARS-CoV-2 infection, neurological complications during disease progression, and in long-term neurological complications, in young adults with a mild-to-moderate disease course.
2. Materials and Methods
2.1. Participants
A sample of university students (N = 336; mean age ± SD: 21.3 ± 1.2 years; age range: 18–25 years; 77% women) from the Volga–Ural region of Russia was collected during the first three waves of the COVID-19 pandemic. All participants were of European ancestry (22% Russians, 61% Tatars, and 17% of mixed ethnicity). The exclusion criteria were a self-reported individual or family history of any psychiatric disorder. The following data were obtained from the participants: sex, ethnic background, SARS-CoV-2 infection in anamnesis, dates of coronaviral infection, clinical symptoms accompanying the disease course, and neurological complications that appeared within 6 months and were maintained over a prolonged period after COVID-19 recovery (up to one year). Disease susceptibility (cases) was assigned based on a positive PCR test or a reported suspected COVID-19 illness. All cases indicated a mild (absence of any shortness of breath, dyspnea, or abnormal chest imaging) or moderate (individuals with a febrile temperature lasting for at least one week or lower respiratory tract inflammation but SpO2 > 94%) disease course without hospitalization, as previously classified [23]. Subjects were classified as resistant to SARS-CoV-2 infection (controls) if they reported an absence of COVID-19 infection during the first three waves of the COVID-19 pandemic prior to their individual vaccination against SARS-CoV-2. Information regarding the clinical symptoms accompanying coronaviral disease was obtained for disease cases preceding individual vaccination. Neurological symptoms included severe headache, memory problems, anosmia/dysosmia, ageusia/dysgeusia, insomnia and sleep problems, anxiety, and depression manifested during the course of the disease, or later after recovery, and maintained for a period of at least one year.
The study was approved by the Biological Ethics Committee at the Institute of Biochemistry and Genetics—Subdivision of the Ufa Federal Research Centre of the Russian Academy of Sciences (Ufa, Russia) (protocol code 19, date of approval, 25 November 2021). Written informed consent was obtained from all participants after they were acquainted with the procedures. All participants were informed about the voluntary and confidential nature of their participation. All procedures performed were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
2.2. SNPs Selection and Genotyping
Genomic DNA was extracted from the leukocytes of enrolled volunteers using a standard phenol–chloroform technique. We selected the top SNPs reported in two major meta-analyses from the COVID-19 HGI describing the genetic impact on COVID-19 susceptibility, hospitalization, and critical illness [5,24] and one Mendelian randomization study linking COVID-19 and cognitive impairment [11]. We selected significant genetic variants from the MR study, including NR1H2 rs1405655, MUC5B rs35705950, SLC6A20 rs2531743, KEAP1 rs45524632, and ABO rs635634. A final set of 25 SNPs from these studies was formed based on their presence in the Infinium Global Screening Array-24 v.3.0 Kit (Illumina, San Diego, California, USA), which was initially used for genotyping on the Illumina IScan platform (Genotek, Russia). For several SNPs identified by HGI, that were absent in the genotyping array, we obtained data on the genotypes of the closest proxy SNPs (https://ldlink.nci.nih.gov, accessed on 20 October 2022, r2 > 0.7) present in the 1000 Genomes Project (www.ldlink.nci.nih.gov (accessed on 20 October 2022)). A complete list of SNPs examined within the framework of this study and their relevance to the genes is presented in the Supplementary Information (Table S1). The chosen SNPs resided in the SMRR1, IVNS1ABP, SLC6A20, LZTFL1, HLA-G, CCHCR1, NOTCH4, ABO, MUC5B, OAS1, OAS3, FURIN, DPP9, TYK2, KEAP1, APOE, NR1H2, and TMPRSS2 genes. Some of the SNPs overlapped among the studies [5,11,24].
2.3. Statistical Analysis
To test for differences in the distribution of COVID19-related outcomes between the sexes, we used a chi-square test. A correlation analysis was conducted between the phenotypes using Spearman’s rank correlation coefficient.
A series of logistic regression analyses were performed to address the association of COVID-19 GWAS SNPs with the three examined disease-related outcomes in mild-to-moderate disease cases. In the regression models, reported previous infection, neurological symptoms accompanying infection, and reported neurological complaints that appeared or were maintained within one year of SARS-CoV-2 infection were included as independent outcomes. In the statistical models, the genotyping data on the investigated SNPs served as the independent variables. In addition, we examined both log-additive and dominant logistic regression models, which evaluate the allele effect in a dose-dependent (AA vs. aA vs. aa) and a dominant manner (AA + aA vs. aa), respectively. For the SNPs with a significant effect on COVID-19 outcomes, we calculated ORs within 95% confidence intervals. To estimate the effect of multiple gene variants on COVID-19 outcomes, we designed logistic regression models with the inclusion of all examined SNPs, followed by the selection of the best prediction model based on the backward elimination procedure, which was chosen on the basis of the best Akaike information criterion (AIC) and p-value in R [25]. All statistical analyses were carried out with R v.4.1.2 and PLINK v.1.9.
3. Results
3.1. Statistical Analysis between Clinical and Demographic Data
The clinical characteristics of the examined sample were as follows: history of COVID-19 infection (67%), accompanying neurological symptoms (70% of cases) and anosmia (65% of cases) within the disease course, and long-term neurological consequences (36% of cases). In the first stage of the analysis, we examined whether there were sex-specific differences in three COVID-19-related outcomes: COVID-19 susceptibility, the presence of neurological complications accompanying the disease course, and those maintained for a prolonged period (within at least one year) after recovery. No statistically significant differences in either disease susceptibility or accompanying and future neurological symptoms (including anosmia) were identified between the sexes. However, there was a trend toward a difference in infection susceptibility between men and women (χ2 = 3.27, p = 0.07), with a higher infection prevalence in men compared to women.
From the correlation analysis, we observed a significant positive correlation between the presence of neurological symptoms during COVID-19 illness and those that appeared/were maintained within a prolonged period after recovery (r = 0.44, p < 0.01), as well as between accompanying anosmia and later neurological consequences (r = 0.45, p < 0.01).
3.2. Association Analysis between SNPs and COVID-19 Outcomes
As a primary step, we checked all analyzed SNPs for their congruence with the Hardy–Weinberg equilibrium (HWE). SNPs that failed the HWE check were excluded from further association analysis (rs61735789, rs117696554, and rs2838046 in the TMPRSS2 gene, rs657152 and rs635634 in the ABO gene).
Our statistical analysis detected an association between rs1894401 in the FURIN gene and COVID-19 infection (β = 2.74, p = 0.006). Individuals carrying the rs1894401 G-allele (GG + GA genotype) had a higher risk of infection than those carrying the AA genotype (OR = 2.21, 95% CI 1.25–3.88) (Figure 1). Another statistically significant result was obtained for rs1405655 residing in the NR1H2 gene, where the rs1405655 C-allele was related to an increased risk of manifesting neurological symptoms during COVID-19 illness in a dose-dependent manner (β = 2.37, p = 0.017, OR = 1.55, 95% CI 1.08–2.23). Furthermore, rs2531743 in the SLC6A20 gene was associated with long-term neurological complaints after recovery from COVID-19 illness (β = 1.99, p = 0.045). To be more precise, carriers of the rs2531743 A-allele had a higher risk of manifesting later neurological symptoms than those bearing the GG genotype (OR = 1.99, 95% CI 1.01–3.91) (Table 1, Figure 1).
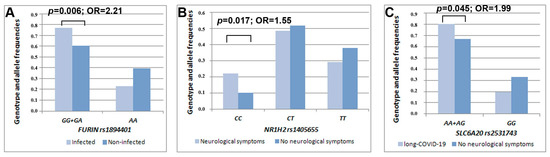
Figure 1.
Distribution of the genotype/allele frequencies of FURIN rs1894401 depending on COVID-19 infection susceptibility (A), NR1H2 rs1405655 depending on the presence/absence of accompanying neurological symptoms (B), and SLC6A20 rs2531743 depending on the presence/absence of neurological consequences over a prolonged period (C) in the examined sample of mild-to-moderate COVID-19 cases.

Table 1.
Characteristics of the study sample.
We also observed a trend for the association between the rs4290734 G-allele residing in the TMPRSS2 gene (β = 1.76, p = 0.078) and the presence of neurological symptoms in our cohort of mild COVID-19 cases (Table 2).

Table 2.
Logistic regression analysis (log-additive models) demonstrating the effects of examined SNPs on COVID-19-related outcomes.
In order to prioritize genetic variants by their significance with respect to COVID-19 susceptibility and neurological symptoms, we examined the combined effect of various SNPs via a series of logistic regressions (under a general linear model procedure). Initially, all SNPs and sexes were included as potential covariates. As a result of backward elimination, the final model predicting enhanced SARS-CoV-2 infection susceptibility included the FURIN rs1894401 AA genotype compared to the GG + GA genotypes (β = 0.85, p = 0.0038, OR = 2.36, 95% CI 1.31–4.24) and SLC6A20 rs2531743 GG-genotype compared to AA+AG genotypes (β = 0.66, p = 0.026, OR = 1.94, 95% CI 1.08–3.49) (Table 3). The best model for predicting the occurrence of neurological symptoms accompanying the COVID-19 disease course consisted of the NR1H2 rs1405655 TT genotype compared to the CC genotype (β = 1.09, p = 0.0059, OR = 2.99, 95% CI 1.39–6.69) and the TMPRSS2 rs4290734 AA genotype compared to the AG genotype (β = 0.71, p = 0.0099, OR = 2.03, 95% CI 1.19–3.50) (Table 3). The best model for predicting the development of neurological complications remaining within one year of disease consisted of the FURIN rs1894401 AA genotype compared to the GG + GA genotypes (β = 0.58, p = 0.085, OR = 1.79, 95% CI 0.93–3.61) and the SLC6A20 rs2531743 GG genotype compared to the AA + AG genotypes (β = 0.75, p = 0.031, OR = 2.12, 95% CI 1.10–4.35) (Table 3). Although FURIN rs1894401 demonstrated a non-significant effect in the last model (p < 0.05), it had to be controlled for in the regression model based on the backward elimination procedure.

Table 3.
Logistic regression models demonstrating the combined effects of the examined SNPs on COVID-19 susceptibility and neurological symptoms.
4. Discussion
Our analysis of the clinical manifestations of COVID-19 in mild and moderate cases in young adults points to the presence of positive correlations between neurological complications during the disease course and future neurological impairments, including severe headache, memory deficit, concentration problems, dysosmia, dysgeusia, insomnia/compromised sleep, anxiety, and depression. Even the presence of anosmia, which is a frequent clinical manifestation of COVID-19, was correlated with the mentioned set of neurological problems later in life in our sample. As previously suggested, such disturbances in the olfactory system could be related to long-term changes in the brain [26]. To date, an increased number of mental problems and neuropsychiatric complications (schizophrenia, affective disorders, Alzheimer’s disease, PTSD, etc.) attributed to COVID-19 illness over a prolonged period have been observed in long COVID-19 cases (for review, see [17]). However, long-term neurological sequelae resulting from SARS-CoV-2 infection have been shown to persist after one year, even in children and adolescents [18]. In this regard, the identification of biological markers (including genetic variants) associated with neurological complaints in younger individuals is of relevance.
In the present preliminary study analyzing 25 SNPs, we confirmed the impact of genetic variants in the FURIN and SLC6A20 genes on reported SARS-CoV-2 infection susceptibility. Moreover, SNPs in the NR1H2, TMPRSS2, and SLC6A20 genes, which were previously linked to COVID-19 hospitalization and cognitive decline, were related to neurological symptoms accompanying the COVID-19 disease course, as well as those remaining over a long-term period after recovery in our examined group of young adults.
We selected rs1894401 in the FURIN gene as a proxy to rs6226 (c.1851G > C), which was examined in a previous meta-analysis of COVID-19 susceptibility [24]. Initially, this SNP was detected via whole-exome sequencing in a Spanish population of patients with familial multiple sclerosis [10]. The observed implication of the FURIN gene in COVID-19 susceptibility is not surprising, since it encodes a proprotein convertase that activates various regulatory proteins, including the SARS-CoV-2 spike (S) protein. Furin, together with TMPRSS2 (transmembrane protease serine 2), is responsible for the cleavage of the SARS-CoV-2 S protein, which represents the SARS-CoV-2 co-receptor, regulates its entry into host cells [27], and is related to an enhanced risk of infection [28]. Several studies have examined the impact of FURIN SNPs on COVID-19-related phenotypes. It was demonstrated that alleles associated with higher protein expression (rs6224 T and rs4702 A) were the markers of COVID-19-implicated death [29]. Interestingly, the rs1894401 G-allele is correlated with the rs6224 risky T-allele (strongly linked, r2 = 0.941) and the rs4702 A-allele (r2 = 0.645). Therefore, we suggest that the rs1894401 G-allele, which was associated with an increased risk of SARS-CoV-2 susceptibility in the present study, is also likely linked to the overexpression of the FURIN gene.
A previous Mendelian randomization study demonstrated the presence of a positive genetic correlation between the dementia-like phenotype and COVID-19 hospitalization [11]. This observation is in line with our findings, which identified an association between rs1405655 in the NR1H2 gene and the presence of neurological symptoms during the COVID-19 disease course. It should be noted that this SNP was selected as significant from a GWAS-based meta-analysis of three COVID-19-related phenotypes: critical illness, disease susceptibility, and hospitalization [5]. In the meta-analysis of data from 49,562 cases and two million controls, the rs1405655 C-allele was confirmed to be associated with disease severity at a genome-wide association level. Our findings also indicated that the rs1405655 C-allele was associated with an increased risk of developing neurological complications during COVID-19 progression, even in young adults with a mild-to-moderate disease course. In turn, the association observed in the present study is not surprising, as statistically significant genetic correlations have been indicated between COVID-19 hospitalization and several mental health issues, such as depression, insomnia, and ADHD [5]. At the molecular level, the NR1H2 gene (also known as LXRB) encodes liver X receptor beta, which belongs to a subfamily of nuclear receptors regulating macrophage function, lipid homeostasis, and inflammation. It was shown that LXRβ has a functional implication in the development of cognitive impairments (such as Alzheimer’s disease) partly due to its key role in Aβ accumulation and cholesterol homeostasis [30]. Furthermore, the NR1H2 gene is located in the same high-LOD score region for AD as the ApoE gene, whose role in the development of AD has been reported in numerous studies and was confirmed in a recent multiomics study [31]. Association studies examining the NR1H2 gene in AD patients seem to be in line with our findings, as the rs1405655 C-allele was shown to segregate in families with this cognitive impairment [30]. As a result, our findings suggest that one of the molecular pathways associated with the manifestation of neurological symptoms during mild or moderate COVID-19 illness is related to impaired cholesterol homeostasis and the downregulation of NR1H2 gene expression.
The TMPRSS2 gene, which is responsible for SARS-CoV-2 penetration into the cell [3], has been repeatedly implicated in COVID-19-related outcomes [10,24,32]. Nevertheless, in the present study we observed a combined effect of TMPRSS2 rs4290734 with NR1H2 rs1405655 on accompanying neurological complications. A previous study identified a proxy SNP (TMPRSS2 rs17854725 or c.879T > C) in a whole-exome scan of COVID-19 patients with familial multiple sclerosis [10], pointing to the possible involvement of the TMPRSS2 gene in cognitive-related phenotypes.
The final finding of the present study was the association of rs2531743—located upstream of the SLC6A20 gene (also known as SIT1)—with COVID-19 susceptibility (as a combined effect with FURIN rs1894401) and neurological complications, which appeared in six months after COVID-19 recovery and was maintained within at least one year. Initially, we selected this genetic variant as part of a set of SNPs in the MR study, which demonstrated a causal relationship between COVID-19 hospitalization and AD development [11]. A recent meta-analysis also demonstrated the role of the SLC6A20 gene (based on rs11385942) in severe COVID-19 outcomes [24]. The SLC6A20 gene encodes a protein that is involved in the transport of amino acids through cell membranes and is responsible for SARS-CoV-2 entry into the cell [32]. To be more precise, rs2531743 is located at 3p21.31, which has been previously linked to COVID-19 outcomes in eleven GWAS. SLC6A20 has been shown to interact with ACE2 in the membrane of small intestinal enterocytes, suggesting a relationship with COVID-19 illness [23]. Recent research has also demonstrated a link between SARS-CoV-2 sensitivity and SLC6A20 gene variants in Vietnamese [23] and Native American [33] populations. In addition, it was suggested that SLC6A20 is a novel regulator of glycine levels [34]. This molecule is known for its impact on the central nervous system [35] and for having a beneficial effect on the proinflammatory cytokine secretion induced by SARS-CoV-2 infection [34].
The main strength of our preliminary study is the simultaneous analysis of a large set of COVID-19-related SNPs in a sample homogenous in age, level of education, and place of residence. However, the present study has several limitations. First, since the sample size was moderate, the observed associations are preliminary and have to be interpreted with caution. Second, since the major hypothesis of our study was to test for the relevance of SNPs with respect to neurological symptoms, we focused mainly on SNPs previously examined in the MR study linking COVID-19 illness to cognitive impairments. Third, we could not rule out whether the same direction of the SNPs’ effect would be present in individuals with a severe disease course and/or other age groups. Finally, due to the preliminary nature of our research, we did not perform any correction for multiple testing. Therefore, we cannot exclude the possibility of false-positive associations. Future directions in this regard include a replication study employing a larger sample size of the same population (young adults and children). In turn, it is of particular interest in future research to investigate the presence of neurological complications in the examined cohort under a longitudinal paradigm.
5. Conclusions
In summary, the present study, based on previous GWAS meta-analyses of COVID-19 susceptibility, hospitalization, and disease severity, partially replicated the implication of a genetic variant in the FURIN gene in SARS-CoV-2 susceptibility. Moreover, SNPs in the NR1H2 and SLC6A20 genes, which have been previously linked to COVID-19 hospitalization and AD, were related to neurological sequelae accompanying even a mild-to-moderate disease course, and those that appeared within six months and were maintained over a long-term period after recovery, in young adults.
Our findings indicate that alleles associated with the overexpression of the FURIN gene were responsible for higher susceptibility to SARS-CoV-2 infection in a general cohort of young adults. In addition, alleles linked to the downregulation of the NR1H2 gene and impaired cholesterol homeostasis were found to contribute to the manifestation of neurological symptoms during mild-to-moderate COVID-19 illness. Finally, the indirect implication of the SLC6A20 and FYCO1 genes in the occurrence or maintenance of neuropsychiatric symptoms as long-term post-COVID-19 complications was bolstered by the reported regulation of the glycine-linked functioning of the central nervous system and proinflammatory cytokine secretion by SLC6A20.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jpm13010123/s1, Table S1. A set of SNPs selected for the association analysis of COVID-19-related phenotypes in the present study. References [5,11,24] are cited in the supplementary materials.
Author Contributions
Conceptualization, A.K. (Anastasiya Kazantseva) and E.K.; methodology, A.K. (Anastasiya Kazantseva), R.E. and Y.D.; validation, R.E. and Z.T.; formal analysis, A.K. (Anastasiya Kazantseva); investigation, A.K. (Anastasiya Kazantseva), R.E. and Z.T.; resources, A.K. (Anastasiya Kazantseva), R.E., Z.T., R.M., S.M., A.K. (Alexandra Karunas) and Y.D.; writing—original draft preparation, A.K. (Anastasiya Kazantseva); writing—review and editing, A.K. (Anastasiya Kazantseva); visualization, A.K. (Anastasiya Kazantseva); supervision, E.K.; project administration, A.K. (Alexander Kanapin) and E.K.; funding acquisition, A.K. (Alexander Kanapin), S.M. and E.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Ministry of Science and Higher Education of the Republic of Bashkortostan (agreement no. 1, 2 December 2022) in respect of the collection of biological materials and clinical data; by the Russian Science Foundation, grant number 17-78-30028, in respect of statistical analysis; and by the Ministry of Science and Higher Education of Russian Federation, grant number 075-15-2021-595, in respect of genotyping. In the study, DNA samples from the “Collection of Biological Materials of Human Beings” of the IBG UFRC RAS were used, supported by the Program of Bioresource Collections of the FASO of Russia, grant number 007-030164/2.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Biological Ethics Committee of the Institute of Biochemistry and Genetics—Subdivision of the Ufa Federal Research Centre of Russian Academy of Sciences (Ufa, Russia) (protocol code 19 and date of approval 25 November 2021).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The datasets used and/or analyzed in the current study are available from the corresponding author on reasonable request.
Acknowledgments
We would like to thank the participants of the study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Williamson, E.J.; Walker, A.J.; Bhaskaran, K.; Bacon, S.; Bates, C.; Morton, C.E.; Curtis, H.J.; Mehrkar, A.; Evans, D.; Inglesby, P.; et al. Factors Associated with COVID-19-Related Death Using OpenSAFELY. Nature 2020, 584, 430–436. [Google Scholar] [CrossRef]
- Brola, W.; Wilski, M. Neurological Consequences of COVID-19. Pharmacol. Rep. 2022, 74, 1208–1222. [Google Scholar] [CrossRef] [PubMed]
- Mustafin, R.; Kazantseva, A.; Kovas, Y.; Khusnutdinova, E. Role of Retroelements in the Development of COVID-19 Neurological Consequences. Russ. Open Med. J. 2022, 11, 313. [Google Scholar] [CrossRef]
- Zguro, K.; Fallerini, C.; Fava, F.; Furini, S.; Renieri, A. Host Genetic Basis of COVID-19: From Methodologies to Genes. Eur. J. Hum. Genet. 2022, 30, 899–907. [Google Scholar] [CrossRef]
- Mapping the Human Genetic Architecture of COVID-19. Nature 2021, 600, 472–477. [CrossRef]
- Ellinghaus, D.; Degenhardt, F.; Bujanda, L.; Buti, M.; Albillos, A.; Invernizzi, P.; Fernández, J.; Prati, D.; Baselli, G.; Asselta, R.; et al. Genomewide Association Study of Severe COVID-19 with Respiratory Failure. N. Engl. J. Med. 2020, 383, 1522–1534. [Google Scholar] [CrossRef] [PubMed]
- Roberts, G.H.L.; Park, D.S.; Coignet, M.V.; McCurdy, S.R.; Knight, S.C.; Partha, R.; Rhead, B.; Zhang, M.; Berkowitz, N.; Haug Baltzell, A.K.; et al. AncestryDNA COVID-19 Host Genetic Study Identifies Three Novel Loci. medRxiv 2020. [Google Scholar] [CrossRef]
- Pairo-Castineira, E.; Clohisey, S.; Klaric, L.; Bretherick, A.D.; Rawlik, K.; Pasko, D.; Walker, S.; Parkinson, N.; Fourman, M.H.; Russell, C.D.; et al. Genetic Mechanisms of Critical Illness in COVID-19. Nature 2021, 591, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Shcherbak, S.G.; Changalidi, A.I.; Barbitoff, Y.A.; Anisenkova, A.Y.; Mosenko, S.V.; Asaulenko, Z.P.; Tsay, V.V.; Polev, D.E.; Kalinin, R.S.; Eismont, Y.A.; et al. Identification of Genetic Risk Factors of Severe COVID-19 Using Extensive Phenotypic Data: A Proof-of-Concept Study in a Cohort of Russian Patients. Genes 2022, 13, 534. [Google Scholar] [CrossRef]
- Torre-Fuentes, L.; Matías-Guiu, J.; Hernández-Lorenzo, L.; Montero-Escribano, P.; Pytel, V.; Porta-Etessam, J.; Gómez-Pinedo, U.; Matías-Guiu, J.A. ACE2, TMPRSS2, and Furin Variants and SARS-CoV-2 Infection in Madrid, Spain. J. Med. Virol. 2021, 93, 863–869. [Google Scholar] [CrossRef]
- Baranova, A.; Cao, H.; Zhang, F. Causal Effect of COVID-19 on Alzheimer’s Disease: A Mendelian Randomization Study. J. Med. Virol. 2022, 95, e28107. [Google Scholar] [CrossRef] [PubMed]
- Kazantseva, A.V.; Enikeeva, R.F.; Davydova, Y.D.; Mustafin, R.N.; Takhirova, Z.R.; Malykh, S.B.; Lobaskova, M.M.; Tikhomirova, T.N.; Khusnutdinova, E.K. The Role of the KIBRA and APOE Genes in Developing Spatial Abilities in Humans. Vavilovskii Zhurnal Genet. Selektsii 2021, 25, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, M.R.; Poland, G.A.; Graeber, A.C.W. Does Apolipoprotein E Genotype Predict COVID-19 Severity? QJM 2020, 113, 529–530. [Google Scholar] [CrossRef] [PubMed]
- Del Ser, T.; Fernández-Blázquez, M.A.; Valentí, M.; Zea-Sevilla, M.A.; Frades, B.; Alfayate, E.; Saiz, L.; Calero, O.; García-López, F.J.; Rábano, A.; et al. Residence, Clinical Features, and Genetic Risk Factors Associated with Symptoms of COVID-19 in a Cohort of Older People in Madrid. Gerontology 2021, 67, 281–289. [Google Scholar] [CrossRef]
- Ji, H.-L.; Zhao, R.; Matalon, S.; Matthay, M.A. Elevated Plasmin(Ogen) as a Common Risk Factor for COVID-19 Susceptibility. Physiol. Rev. 2020, 100, 1065–1075. [Google Scholar] [CrossRef]
- Yuki, K.; Fujiogi, M.; Koutsogiannaki, S. COVID-19 Pathophysiology: A Review. Clin. Immunol. 2020, 215, 108427. [Google Scholar] [CrossRef]
- Zawilska, J.B.; Kuczyńska, K. Psychiatric and Neurological Complications of Long COVID. J. Psychiatr. Res. 2022, 156, 349–360. [Google Scholar] [CrossRef]
- Savino, R.; Polito, A.N.; Arcidiacono, G.; Poliseno, M.; Lo Caputo, S. Neuropsychiatric Disorders in Pediatric Long COVID-19: A Case Series. Brain Sci. 2022, 12, 514. [Google Scholar] [CrossRef]
- Glessner, J.T.; Chang, X.; Mentch, F.; Qu, H.; Abrams, D.J.; Thomas, A.; Sleiman, P.M.A.; Hakonarson, H. COVID-19 in Pediatrics: Genetic Susceptibility. Front. Genet. 2022, 13, 928466. [Google Scholar] [CrossRef]
- Schulert, G.S.; Blum, S.A.; Cron, R.Q. Host Genetics of Pediatric SARS-CoV-2 COVID-19 and Multisystem Inflammatory Syndrome in Children. Curr. Opin. Pediatr. 2021, 33, 549–555. [Google Scholar] [CrossRef]
- Singh, B.; Lant, S.; Cividini, S.; Cattrall, J.W.; Goodwin, L.C.; Benjamin, L.; Michael, B.D.; Khawaja, A.; Matos, A.D.M.B.; Alkeridy, W.; et al. Prognostic Indicators and Outcomes of Hospitalised COVID-19 Patients with Neurological Disease: An Individual Patient Data Meta-Analysis. PLoS ONE 2022, 17, e0263595. [Google Scholar] [CrossRef]
- Mattioli, F.; Stampatori, C.; Righetti, F.; Sala, E.; Tomasi, C.; De Palma, G. Neurological and Cognitive Sequelae of COVID-19: A Four Month Follow-Up. J. Neurol. 2021, 268, 4422–4428. [Google Scholar] [CrossRef]
- Nhung, V.P.; Ton, N.D.; Ngoc, T.T.B.; Thuong, M.T.H.; Hai, N.T.T.; Oanh, K.T.P.; Hien, L.T.T.; Thach, P.N.; Hai, N.V.; Ha, N.H. Host Genetic Risk Factors Associated with COVID-19 Susceptibility and Severity in Vietnamese. Genes 2022, 13, 1884. [Google Scholar] [CrossRef] [PubMed]
- de Rojas, I.; Hernández, I.; Montrreal, L.; Quintela, I.; Calero, M.; Royo, J.L.; Huerto Vilas, R.; González-Pérez, A.; Franco-Macías, E.; Macías, J.; et al. Genomic Characterization of Host Factors Related to SARS-CoV-2 Infection in People with Dementia and Control Populations: The GR@ACE/DEGESCO Study. J. Pers. Med. 2021, 11, 1318. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: www.R-project.org (accessed on 20 October 2022).
- Boscolo-Rizzo, P.; Menegaldo, A.; Fabbris, C.; Spinato, G.; Borsetto, D.; Vaira, L.A.; Calvanese, L.; Pettorelli, A.; Sonego, M.; Frezza, D.; et al. Six-Month Psychophysical Evaluation of Olfactory Dysfunction in Patients with COVID-19. Chem. Senses 2021, 46, bjab006. [Google Scholar] [CrossRef] [PubMed]
- Achom, A.; Das, R.; Pakray, P. An Improved Fuzzy Based GWO Algorithm for Predicting the Potential Host Receptor of COVID-19 Infection. Comput. Biol. Med. 2022, 151, 106050. [Google Scholar] [CrossRef]
- Drak Alsibai, K. Expression of Angiotensin-Converting Enzyme 2 and Proteases in COVID-19 Patients: A Potential Role of Cellular FURIN in the Pathogenesis of SARS-CoV-2. Med. Hypotheses 2020, 143, 109893. [Google Scholar] [CrossRef] [PubMed]
- Coto, E.; Albaiceta, G.M.; Amado-Rodríguez, L.; García-Clemente, M.; Cuesta-Llavona, E.; Vázquez-Coto, D.; Alonso, B.; Iglesias, S.; Melón, S.; Alvarez-Argüelles, M.E.; et al. FURIN Gene Variants (Rs6224/Rs4702) as Potential Markers of Death and Cardiovascular Traits in Severe COVID-19. J. Med. Virol. 2022, 94, 3589–3595. [Google Scholar] [CrossRef]
- Adighibe, O.; Arepalli, S.; Duckworth, J.; Hardy, J.; Wavrant-De Vrièze, F. Genetic Variability at the LXR Gene (NR1H2) May Contribute to the Risk of Alzheimer’s Disease. Neurobiol. Aging 2006, 27, 1431–1434. [Google Scholar] [CrossRef]
- Heath, L.; Earls, J.C.; Magis, A.T.; Kornilov, S.A.; Lovejoy, J.C.; Funk, C.C.; Rappaport, N.; Logsdon, B.A.; Mangravite, L.M.; Kunkle, B.W.; et al. Manifestations of Alzheimer’s Disease Genetic Risk in the Blood Are Evident in a Multiomic Analysis in Healthy Adults Aged 18 to 90. Sci. Rep. 2022, 12, 6117. [Google Scholar] [CrossRef]
- Ferreira, L.C.; Gomes, C.E.M.; Rodrigues-Neto, J.F.; Jeronimo, S.M.B. Genome-Wide Association Studies of COVID-19: Connecting the Dots. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2022, 106, 105379. [Google Scholar] [CrossRef] [PubMed]
- Pastana, L.F.; Silva, T.A.; Gellen, L.P.A.; Vieira, G.M.; de Assunção, L.A.; Leitão, L.P.C.; da Silva, N.M.; Coelho, R.D.C.C.; de Alcântara, A.L.; Vinagre, L.W.M.S.; et al. The Genomic Profile Associated with Risk of Severe Forms of COVID-19 in Amazonian Native American Populations. J. Pers. Med. 2022, 12, 554. [Google Scholar] [CrossRef] [PubMed]
- Semiz, S. SIT1 Transporter as a Potential Novel Target in Treatment of COVID-19. Biomol. Concepts 2021, 12, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Botto, R.; Callai, N.; Cermelli, A.; Causarano, L.; Rainero, I. Anxiety and Depression in Alzheimer’s Disease: A Systematic Review of Pathogenetic Mechanisms and Relation to Cognitive Decline. Neurol. Sci. 2022, 43, 4107–4124. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).