Abstract
The complexity of lung adenocarcinoma (LUAD), the development of which involves many interacting biological processes, makes it difficult to find therapeutic biomarkers for treatment. FK506-binding proteins (FKBPs) are composed of 12 members classified as conservative intracellular immunophilin family proteins, which are often connected to cyclophilin structures by tetratricopeptide repeat domains and have peptidyl prolyl isomerase activity that catalyzes proline from residues and turns the trans form into the cis form. Since FKBPs belong to chaperone molecules and promote protein folding, previous studies demonstrated that FKBP family members significantly contribute to the degradation of damaged, misfolded, abnormal, and foreign proteins. However, transcript expressions of this gene family in LUAD still need to be more fully investigated. In this research, we adopted high-throughput bioinformatics technology to analyze FKBP family genes in LUAD to provide credible information to clinicians and promote the development of novel cancer target drugs in the future. The current data revealed that the messenger (m)RNA levels of FKBP2, FKBP3, FKBP4, FKBP10, FKBP11, and FKBP14 were overexpressed in LUAD, and FKBP10 had connections to poor prognoses among LUAD patients in an overall survival (OS) analysis. Based on the above results, we selected FKBP10 to further conduct a comprehensive analysis of the downstream pathway and network. Through a DAVID analysis, we found that FKBP10 was involved in mitochondrial electron transport, NADH to ubiquinone transport, mitochondrial respiratory chain complex I assembly, etc. The MetaCore pathway analysis also indicated that FKBP10 was involved in "Ubiquinone metabolism", "Translation_(L)-selenoaminoacid incorporation in proteins during translation", and "Transcription_Negative regulation of HIF1A function". Collectively, this study revealed that FKBP family members are both significant prognostic biomarkers for lung cancer progression and promising clinical therapeutic targets, thus providing new targets for treating LUAD patients.
1. Introduction
In its advanced stages, lung cancer (LC) is the one of the most lethal types of cancer and is responsible for significant mortality worldwide. The 5-year overall survival (OS) of patients with LC ranges from 5% to 17.7% [1]. Depending on the histology, LC includes small cell lung carcinoma (SCLC) and non-SCLC (NSCLC) subtypes. NSCLC is further divided into three secondary categories: lung adenocarcinoma (LUAD), squamous cell lung carcinoma (SCLC), and large cell lung carcinoma (LCLC). However, the proportion of LUAD is up to 40%, thereby representing the main form of LC. The leading cause of LUAD, which originates from the small airway epithelium and the secretion of mucus and other substances from type II alveolar cells, is smoking. Additionally, LUAD easily occurs in the periphery of the lungs. Due to its early spread, aggression, and ability to easily metastasize, the survival rate of LUAD is less than 5 years [2,3,4]. Although several therapies such as chemotherapy and surgical resection are used to decrease mortality, there are still dilemmas including late detection for approximately 75% of LUAD patients, as well as high recurrence rates and poor prognoses [5,6]. Therefore, it is imperative to explore potential biomarkers and develop novel targeted cancer therapies for future application [7,8,9,10,11].
FK506-binding proteins (FKBPs) include 12 members and are classified as conservative intracellular immunophilin family proteins. FKBPs are often connected to cyclophilin structures via tetratricopeptide repeat domains and have peptidyl prolyl isomerase activity, which catalyzes proline from residues to turn the trans form into the cis form [12,13,14,15]. FKBPs are chaperone molecules and promote protein folding [16]. FKBP family members, moreover, are tied to immunosuppressants such as FK506, rapamycin, and cyclosporin A to induce multiple physiological responses [12]. Thus, these proteins are related to numerous human malignancies. As an example, FKBP2 is overexpressed in the hypoxic environment of glioblastoma multiforme and triggers tumor metastasis [17]. FKBP3 was reported to be closely associated with colorectal cancer [18] and NSCLC [19]. FKBP4 expression was also found to be upregulated in breast cancer [20], in NSCLC by activating the Akt-mammalian target of rapamycin (mTOR) signaling pathway [21], and in colorectal cancer in males [22]. Notably, FKBP5 may become a prognostic indicator in pancreatic cancer [23]. The methylation of FKBP6 was found to serve as a novel biomarker in cervical cancer [24]. Previous studies reported that transcription levels of FKBP7 were increased in prostate cancer [25] and melanomas [26]. FKBP9 was proven to be related to poor prognoses in prostate cancer [27] and gliomas [28]. Transcriptional levels of FKBP10 and FKBP11 are upregulated in renal cell carcinoma tissues compared to normal tissues [13], and FKBP14 is involved in various types of human tumors such as gastric cancer [29,30], human cervical cancers [31], ovarian cancer [32], and osteosarcomas [33]. In addition, FKBP15 was verified to be upregulated in breast cancer patients [34]. Although the FKBP gene family has been studied in a variety of human malignancies, a comprehensive analysis including individual gene expression levels, genetic variations, immune infiltration in the tumor microenvironment (TME), and biological mechanisms of FKBP family members in LUAD has not been elaborated [35,36,37,38,39,40].
Recent epidemiologic studies indicated that lung cancer remains one of the most fatal malignancies, despite remarkable improvements that have been made in medical and surgical approaches. Indeed, shortages of highly sensitive screening tests, delays in early screening, and high probabilities of drug resistance and chemoresistance have resulted in increased risks of metastasis and relapse, as well as a meager survival rate for NSCLC patients. Therefore, identifying specific key molecular pathways and highly sensitive biomarkers for NSCLC is urgently needed to formulate effective treatments through personalized medicine [41].
2. Materials and Methods
2.1. Oncomine Gene Analysis for Expression Levels of FKBP Family Members in LUAD
The Oncomine platform (http://www.oncomine.org/, accessed on 22 November 2021) is an online cancer microarray bioinformatics database established to display transcriptional levels of target cancer and normal specimens from 715 datasets [42,43,44]. In this research, we analyzed individual mRNA expression levels of FKBP family members in various types of cancer via Oncomine with the setting of p < 0.01, fold change of 1.5, and gene rank in the top 10%.
2.2. Gene Expression Profiling Interactive Analysis (GEPIA) 2 Analysis for Clinicpathological States of FKBP Family Members
GEPIA 2 (http://gepia2.cancer-pku.cn/#index/, accessed on 22 November 2021) is a beneficial bioinformatics database that offers precise analyses of transcriptional levels of mRNA expressions for 8587 normal tissues and 9736 cancer samples from The Genotype-Tissue Expression (GTEx) project (https://www.gtexportal.org/home/, accessed on 22 November 2021) and The Cancer Genome Atlas (TCGA) (https://tcga-data.nci.nih.gov/tcga/, accessed on 22 November 2021) [43,45,46]. GEPIA 2 supplies major functions including gene expression analyses, gene correlation analyses, survival analyses, similar gene predictions, and dimensionality reduction analyses. In the present study, GEPIA 2 was used to explore gene expressions in different stages and for normal/cancer tissue comparisons.
2.3. Kaplan-Meier (KM) Plotter Survival Assessment of FKBP Gene Family Members
The KM plotter (http://kmplot.com/analysis/, accessed on 30 November 2021) is a visual bioinformatics database containing up to 54,000 genes in 21 cancer types including lung, ovarian, breast, and gastric cancers [47]. This public database can be used to conduct meta-analyses with TCGA, Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/, accessed on 30 November 2021), and European Genome-phenome Archive (EGA) (https://ega-archive.org/, accessed on 30 November 2021) [48]. We adopted a pan-cancer platform to analyze the prognostic merits of transcriptional levels of individual FKBP family members in LUAD patients (n = 513) for OS and relapse-free survival (RFS) with the KM plotter choosing median values, a p log rank of <0.05, and hazard ratios (HRs) of >1.
2.4. cBioPortal Analysis of Genetic Alterations of FKBP Family Members in LUAD
cBioPortal (http://cbioportal.org/, accessed on 30 November 2021) is an online resource that integrates several cancer-related databases to analyze genetic alterations, DNA methylation, copy number changes, etc. [49,50,51] from more than 5000 cancer specimens in 20 cancer studies [44]. In this study, we explored genetic alterations of the FKBP gene family in LUAD with 503 complete samples from TCGA in cBioPortal.
2.5. Gene MANIA Was Used to Build Gene-Gene Interactions (GGIs) and Explore Their Functions
Gene MANIA (http://www.genemania.org/, accessed on 4 December 2021) is a versatile tool for predicting gene functions, analyzing gene lists, and recognizing the most interrelated genes such as Homo sapiens, based on more than 800 connections [52,53]. We examined GGI networks and functions of FKBP family numbers by Gene MANIA.
2.6. STRING Analysis of the FKBP Gene Family and Other Associations of Expressed Proteins
The purpose of the STRING informatics tool (https://string-db.org/, accessed on 4 December 2021) is to gather and integrate accessible sources of protein-protein interactions (PPIs) and conduct computational forecasts. The aim of this tool is to achieve a comprehensive analysis [54]. The newest version of STRING (11.5) for organisms, renewed on 12 August 2021, is nearly triple the size of the older version (11.0b) and covers 1,409,467,592,464 proteins and 20,052,394,042 interactions.
2.7. Database for Annotation, Visualization and Integrated Discovery (DAVID) and MetaCore Analysis of Coexpressions of FKBP Family Members
DAVID 6.8 (https://david.ncifcrf.gov/, accessed on 4 December 2021) provides a platform to facilitate analysis of gene lists of interest [55], and data visualization through online platform (http://www.bioinformatics.com.cn/srplot, accessed on 4 December 2021). These platform consists of the Kyoto Encyclopedia of Genes and Genomes (KEGG) and gene ontology (GO) and is composed of molecular functions (MFs), biological processes (BPs), and cellular components (CCs) [56]. The goal of KEGG is to assign biological functions to genes and genomes [57], while GO offers information about gene products, processes, and functions [58]. Together with the MetaCore platform, we mapped the intersection between these two sets of data in terms of related pathways and involved networks. A p value of <0.05 was considered significant, as previously described.
2.8. Tumor Immune Estimation Resource (TIMER) 1.0 Comprehensive Investigation of Components of Immune Cell Infiltration of FKBP Gene Family Members in LUAD
TIMER 1.0 (http://timer.comp-genomics.org/, accessed on 4 December 2021) is a convenient server for the analysis and visualization of associations of target genes and related immune cells between tumor and normal samples from 10,897 samples in 32 cancer types [59,60]. In this research, we analyzed correlations of different FKBP family members in LUAD with the enrichment of immune cell infiltrates, including B cells, cluster of differentiation 8-positive (CD8+) T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells (DCs).
2.9. Statistical Analysis
We utilized TCGA Pan-Cancer Atlas, a dataset from cBioPortal, to obtain patient data and query the effects of the expressions of different FKBP family members on OS. For the survival analysis, a KM plotter was applied, with all default settings, and RFS was preferred, with automatic cutoff values and J best probe set. All possible cutoff values between the lower and upper quartiles were determined, and the best presenting threshold was subsequently used as the cutoff [51]. A log-rank p value of <0.05 was considered statistically significant.
3. Results
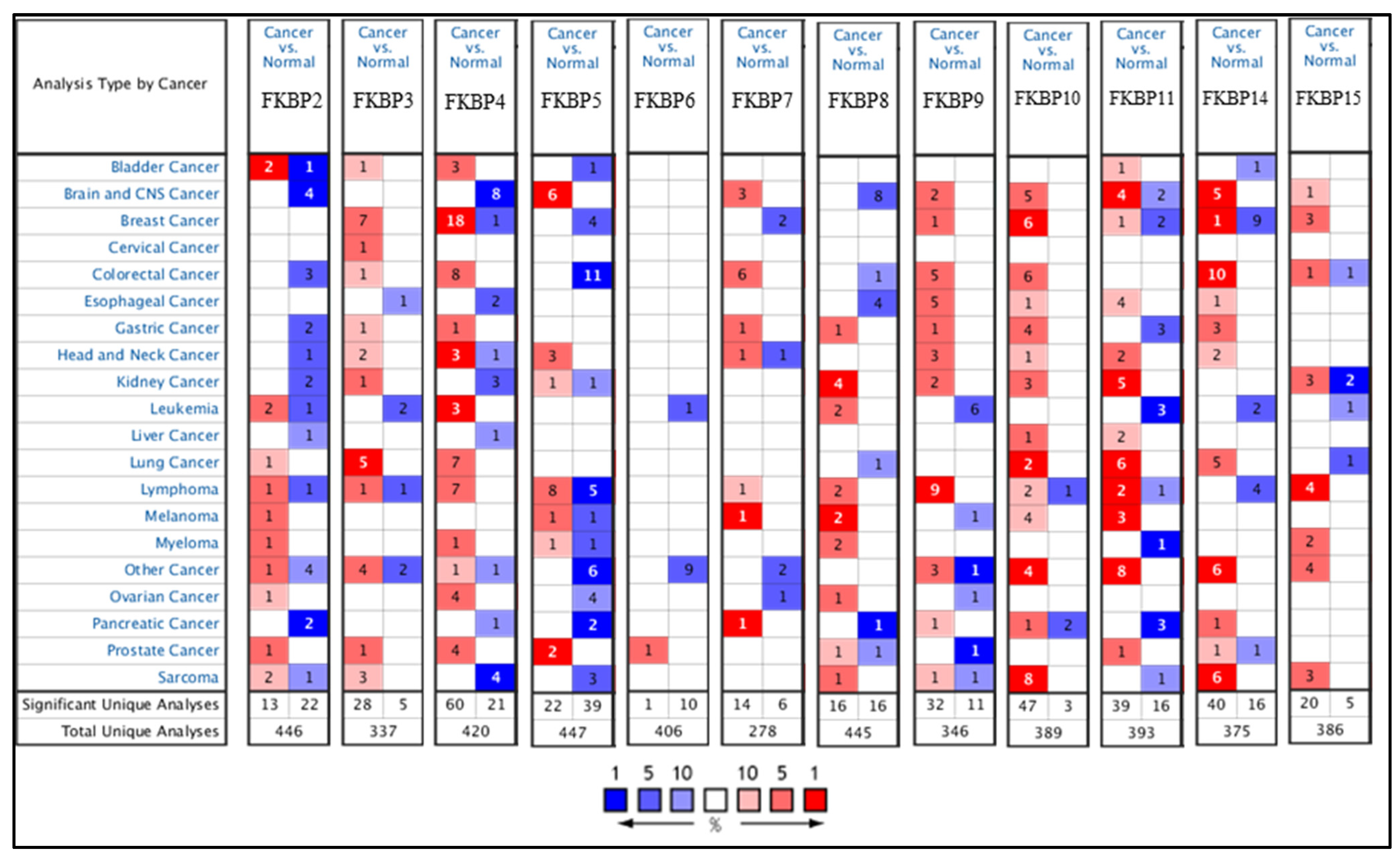
Transcriptional levels of FKBP family members were linked to various cancers due to abnormal expressions, but no publications have mentioned the connection between FKBP family genes and LUAD. In the present study, we compared levels of FKBP transcripts in different tumor tissues and normal samples by utilizing an Oncomine analysis (Figure 1). Findings indicated that mRNA levels of FKBP2, FKBP3, FKBP4, FKBP10, FKBP11, and FKBP14 were overexpressed in LUAD tissues compared to healthy tissues. In the datasets from Stearmen et al. [61], FKBP2 expression was elevated in LUAD patients, with a fold change of 1.520 (p = 2.94 × 10−5) (Supplementary Table S1). In three datasets, transcriptional levels of FKBP3 in tumor tissues were higher than those in normal samples. In the LUAD datasets of Su et al. [62], FKBP3 mRNA was significantly overexpressed in LUAD tissues, with a fold change of 1.761 (p = 8.06 × 10−9), and in addition, datasets from Landi et al. [63] revealed that the increase in transcriptional levels of FKBP3 in LUAD was invalid, with a fold change of 1.724 (p = 1.00 × 10−16). Additionally, Hou et al.’s datasets [64] showed upregulated fold changes of 1.600 (p = 9.13 × 10−12), 1.783 (p = 3.84 × 10−12), and 1.810 (p = 6.73 × 10−7) of FKBP3 in LUAD, SCLC, and LCLC, respectively. In LUAD datasets of Stearman et al. [61] and Beer et al. [65], compared to normal tissues, FKBP4 mRNA was obviously higher with fold changes of 1.715 (p = 2.02 × 10−7) and 1.523 (p = 6.99 × 10−6), respectively. The SCLC datasets of Wachi et al. [66] stated that FKBP4 was overexpressed by a fold change of 2.085 (p = 3.19 × 10−4). In Hou et al.’s datasets [64], FKBP4 was also discovered to be upregulated in LCLC, SCLC, and LUAD with respective fold changes of 3.607 (p = 3.84 × 10−8), 2.765 (p = 9.11 × 10−15), and 2.082 (p = 3.46 × 10−9). Transcription levels of FKBP4 were found to be increased in SCLC patients in Bhattacharjee et al.’s datasets [67], with a fold change of 3.530 (p = 0.002). In Garber et al.’s datasets [68], FKBP10 expression was also reported to be higher in LCLC and SCLC, with respective fold changes of 4.332 (p = 2.17 × 10−4) and 2.417 (p = 0.003). FKBP11 was overexpressed in LUAD, with fold changes of 2.826 (p = 2.20 × 10−6), 2.031 (p = 2.81 × 10−7), 2.114 (p = 1.02 ×10 −13), 2.340 (p = 1.82 × 10−9), 2.002 (p = 2.05 × 10−10), and 1.740 (p = 2.53 × 10−9) in the datasets of Garber et al. [68], Su et al. [62], Landi et al. [63], Hou et al. [64], Okayama et al. [69], and Selatmat et al. [70], respectively. Transcriptional levels of FKBP14 were upregulated in LUAD, with fold changes of 1.948 (p = 1.37 × 10−6), 1.781 (p = 0.002), 1.722 (p = 1.33 × 10−8), and 3.691 (p = 1.27 × 10−10) in datasets of Su et al. [62], Garber et al. [68], Hou et al. [64], and Selamat et al. [70], respectively. In SCLC, FKBP14 expression was elevated as was proven by Hou et al.’s [64] datasets. Furthermore, expressions of FKBP8 and FKBP15 were lower in LUAD in Bhattacharjee et al.’s datasets [67], with a fold change of −4.273 (p = 6.80 × 10−5), and a lower LCLC in Hou et al.’s datasets [64], with a fold change of −1.839 (p = 4.87 × 10−13). However, mRNA expression levels of FKBP5, FKBP6, FKBP7, and FKBP9 exhibited no significant differences between lung cancer tissues and healthy samples (Supplementary Table S1). In addition, we also analyzed the expressions of FKBP family members in cancer and normal tissues and pathological features in LUAD using GEPIA 2 (Supplementary Figure S1).
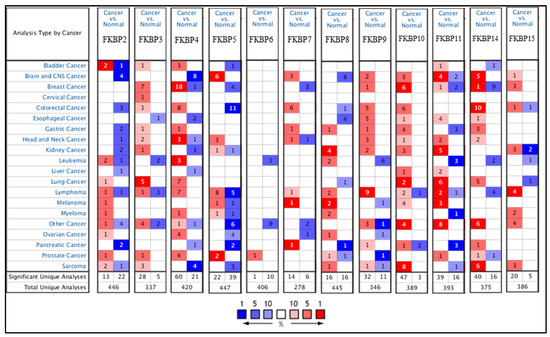
Figure 1.
The mRNA expression levels of FKBP family members in various cancer types analyzed using the Oncomine database. Transcriptional levels of FKBP2, FKBP3, FKBP4, FKBP10, FKBP11, and FKBP14 were higher in lung cancer samples than in normal samples. Red and blue cells, respectively, represent statistically upregulated and downregulated mRNA expression levels of FKBP family members. The threshold was set to a fold change of p < 0.05; fold change > 1.5 p = 0.05; gene rank top 10%.
3.1. Survival Analysis and Prognostic Values of FKBP Family Members in LUAD
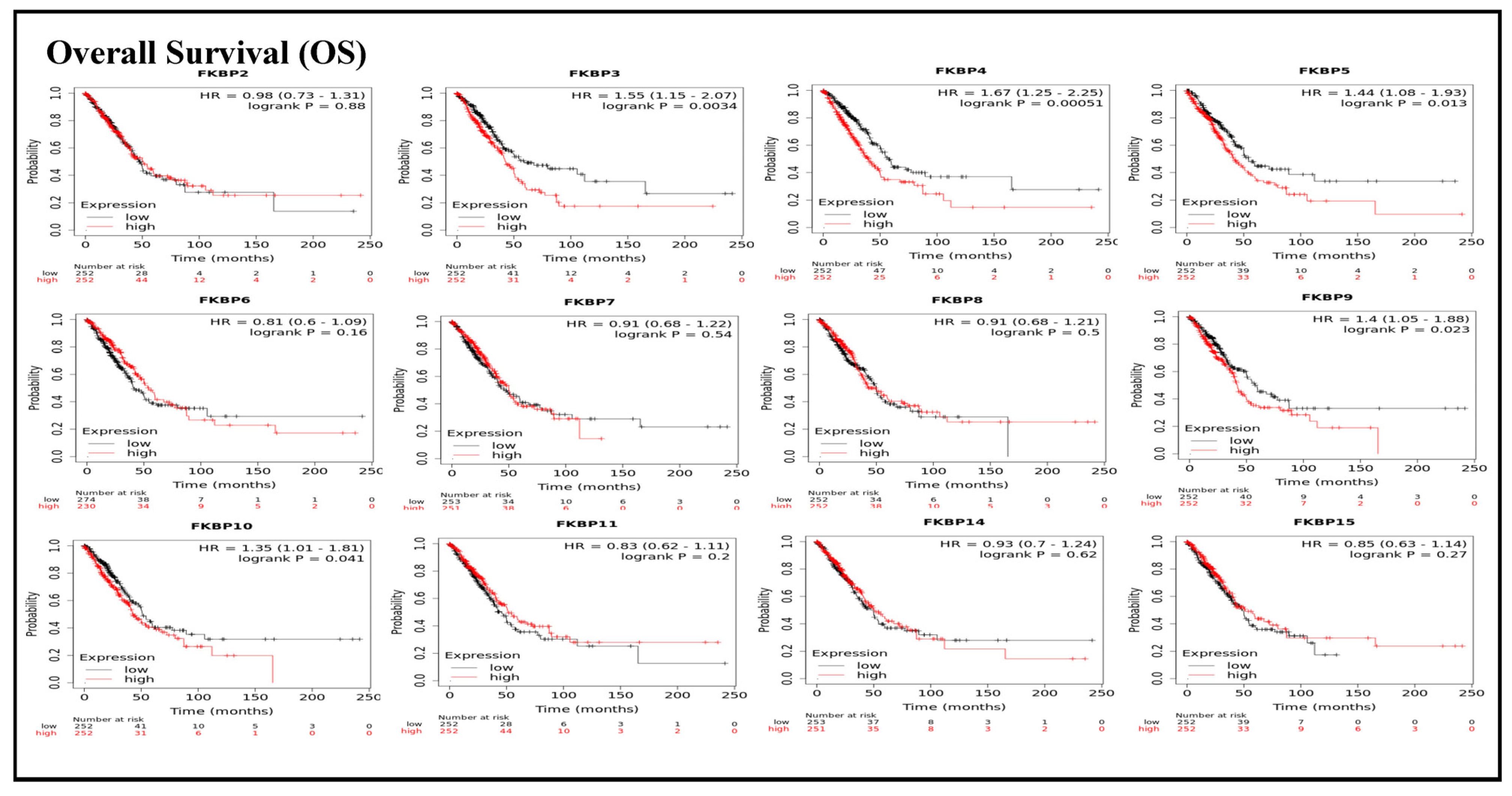
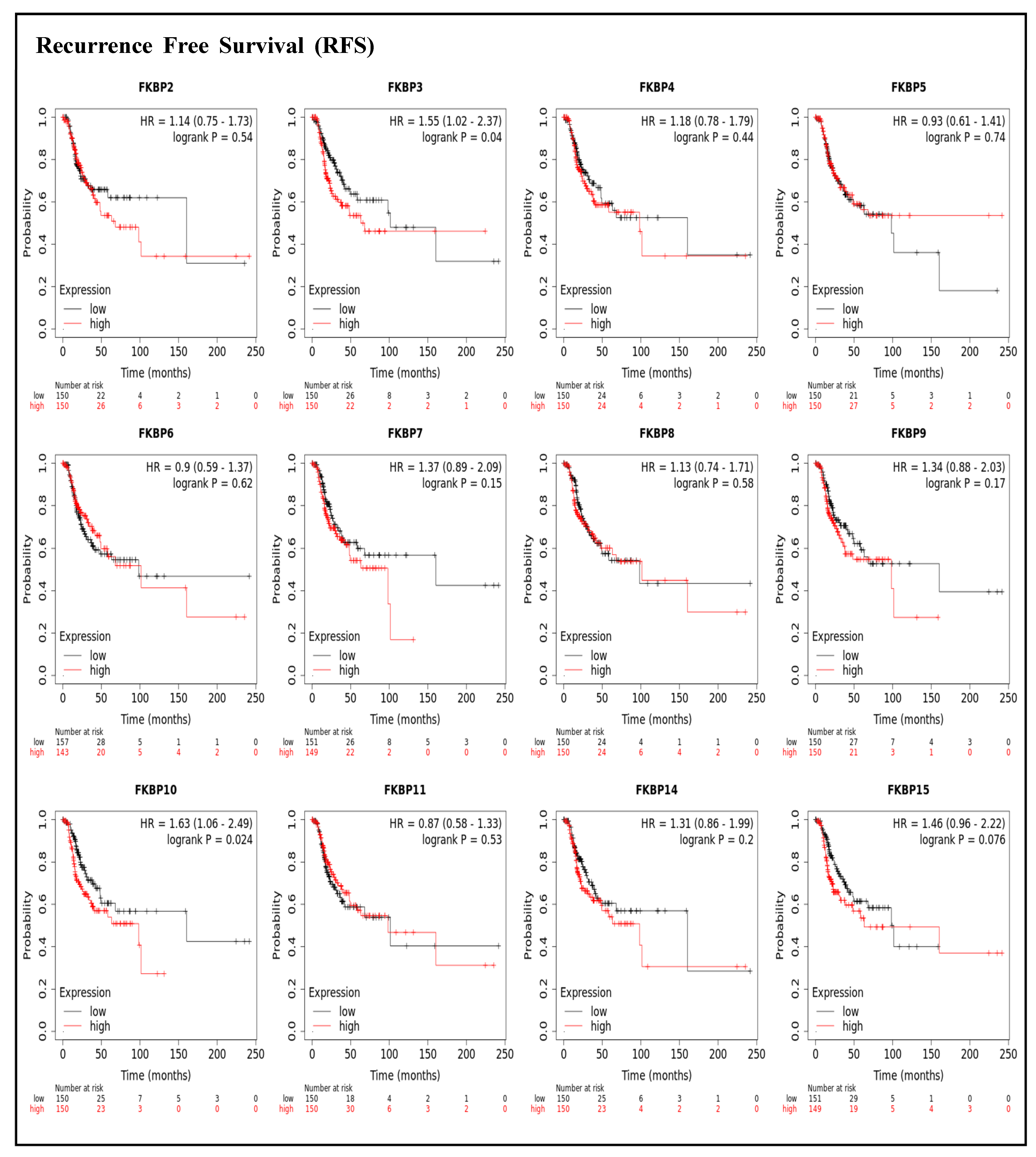
To assess associations between distinct transcriptional levels of FKBP family members and survival rates of LUAD, we used the KM plotter database, and preferentially analyzed OS (Figure 2). Outcomes revealed that five members of the FKBP gene family had correlations with poor prognoses among LUAD patients in the OS analysis including FKBP3 (p = 0.0034; hazard ratio (HR) = 1.55), FKBP4 (p = 0.00051; HR = 1.67), FKBP5 (p = 0.013; HR = 1.44), FKBP9 (p = 0.023; HR = 1.4), and FKBP10 (p = 0.041; HR = 1.35). However, transcriptional levels of FKBP2, FKBP6, FKBP7, FKBP8 FKBP11, FKBP14, and FKBP15 were not linked to the OS of LUAD patients. mRNA expressions of FKBP family memberss in relation to RFS were also analyzed (Figure 3). Low transcriptional levels of FKBP3 (p = 0.04; HR = 1.55) and FKBP10 (p = 0.024; HR = 1.63) in LUAD patients were correlated with a longer RFS, while the remaining FKBP family members were not related to RFS in LUAD samples (Supplementary Table S2).
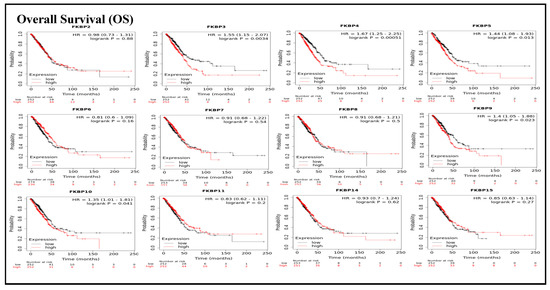
Figure 2.
Prognoses of mRNA expressions of FKBP gene family members in lung adenocarcinoma (LUAD) patients in the overall survival (OS) analysis using the KM plotter database. Low expression levels of FKBP3, FKBP4, FKBP5, FKBP9, and FKBP10 were significant relative to the higher OS values. The setting for filtering LUAD patients was the median level.
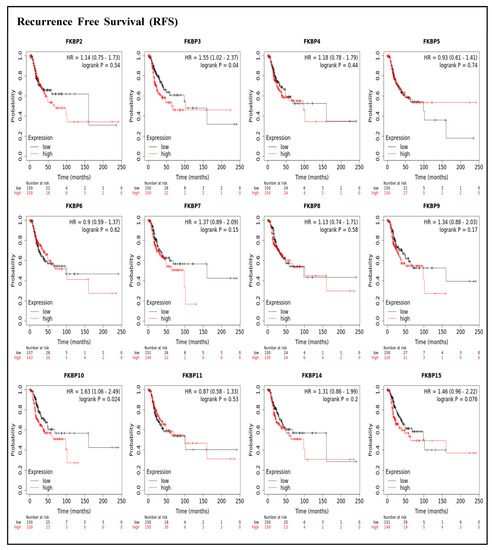
Figure 3.
Prognoses of mRNA expression levels of FKBP gene family members in lung adenocarcinoma (LUAD) patients in terms of a relapse-free survival (RFS) analysis using the KM plotter database. Low expression levels of FKBP3 and FKBP10 were significantly related to greater RFS values. The setting for filtering LUAD patients was the median level.
3.2. Genetic Alteration Analysis of FKBP Family Members in LUAD
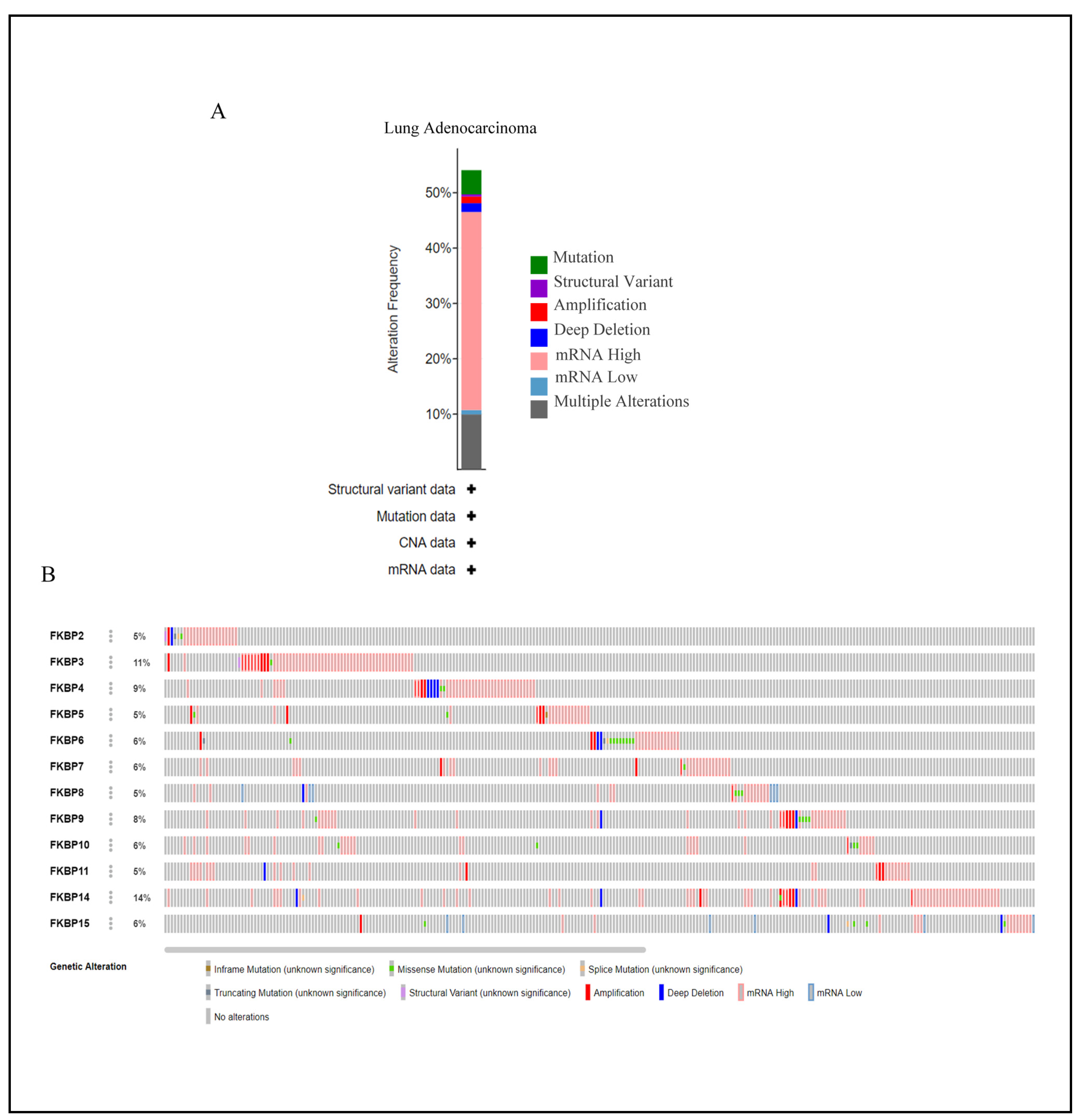
Mutations in FKBP family numbers had large impacts on FK506 binding and its enzymatic functions [71]. Thus, we conducted a visual analysis to gain insights into genetic alterations of the FKBP gene family alongside the cBioPortal bioinformatics tool from TCGA. Results revealed that FKBP family members were altered in 272 instances among 503 LUAD patients, and these alterations included mutations, structural variants, amplifications, deep deletions, mRNA high, mRNA low, and multiple alterations based on the dataset (Figure 4A). The rate of genetic alterations in FKBP family members ranged from 5% to 14% (FKBP2, FKBP5, FKBP8, and FKBP11: 5%; FKBP6, FKBP7, FKBP10, and FKBP15: 6%; FKBP9: 8%; FKBP4: 9%; FKBP3: 11%; and FKBP14: 14%) (Figure 4B).
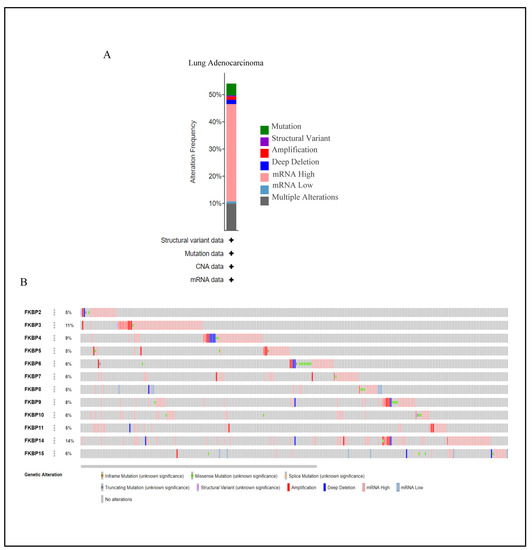
Figure 4.
Analysis of genetic alterations of FKBP family numbers in lung adenocarcinoma (LUAD) samples using the CBioPortal database. (A) Summary of genetic alterations in distinctive FKBP gene family members in LUAD. (B) OncoPrint dataset showing the FKBP family numbers showing alterations of individual FKBPs on queried genes. The horizontal axis represents each LUAD patient from the TCGA database, and the rate of genetic alterations in FKBP family members ranged from 5% to 14%.
3.3. Analysis of GGIs and PPIs and Coexpression of Pathway Abundance of the FKBP Gene Family
Due to genetic diversity, GGIs have impacts on gene functions, relative pathways, and even the development of target drugs [72]. In this study, we analyzed GGI networks of FKBP family members with neighbor genes via GeneMANIA (Supplementary Figure S2A). Results showed that HECTD1, TTC6, and other correlated genes were intensely linked with FKBP family members in shared protein domains (55.30%); coexpression (31.22%); physical interactions (13.16%); predictions (0.23%); co-localization (0.08%); and genetic interactions (0.02%); and functions including drug binding, cis-trans isomerase activity, protein folding, peptidyl-proline medication, isomerase activity, negative regulation of calcium ion transport into the cytosol, and positive regulation of the sequestration of calcium ions. Additionally, because cellular life is based on a complicated network of functional influences among biomolecules [73], we also evaluated the PPIs of FKBP family members in Homo sapiens using the STRING database (Supplementary Figure S2B).
The current data revealed that mRNA levels of FKBP2, FKBP3, FKBP4, FKBP10, FKBP11, and FKBP14 were overexpressed in LUAD, and FKBP10 had connections to poor prognoses among LUAD patients in an OS analysis. Based on the above results, we selected FKBP10 to further conduct a comprehensive analysis of the downstream pathway and network. Next, to deeply analyze coexpressed genes with FKBP10, we downloaded an archive of genes coexpressed with FKBP10 in LUAD and chose data for the first 1000 small p values from cBioPortal before performing DAVID. We analyzed two different aspects. First, GO term enrichment (GOTERM) revealed several FKBP10-correlated pathways, including protein binding (p = 1.6 × 10−16), isomerase activity (p = 8.6 × 10−2), etc. (Supplementary Figure S3, Supplementary Table S3). GOTERM_BPs described biological events in which these coexpressed genes of FKBP10 were involved, including mitochondrial electron transport, NADH to ubiquinone, mitochondrial respiratory chain complex I assembly, etc. (Supplementary Figure S4, Supplementary Table S4). The CCs of genes coexpressed with FKBP10 were also revealed in GOTERM_(CC) (Supplementary Figure S5, Supplementary Table S5). In another aspect of a KEGG analysis, outcomes indicated that there were 325 coexpressed genes (33.8%) in the non-alcoholic fatty liver disease (NAFLD) pathway (Supplementary Figure S6).
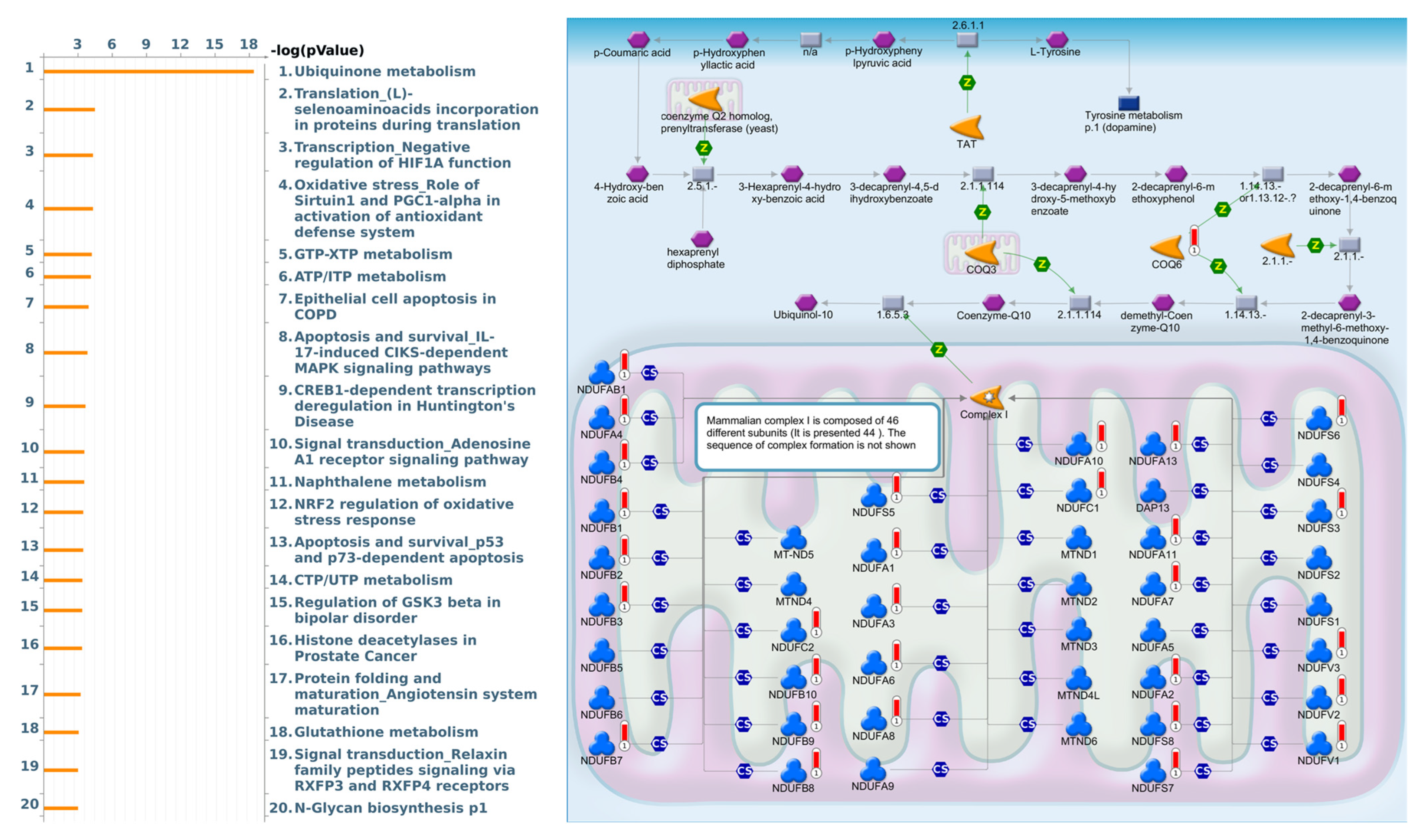
As a result, annotations of almost all BPs obtained from GeneGo Metacore showed that genes coexpressed with FKBP10 participated in several networks and cell-cycle-related pathways such as “Ubiquinone metabolism”, “Translation_(L)-selenoaminoacids incorporation in proteins during translation”, “Transcription_Negative regulation of HIF1A function”, “Oxidative stress_Role of Sirtuin1 and PGC1-alpha in activation of antioxidant defense system”, “GTP-XTP metabolism”, “ATP/ITP metabolism”, “Epithelial cell apoptosis in COPD”, “Apoptosis and survival_IL-17-induced CIKS-dependent MAPK signaling pathways”, “CREB1-dependent transcription deregulation in Huntington’s disease”, and “Signal transduction_Adenosine A1 receptor signaling pathway” (Figure 5, Supplementary Table S6).
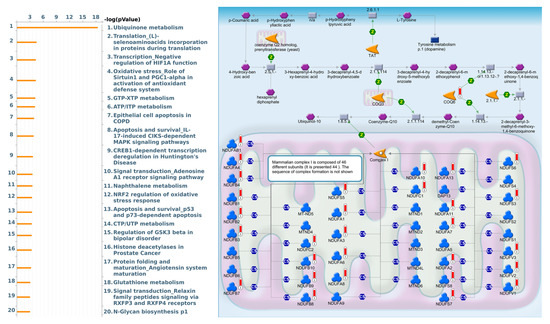
Figure 5.
MetaCore pathway analysis of the coexpression gene network of FKBP family members in lung adenocarcinoma (LUAD) patients. The MetaCore pathway analysis of “biological processes” revealed that “Ubiquinone metabolism”-related pathways were correlated with LUAD development.
3.4. Levels of Immune Cell Infiltration of Different FKBP Family Members in LUAD Patients
The TME is extremely important for the existence of cancer and is composed of vessels, extracellular matrix (ECM), and various immune cells, which favor tumor invasion, proliferation, and metastasis. Therefore, the occurrence of cancer is closely related to immune cells [74,75]. To fully understand the associations between FKBP family members and immune cell infiltration in LUAD, we evaluated the immunological microenvironment using the TIMER database (Supplementary Figure S7). Results indicated that FKBP2 was negatively correlated with CD8+ T cells (r = −0.345; p = 4.28 × 10−15), macrophages (r = −0.26; p = 5.8 × 10−9), neutrophils (r = −0.304; p = 8.41 × 10−12), and DCs (r = −0.283; p = 2.08 × 10−10). Transcriptional levels of FKBP3 were negatively associated with B cells (r = −0.114; p = 1.2 × 10−2), CD8+ T cells (r = −0.112; p = 1.33 × 10−10), CD4+ T cells (r = −0.133; p = 3.48 × 10−3), macrophages (r = −0.194; p = 1.72 × 10−5), neutrophils (r = −0.105; p = 2.16 × 10−2), and DCs (r = −0.167; p = 2.13 × 10−4). FKBP4 expression showed a positive correlation with purity (r = 0.12; p = 7.59 × 10−3) but negative correlations with B cells (r = −0.265; p = 3.34 × 10−9), CD4+ T cells (r = −0.151; p = 8.51 × 10−4), and DCs (r = −0.099; p = 2.84 × 10−2). FKBP5 had a negative association with purity (r = −0.125; p = 5.53 × 10−3) but positive associations with CD8+ T cells (r = 0.242; p = 6.56 × 10−8), macrophages (r = 0.194; p = 1.65 × 10−5), neutrophils (r = 0.219; p = 1.11 × 10−6), and DCs (r = 0.231; p = 2.4 × 10−7). FKBP6 expression was positively correlated with B cells (r = 0.245; p = 4.49 × 10−8), CD4+ T cells (r = 0.18; p = 6.72 × 10−5), macrophages (r = 0.129; p = 4.34 × 10−3), and DCs (r = 0.129; p = 4.39 × 10−3). Correlations of FKBP7 with immune cells were negative in terms of purity (r = −0.102; p = 2.4 × 10−2) and positive in terms of macrophages (r = 0.12; p = 7.91 × 10−3). FKBP8 expression was negatively associated with CD8+ T cells (r = −0.241; p = 7.7 × 10−8) and positively associated with CD4+ T cells (r = 0.302; p = 1.08 × 10−11). FKBP9 expression exhibited a positive association with neutrophils (r = 0.136; p = 2.72 × 10−3). Transcriptional levels of FKBP10 showed a negative link with B cells (r = −0.1; p = 2.8 × 10−2). FKBP11 expression revealed negative associations with purity (r = −0.159; p = 4.05 × 10−4), macrophages (r = −0.164; p = 2.88 × 10−4), neutrophils (r = −0.093; p = 4.09 × 10−2), and DCs (r = −0.19; p = 2.35 × 10−5) and a positive association with B cells (r = 0.129; p = 4.48 × 10−3). FKBP14 expression was negatively related to purity (r = −0.138; p = 2.13 × 10−3) but positively associated with CD8+ T cells (r = 0.114; p = 1.21 × 10−2), macrophages (r = 0.14; p = 2.01 × 10−3), neutrophils (r = 0.242; p = 7.16 × 10−8), and DCs (r = 0.181; p = 5.81 × 10−5). FKBP15 expression was negatively associated with all cells analyzed including purity (r = −0.262; p = 3.47 × 10−9), and positively associated with B cells (r = 0.334; p = 4.53 × 10−14), CD8+ T cells (r = 0.204; p = 5.64 × 10−6), CD4+ T cells (r = 0.535; p = 4.01 × 10−37), macrophages (r = 0.405; p = 1.29 × 10−2), neutrophils (r = 0.581; p = 4.89 × 10−45), and DCs (r = 0.612; p = 1.73 × 10−51). These outcomes revealed that the FKBP gene family plays important roles in immunological effects.
4. Discussion
In this study, we analyzed FKBP mRNA expressions, clinical phase IV, survival rates, genetic variants, and coexpressed genes. Moreover, we also determined correlations of infiltration levels of immune cells and FKBP gene expressions in LUAD. Although direct evidence still needs to be provided as to the biological functions of FKBP, such as cell models or patient tissue samples, to demonstrate the roles of FKBP in LUAD, based on our results, we can provide the concept that FKBP can potentially be a biomarker in LUAD.
By applying advances in high-throughput screening to cancer transcriptome profiling, alterations in the transcriptome patterns of FKBP gene families were found to be significantly associated with several types of malignancies [76,77,78,79,80]. FKBP gene expressions were found to be involved in tumor multi-stage progression along with other tumor-related factors. According to various database analyses, FKBP3, FKBP4, and FKBP10 were closely associated with LUAD. Previous studies determined that FKBP3 is a crucial oncogene in distinct cancers. The combination of FKBP3 and HDAC2 was related to oxaliplatin resistance in colorectal cancer via the PTEN/AKT pathway [18]. Downregulation of FKBP3 suppressed breast cancer [81], and FKBP3 expression was associated with poor survival in LUAD [82,83,84]. Furthermore, FKBP4 was reported to be related to breast cancer [20,85], colorectal cancer [22], prostate cancer [86], and lung cancer [21,87]. FKBP10 was connected with gastric cancer [88], stomach adenocarcinomas [89], papillary thyroid cancer [90], and lung cancer.
Our data were found to be consistent with those in previous research, as current findings indicated that mRNA levels of FKBP2, FKBP3, FKBP4, FKBP10, FKBP11, and FKBP14 were overexpressed in LUAD compared to healthy tissues. The OS analytical results revealed that five members of the FKBP gene family, viz., FKBP3, FKBP4, FKBP5, FKBP9, and FKBP10, had connections with poor prognoses among LUAD patients.
Based on the above results, we selected FKBP10 to further conduct a comprehensive analysis of the downstream pathway and network. Therefore, we explored the characteristics and functions of FKBP10 in more detail and discovered that FKBP10, also called FKBP65 (65-kDa), possesses four cytosolic PPI activities [88,91]. Moreover, downregulation of FKBP10 with collagen VI increased the formation of primary human lung fibroblasts (phLFs) [88]. Additionally, downregulation of FKBP10 suppressed tumorsphere formation by regulating protein translation [92]. Genes coexpressed with FKBP10 in TCGA LUAD were subsequently used to perform a pathway analysis. Through the DAVID analysis, we found FKBP10 to be involved in mitochondrial electron transport, NADH to ubiquinone and mitochondrial respiratory chain complex I assembly, etc. The Metacore pathway analysis also indicated that FKBP10 was involved in “Ubiquinone metabolism”, “Translation_(L)-selenoaminoacids incorporation in proteins during translation”, and “Transcription_Negative regulation of HIF1A function”.
In recent years, cancer immunotherapy, a novel strategy that aims to activate and boost the immune system to directly recognize and eliminate tumor cells, has undergone tremendous developments, and is now regarded as a promising cancer treatment [93,94,95]. Increasing evidence indicates that the immunosuppressive environment mediated by tumor-infiltrating immune cells (TICs), such as regulatory T (Treg) cells and tumor-associated macrophages (TAMs), hinders the delivery of immunotherapies in LUAD. Our data suggested that TICs are strongly correlated with FKBP expressions. Therefore, this analysis of the tumor immune microenvironment could help develop clinical immunotherapies and provide accurate personalized treatment plans for patients.
5. Conclusions
Previous research had not fully explored the roles of FKBP family genes in LUAD. Consequently, this study represents the first work that specifically examined the roles of FKBP members in this disease, prior to providing a more-extensive and -incisive understanding of the potential therapeutic and prognostic value for LUAD patients.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jpm13010049/s1, Supplementary Figure S1: The transcriptional levels of FKBP family member in LDAC and relationship between clinical stages in development of LUAD analyzed by GEPIA 2 database. (A) mRNA expression of FKBP5 in LUAD was greater than normal samples. The q-value cut-off was set to 0.01. (B) The stage plot indicated FKBP4 and FKBP5 were correlated with clinical stages in LUAD known by the height of the white dot in the four stages; Supplementary Figure S2: Analysis of association and abundance of FKBP family members (A) Connections between different FKBP family members and neighbor genes in Homo sapiens by the GeneMANIA database. Each node stands for an individual gene, and the size of the node represents the intensity of the gene-gene interaction (GGI). (B) Protein-protein interaction (PPI) associations between expressed FKBP10 and predicated proteins by the STRING database. The setting for maximum number of interactors to show was no more than 50 interactors; Supplementary Figure S3. Exploration of GOTERM_MF pathways for the coexpression of FKBP10 in lung adenocarcinoma (LUAD) by combining cBioPortal and the DAVID database; Supplementary Figure S4: Exploration of GOTERM_BP pathways for the coexpression of FKBP10 in lung adenocarcinoma (LUAD) by combining cBioPortal and the DAVID database; Supplementary Figure S5. Exploration of GOTERM_CC pathways for the coexpression of FKBP10 in lung adenocarcinoma (LUAD) by combining cBioPortal and the DAVID database; Supplementary Figure S6. Exploration of KEGG pathways for the coexpression of FKBP10 in lung adenocarcinoma (LUAD) by combining cBioPortal and the DAVID database. * Listed genes shown in the diagram are marked by a red star; Supplementary Figure S7. Associations between FKBP family members and immune infiltration consisting of purity, B cells, cluster of differentiation-positive (CD8+) T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells from TCGA in lung adenocarcinoma (LUAD) by the TIMER database. * Means a partial correlation (r), and p < 0.05 indicates a significant difference; Supplementary Table S1: Significant changes in expressions of FKBP family members between different types of lung cancer and normal tissue; Supplementary Table S2: Associations of prognoses with transcription mRNA levels of FKBP family members in patients with lung cancer; Supplementary Table S3: Gene Ontology term enrichment (GOTERM)_ revealed several FKBP10 correlated pathways and molecular function; Supplementary Table S4: Gene Ontology term enrichment (GOTERM)_ revealed several FKBPs correlated pathways and biological process; Supplementary Table S5: Gene Ontology term enrichment (GOTERM)_ revealed several FKBP10 correlated pathways and cellular components; Supplementary Table S6: GeneGo Metacore showed that the co-expressed genes of FKBP10 participated in several networks.
Author Contributions
Conceptualization, C.-Y.W. and W.-J.W.; Methodology, H.D.K.T., D.T.M.X. and C.-F.S.; Formal analysis, C.-C.W. and G.A.; Investigation, C.-C.W. and Y.-H.H.; Data curation, C.-C.W., W.-J.S., C.-Y.W. and W.-J.W.; Writing—original draft, C.-C.W. and C.-Y.W.; Writing—review & editing, W.-J.W.; Funding acquisition, W.-J.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by grants from the Ministry of Science and Technology (MOST) of Taiwan (MOST-110-2320-B-039-068 to W.-J.W. and 109-2320-B-038-009-MY2 to C.-Y.W.), National Science and Technology Council of Taiwan (NSTC-111-2314-B-182A-151 to C.-C.W.), Kaohsiung Chang Gung Memorial Hospital (CMRPG8K1271-3, NMRPG8M0241, CMRPG8M0331, and CMRPG8L0521 to C-C.W.), China Medical University (CMU110-MF-47 to W.-J.W.), Taipei Medical University (TMU-108-AE1-B16 to C.-Y.W.), and the TMU Research Center of Cancer Translational Medicine from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan (DP2-111-21121-01-C-01-01).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Ruiz-Cordero, R.; Devine, W.P. Targeted Therapy and Checkpoint Immunotherapy in Lung Cancer. Surg. Pathol. Clin. 2020, 13, 17–33. [Google Scholar] [CrossRef] [PubMed]
- Denisenko, T.V.; Budkevich, I.N.; Zhivotovsky, B. Cell death-based treatment of lung adenocarcinoma. Cell Death Dis. 2018, 9, 117. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Zou, C.; Zhu, Y.; Luo, Y.; Chen, L.; Lei, Y.; Tang, K.; Sun, Y.; Zhang, W.; Li, S.; et al. HIF-1a-regulated miR-1275 maintains stem cell-like phenotypes and promotes the progression of LUAD by simultaneously activating Wnt/beta-catenin and Notch signaling. Theranostics 2020, 10, 2553–2570. [Google Scholar] [CrossRef] [PubMed]
- Zappa, C.; Mousa, S.A. Non-small cell lung cancer: Current treatment and future advances. Transl. Lung Cancer Res. 2016, 5, 288–300. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Wu, H. CC Chemokine Receptors in Lung Adenocarcinoma: The Inflammation-Related Prognostic Biomarkers and Immunotherapeutic Targets. J. Inflamm. Res. 2021, 14, 267–285. [Google Scholar] [CrossRef] [PubMed]
- Bi, K.W.; Wei, X.G.; Qin, X.X.; Li, B. BTK Has Potential to Be a Prognostic Factor for Lung Adenocarcinoma and an Indicator for Tumor Microenvironment Remodeling: A Study Based on TCGA Data Mining. Front. Oncol. 2020, 10, 424. [Google Scholar] [CrossRef]
- Chung, C.C.; Huang, T.Y.; Chu, H.R.; De Luca, R.; Candelotti, E.; Huang, C.H.; Yang, Y.S.H.; Incerpi, S.; Pedersen, J.Z.; Lin, C.Y.; et al. Heteronemin and tetrac derivatives suppress non-small cell lung cancer growth via ERK1/2 inhibition. Food Chem. Toxicol. 2022, 161, 112850. [Google Scholar] [CrossRef]
- Hsiao, S.H.; Chen, W.T.; Chung, C.L.; Chou, Y.T.; Lin, S.E.; Hong, S.Y.; Chang, J.H.; Chang, T.H.; Chien, L.N. Comparative survival analysis of platinum-based adjuvant chemotherapy for early-stage squamous cell carcinoma and adenocarcinoma of the lung. Cancer Med. 2022, 11, 2067–2078. [Google Scholar] [CrossRef]
- Kuo, K.T.; Lin, C.H.; Wang, C.H.; Pikatan, N.W.; Yadav, V.K.; Fong, I.H.; Yeh, C.T.; Lee, W.H.; Huang, W.C. HNMT Upregulation Induces Cancer Stem Cell Formation and Confers Protection against Oxidative Stress through Interaction with HER2 in Non-Small-Cell Lung Cancer. Int. J. Mol. Sci. 2022, 23, 1663. [Google Scholar] [CrossRef]
- Tseng, P.-C.; Chen, C.-L.; Lee, K.-Y.; Feng, P.-H.; Wang, Y.-C.; Satria, R.D.; Lin, C.-F. Epithelial-to-mesenchymal transition hinders interferon-γ-dependent immunosurveillance in lung cancer cells. Cancer Lett. 2022, 539, 215712. [Google Scholar] [CrossRef]
- Lee, H.C.; Lu, Y.H.; Huang, Y.L.; Huang, S.L.; Chuang, H.C. Air Pollution Effects to the Subtype and Severity of Lung Cancers. Front. Med. 2022, 9, 835026. [Google Scholar] [CrossRef]
- Solassol, J.; Mange, A.; Maudelonde, T. FKBP family proteins as promising new biomarkers for cancer. Curr Opin Pharmacol 2011, 11, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Qin, X.; Fang, J.; Tang, Y.; Fan, Y. Multi-Omics Analysis of the Expression and Prognosis for FKBP Gene Family in Renal Cancer. Front. Oncol. 2021, 11, 697534. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.B.; Hong, Y.; Dhe-Paganon, S.; Yoon, H.S. FKBP family proteins: Immunophilins with versatile biological functions. Neurosignals 2008, 16, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Barik, S. On the role, ecology, phylogeny, and structure of dual-family immunophilins. Cell Stress Chaperones 2017, 22, 833–845. [Google Scholar] [CrossRef] [PubMed]
- Bonner, J.M.; Boulianne, G.L. Diverse structures, functions and uses of FK506 binding proteins. Cell Signal 2017, 38, 97–105. [Google Scholar] [CrossRef]
- Wen, Y.D.; Zhu, X.S.; Li, D.J.; Zhao, Q.; Cheng, Q.; Peng, Y. Proteomics-based prognostic signature and nomogram construction of hypoxia microenvironment on deteriorating glioblastoma (GBM) pathogenesis. Sci. Rep. 2021, 11, 17170. [Google Scholar] [CrossRef]
- Tong, J.; Shen, Y.; Chen, X.; Wang, R.; Hu, Y.; Zhang, X.; Zhang, Z.; Han, L. FKBP3 mediates oxaliplatin resistance in colorectal cancer cells by regulating HDAC2 expression. Oncol. Rep. 2019, 42, 1404–1412. [Google Scholar] [CrossRef]
- Zhu, W.; Li, Z.; Xiong, L.; Yu, X.; Chen, X.; Lin, Q. FKBP3 Promotes Proliferation of Non-Small Cell Lung Cancer Cells through Regulating Sp1/HDAC2/p27. Theranostics 2017, 7, 3078–3089. [Google Scholar] [CrossRef]
- Xiong, H.; Chen, Z.; Zheng, W.; Sun, J.; Fu, Q.; Teng, R.; Chen, J.; Xie, S.; Wang, L.; Yu, X.F.; et al. FKBP4 is a malignant indicator in luminal A subtype of breast cancer. J. Cancer 2020, 11, 1727–1736. [Google Scholar] [CrossRef]
- Meng, W.; Meng, J.; Jiang, H.; Feng, X.; Wei, D.; Ding, Q. FKBP4 Accelerates Malignant Progression of Non-Small-Cell Lung Cancer by Activating the Akt/mTOR Signaling Pathway. Anal. Cell Pathol. 2020, 2020, 6021602. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Gao, N. KDM5D inhibits the transcriptional activation of FKBP4 by suppressing the expression of E2F1 in colorectal cancer in males. Biochem. Pharmacol. 2021, 194, 114814. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Wang, L. FKBP5 as a selection biomarker for gemcitabine and Akt inhibitors in treatment of pancreatic cancer. PLoS ONE 2012, 7, e36252. [Google Scholar] [CrossRef] [PubMed]
- Brebi, P.; Maldonado, L.; Noordhuis, M.G.; Ili, C.; Leal, P.; Garcia, P.; Brait, M.; Ribas, J.; Michailidi, C.; Perez, J.; et al. Genome-wide methylation profiling reveals Zinc finger protein 516 (ZNF516) and FK-506-binding protein 6 (FKBP6) promoters frequently methylated in cervical neoplasia, associated with HPV status and ethnicity in a Chilean population. Epigenetics 2014, 9, 308–317. [Google Scholar] [CrossRef]
- Garrido, M.F.; Martin, N.J.; Bertrand, M.; Gaudin, C.; Commo, F.; El Kalaany, N.; Al Nakouzi, N.; Fazli, L.; Del Nery, E.; Camonis, J.; et al. Regulation of eIF4F Translation Initiation Complex by the Peptidyl Prolyl Isomerase FKBP7 in Taxane-resistant Prostate Cancer. Clin. Cancer Res. 2019, 25, 710–723. [Google Scholar] [CrossRef]
- Hagedorn, M.; Siegfried, G.; Hooks, K.B.; Khatib, A.M. Integration of zebrafish fin regeneration genes with expression data of human tumors in silico uncovers potential novel melanoma markers. Oncotarget 2016, 7, 71567–71579. [Google Scholar] [CrossRef]
- Jiang, F.N.; Dai, L.J.; Yang, S.B.; Wu, Y.D.; Liang, Y.X.; Yin, X.L.; Zou, C.Y.; Zhong, W.D. Increasing of FKBP9 can predict poor prognosis in patients with prostate cancer. Pathol. Res. Pract. 2020, 216, 152732. [Google Scholar] [CrossRef]
- Xu, H.; Liu, P.; Yan, Y.; Fang, K.; Liang, D.; Hou, X.; Zhang, X.; Wu, S.; Ma, J.; Wang, R.; et al. FKBP9 promotes the malignant behavior of glioblastoma cells and confers resistance to endoplasmic reticulum stress inducers. J. Exp. Clin. Cancer Res. 2020, 39, 44. [Google Scholar] [CrossRef]
- Ghoorun, R.A.; Wu, X.H.; Chen, H.L.; Ren, D.L.; Wu, X.B. Prognostic Significance of FKBP14 in Gastric Cancer. Onco. Targets Ther. 2019, 12, 11567–11577. [Google Scholar] [CrossRef]
- Wang, R.; Fang, H.; Fang, Q. Downregulation of FKBP14 by RNA interference inhibits the proliferation, adhesion and invasion of gastric cancer cells. Oncol. Lett. 2017, 13, 2811–2816. [Google Scholar] [CrossRef]
- Sun, L.Y.; Tao, J.Z.; Yan, B.; Lin, J.S. Inhibitory effects of FKBP14 on human cervical cancer cells. Mol. Med. Rep. 2017, 16, 4265–4272. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lu, M.; Miao, Y.; Qi, L.; Bai, M.; Zhang, J.; Feng, Y. RNAi-Mediated Downregulation of FKBP14 Suppresses the Growth of Human Ovarian Cancer Cells. Oncol. Res. 2016, 23, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Qi, X.J.; Liu, H.Z.; Su, H. MiR-361 inhibits osteosarcoma cell lines invasion and proliferation by targeting FKBP14. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 79–86. [Google Scholar] [CrossRef]
- Ibusuki, M.; Fu, P.; Yamamoto, S.; Fujiwara, S.; Yamamoto, Y.; Honda, Y.; Iyama, K.; Iwase, H. Establishment of a standardized gene-expression analysis system using formalin-fixed, paraffin-embedded, breast cancer specimens. Breast Cancer 2013, 20, 159–166. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets--update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.C.; Liu, T.P.; Yang, P.M. CDKN2A-Inactivated Pancreatic Ductal Adenocarcinoma Exhibits Therapeutic Sensitivity to Paclitaxel: A Bioinformatics Study. J. Clin. Med. 2020, 9, 4019. [Google Scholar] [CrossRef]
- Lin, T.Y.; Wang, P.W.; Huang, C.H.; Yang, P.M.; Pan, T.L. Characterizing the Relapse Potential in Different Luminal Subtypes of Breast Cancers with Functional Proteomics. Int. J. Mol. Sci. 2020, 21, 6077. [Google Scholar] [CrossRef]
- Liu, L.W.; Hsieh, Y.Y.; Yang, P.M. Bioinformatics Data Mining Repurposes the JAK2 (Janus Kinase 2) Inhibitor Fedratinib for Treating Pancreatic Ductal Adenocarcinoma by Reversing the KRAS (Kirsten Rat Sarcoma 2 Viral Oncogene Homolog)-Driven Gene Signature. J. Pers. Med. 2020, 10, 130. [Google Scholar] [CrossRef]
- Yang, P.M.; Hsieh, Y.Y.; Du, J.L.; Yen, S.C.; Hung, C.F. Sequential Interferon β-Cisplatin Treatment Enhances the Surface Exposure of Calreticulin in Cancer Cells via an Interferon Regulatory Factor 1-Dependent Manner. Biomolecules 2020, 10, 643. [Google Scholar] [CrossRef]
- Yang, P.M.; Lin, L.S.; Liu, T.P. Sorafenib Inhibits Ribonucleotide Reductase Regulatory Subunit M2 (RRM2) in Hepatocellular Carcinoma Cells. Biomolecules 2020, 10, 117. [Google Scholar] [CrossRef]
- Jiang, W.; Cai, G.; Hu, P.C.; Wang, Y. Personalized medicine in non-small cell lung cancer: A review from a pharmacogenomics perspective. Acta Pharm. Sin. B 2018, 8, 530–538. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhao, X.; Yuan, Y.; Jing, J.J. The expression patterns and the diagnostic/prognostic roles of PTPN family members in digestive tract cancers. Cancer Cell Int. 2020, 20, 238. [Google Scholar] [CrossRef]
- Zhou, J.; Wu, G.; Tong, Z.; Sun, J.; Su, J.; Cao, Z.; Luo, Y.; Wang, W. Prognostic relevance of SMC family gene expression in human sarcoma. Aging 2020, 13, 1473–1487. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.W.; Hsieh, Y.H.; Ku, S.C.; Shen, W.J.; Anuraga, G.; Khoa Ta, H.D.; Lee, K.H.; Lee, Y.C.; Lin, C.H.; Wang, C.Y.; et al. Potential Prognostic Biomarkers of OSBPL Family Genes in Patients with Pancreatic Ductal Adenocarcinoma. Biomedicines 2021, 9, 1601. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Hu, C.; Fan, Z.J.; Shen, G.L. Transcript levels of keratin 1/5/6/14/15/16/17 as potential prognostic indicators in melanoma patients. Sci. Rep. 2021, 11, 1023. [Google Scholar] [CrossRef]
- Wang, C.-C.; Shen, W.-J.; Anuraga, G.; Khoa Ta, H.D.; Xuan, D.T.M.; Chen, S.-T.; Shen, C.-F.; Jiang, J.-Z.; Sun, Z.; Wang, C.-Y.; et al. Novel Potential Therapeutic Targets of PTPN Families for Lung Cancer. J. Pers. Med. 2022, 12, 1947. [Google Scholar] [CrossRef]
- Li, Q.; Pan, Y.; Cao, Z.; Zhao, S. Comprehensive Analysis of Prognostic Value and Immune Infiltration of Chromobox Family Members in Colorectal Cancer. Front. Oncol. 2020, 10, 582667. [Google Scholar] [CrossRef]
- Ta, H.D.K.; Wang, W.J.; Phan, N.N.; An Ton, N.T.; Anuraga, G.; Ku, S.C.; Wu, Y.F.; Wang, C.Y.; Lee, K.H. Potential Therapeutic and Prognostic Values of LSM Family Genes in Breast Cancer. Cancers 2021, 13, 4902. [Google Scholar] [CrossRef]
- Lin, S.; Cao, C.; Meng, Y.; Wu, P.; Gao, P.; Zhi, W.; Peng, T.; Wu, P.; Gui, L. Comprehensive analysis of the value of RAB family genes in prognosis of breast invasive carcinoma. Biosci. Rep. 2020, 40, BSR20201103. [Google Scholar] [CrossRef]
- Huang, Y.L.; Ning, G.; Chen, L.B.; Lian, Y.F.; Gu, Y.R.; Wang, J.L.; Chen, D.M.; Wei, H.; Huang, Y.H. Promising diagnostic and prognostic value of E2Fs in human hepatocellular carcinoma. Cancer Manag. Res. 2019, 11, 1725–1740. [Google Scholar] [CrossRef]
- Ramezani, M.; Baharzadeh, F.; Almasi, A.; Sadeghi, M. A Systematic Review and Meta-Analysis: Evaluation of the beta-Human Papillomavirus in Immunosuppressed Individuals with Cutaneous Squamous Cell Carcinoma. Biomedicine 2020, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Montojo, J.; Zuberi, K.; Rodriguez, H.; Kazi, F.; Wright, G.; Donaldson, S.L.; Morris, Q.; Bader, G.D. GeneMANIA Cytoscape plugin: Fast gene function predictions on the desktop. Bioinformatics 2010, 26, 2927–2928. [Google Scholar] [CrossRef] [PubMed]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Liu, S.; Xie, X.; Lei, H.; Zou, B.; Xie, L. Identification of Key circRNAs/lncRNAs/miRNAs/mRNAs and Pathways in Preeclampsia Using Bioinformatics Analysis. Med. Sci. Monit. 2019, 25, 1679–1693. [Google Scholar] [CrossRef]
- Guo, Y.; He, Y. Comprehensive analysis of the expression of SLC30A family genes and prognosis in human gastric cancer. Sci. Rep. 2020, 10, 18352. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Gene Ontology, C. Gene Ontology Consortium: Going forward. Nucleic Acids Res. 2015, 43, D1049–D1056. [Google Scholar] [CrossRef]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef]
- Pan, J.H.; Zhou, H.; Cooper, L.; Huang, J.L.; Zhu, S.B.; Zhao, X.X.; Ding, H.; Pan, Y.L.; Rong, L. LAYN Is a Prognostic Biomarker and Correlated With Immune Infiltrates in Gastric and Colon Cancers. Front. Immunol. 2019, 10, 6. [Google Scholar] [CrossRef] [PubMed]
- Stearman, R.S.; Dwyer-Nield, L.; Zerbe, L.; Blaine, S.A.; Chan, Z.; Bunn, P.A., Jr.; Johnson, G.L.; Hirsch, F.R.; Merrick, D.T.; Franklin, W.A.; et al. Analysis of orthologous gene expression between human pulmonary adenocarcinoma and a carcinogen-induced murine model. Am. J. Pathol. 2005, 167, 1763–1775. [Google Scholar] [CrossRef] [PubMed]
- Su, L.J.; Chang, C.W.; Wu, Y.C.; Chen, K.C.; Lin, C.J.; Liang, S.C.; Lin, C.H.; Whang-Peng, J.; Hsu, S.L.; Chen, C.H.; et al. Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genom. 2007, 8, 140. [Google Scholar] [CrossRef] [PubMed]
- Landi, M.T.; Dracheva, T.; Rotunno, M.; Figueroa, J.D.; Liu, H.; Dasgupta, A.; Mann, F.E.; Fukuoka, J.; Hames, M.; Bergen, A.W.; et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS ONE 2008, 3, e1651. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Aerts, J.; den Hamer, B.; van Ijcken, W.; den Bakker, M.; Riegman, P.; van der Leest, C.; van der Spek, P.; Foekens, J.A.; Hoogsteden, H.C.; et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS ONE 2010, 5, e10312. [Google Scholar] [CrossRef] [PubMed]
- Beer, D.G.; Kardia, S.L.; Huang, C.C.; Giordano, T.J.; Levin, A.M.; Misek, D.E.; Lin, L.; Chen, G.; Gharib, T.G.; Thomas, D.G.; et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat. Med. 2002, 8, 816–824. [Google Scholar] [CrossRef]
- Wachi, S.; Yoneda, K.; Wu, R. Interactome-transcriptome analysis reveals the high centrality of genes differentially expressed in lung cancer tissues. Bioinformatics 2005, 21, 4205–4208. [Google Scholar] [CrossRef]
- Bhattacharjee, A.; Richards, W.G.; Staunton, J.; Li, C.; Monti, S.; Vasa, P.; Ladd, C.; Beheshti, J.; Bueno, R.; Gillette, M.; et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc. Natl. Acad. Sci. USA 2001, 98, 13790–13795. [Google Scholar] [CrossRef]
- Garber, M.E.; Troyanskaya, O.G.; Schluens, K.; Petersen, S.; Thaesler, Z.; Pacyna-Gengelbach, M.; van de Rijn, M.; Rosen, G.D.; Perou, C.M.; Whyte, R.I.; et al. Diversity of gene expression in adenocarcinoma of the lung. Proc. Natl. Acad. Sci. USA 2001, 98, 13784–13789. [Google Scholar] [CrossRef]
- Okayama, H.; Kohno, T.; Ishii, Y.; Shimada, Y.; Shiraishi, K.; Iwakawa, R.; Furuta, K.; Tsuta, K.; Shibata, T.; Yamamoto, S.; et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012, 72, 100–111. [Google Scholar] [CrossRef]
- Selamat, S.A.; Chung, B.S.; Girard, L.; Zhang, W.; Zhang, Y.; Campan, M.; Siegmund, K.D.; Koss, M.N.; Hagen, J.A.; Lam, W.L.; et al. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome. Res. 2012, 22, 1197–1211. [Google Scholar] [CrossRef]
- Juvvadi, P.R.; Bobay, B.G.; Gobeil, S.M.C.; Cole, D.C.; Venters, R.A.; Heitman, J.; Spicer, L.D.; Steinbach, W.J. FKBP12 dimerization mutations effect FK506 binding and differentially alter calcineurin inhibition in the human pathogen Aspergillus fumigatus. Biochem. Biophys. Res. Commun. 2020, 526, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, A.; Lu, T.P. Gene-gene interaction: The curse of dimensionality. Ann. Transl. Med. 2019, 7, 813. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Zuo, S.; Wei, M.; Wang, S.; Dong, J.; Wei, J. Pan-Cancer Analysis of Immune Cell Infiltration Identifies a Prognostic Immune-Cell Characteristic Score (ICCS) in Lung Adenocarcinoma. Front. Immunol. 2020, 11, 1218. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, M.; Ng, D.M.; Haleem, M.; Yi, T.; Hu, S.; Zhu, H.; Zhao, G.; Liao, Q. Multi-omics Data Analyses Construct TME and Identify the Immune-Related Prognosis Signatures in Human LUAD. Mol. Ther. Nucleic Acids 2020, 21, 860–873. [Google Scholar] [CrossRef] [PubMed]
- Thorat, M.A.; Balasubramanian, R. Breast cancer prevention in high-risk women. Best Pract. Res. Clin. Obstet. Gynaecol. 2020, 65, 18–31. [Google Scholar] [CrossRef]
- Lin, C.Y.; Lee, C.H.; Chuang, Y.H.; Lee, J.Y.; Chiu, Y.Y.; Wu Lee, Y.H.; Jong, Y.J.; Hwang, J.K.; Huang, S.H.; Chen, L.C.; et al. Membrane protein-regulated networks across human cancers. Nat. Commun. 2019, 10, 3131. [Google Scholar] [CrossRef]
- Tsai, H.T.; Huang, C.S.; Tu, C.C.; Liu, C.Y.; Huang, C.J.; Ho, Y.S.; Tu, S.H.; Tseng, L.M.; Huang, C.C. Multi-gene signature of microcalcification and risk prediction among Taiwanese breast cancer. Sci. Rep. 2020, 10, 18276. [Google Scholar] [CrossRef]
- Nguyen, H.D.; Liao, Y.C.; Ho, Y.S.; Chen, L.C.; Chang, H.W.; Cheng, T.C.; Liu, D.; Lee, W.R.; Shen, S.C.; Wu, C.H.; et al. The α9 Nicotinic Acetylcholine Receptor Mediates Nicotine-Induced PD-L1 Expression and Regulates Melanoma Cell Proliferation and Migration. Cancers 2019, 11, 1991. [Google Scholar] [CrossRef]
- Lee, K.L.; Kuo, Y.C.; Ho, Y.S.; Huang, Y.H. Triple-Negative Breast Cancer: Current Understanding and Future Therapeutic Breakthrough Targeting Cancer Stemness. Cancers 2019, 11, 1334. [Google Scholar] [CrossRef]
- Liu, P.; Xie, X.; Yang, A.; Kong, Y.; Allen-Gipson, D.; Tian, Z.; Zhou, L.; Tang, H.; Xie, X. Melatonin Regulates Breast Cancer Progression by the lnc010561/miR-30/FKBP3 Axis. Mol. Ther. Nucleic Acids 2020, 19, 765–774. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; Zhao, B.; Wang, Y.; Li, L.; Li, J.; Li, X.; Chang, L.; Chen, Q.; Liao, Z. Construction of the optimization prognostic model based on differentially expressed immune genes of lung adenocarcinoma. BMC Cancer 2021, 21, 213. [Google Scholar] [CrossRef]
- Wei, B.; Kong, W.; Mou, X.; Wang, S. Comprehensive analysis of tumor immune infiltration associated with endogenous competitive RNA networks in lung adenocarcinoma. Pathol. Res. Pract. 2019, 215, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yang, B.; Li, X.; Ren, H.; Zhang, L.; Li, L.; Li, W.; Wang, X.; Zhou, H.; Zhang, W. Identification of Prognostic Factors Related to Super Enhancer-Regulated ceRNA Network in Metastatic Lung Adenocarcinoma. Int. J. Gen. Med. 2021, 14, 6261–6275. [Google Scholar] [CrossRef]
- Mange, A.; Coyaud, E.; Desmetz, C.; Laurent, E.; Beganton, B.; Coopman, P.; Raught, B.; Solassol, J. FKBP4 connects mTORC2 and PI3K to activate the PDK1/Akt-dependent cell proliferation signaling in breast cancer. Theranostics 2019, 9, 7003–7015. [Google Scholar] [CrossRef] [PubMed]
- Federer-Gsponer, J.R.; Quintavalle, C.; Muller, D.C.; Dietsche, T.; Perrina, V.; Lorber, T.; Juskevicius, D.; Lenkiewicz, E.; Zellweger, T.; Gasser, T.; et al. Delineation of human prostate cancer evolution identifies chromothripsis as a polyclonal event and FKBP4 as a potential driver of castration resistance. J. Pathol. 2018, 245, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Jia, S.; Li, L.; Xie, L.; Zhang, W.; Zhu, T.; Qian, B. Transcriptome Based Estrogen Related Genes Biomarkers for Diagnosis and Prognosis in Non-small Cell Lung Cancer. Front. Genet. 2021, 12, 666396. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Zhao, K.; Zhu, J.H.; Chen, G.; Qin, X.G.; Chen, J.Q. Comprehensive evaluation of FKBP10 expression and its prognostic potential in gastric cancer. Oncol. Rep. 2019, 42, 615–628. [Google Scholar] [CrossRef]
- Wang, R.G.; Zhang, D.; Zhao, C.H.; Wang, Q.L.; Qu, H.; He, Q.S. FKBP10 functioned as a cancer-promoting factor mediates cell proliferation, invasion, and migration via regulating PI3K signaling pathway in stomach adenocarcinoma. Kaohsiung J. Med. Sci. 2020, 36, 311–317. [Google Scholar] [CrossRef]
- Sarquis, M.; Moraes, D.C.; Bastos-Rodrigues, L.; Azevedo, P.G.; Ramos, A.V.; Reis, F.V.; Dande, P.V.; Paim, I.; Friedman, E.; De Marco, L. Germline Mutations in Familial Papillary Thyroid Cancer. Endocr. Pathol. 2020, 31, 14–20. [Google Scholar] [CrossRef]
- Coss, M.C.; Winterstein, D.; Sowder, R.C., 2nd; Simek, S.L. Molecular cloning, DNA sequence analysis, and biochemical characterization of a novel 65-kDa FK506-binding protein (FKBP65). J. Biol. Chem. 1995, 270, 29336–29341. [Google Scholar] [CrossRef] [PubMed]
- Ramadori, G.; Konstantinidou, G.; Venkateswaran, N.; Biscotti, T.; Morlock, L.; Galie, M.; Williams, N.S.; Luchetti, M.; Santinelli, A.; Scaglioni, P.P.; et al. Diet-induced unresolved ER stress hinders KRAS-driven lung tumorigenesis. Cell Metab. 2015, 21, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Doroshow, D.B.; Sanmamed, M.F.; Hastings, K.; Politi, K.; Rimm, D.L.; Chen, L.; Melero, I.; Schalper, K.A.; Herbst, R.S. Immunotherapy in non–small cell lung cancer: Facts and hopes. Clin. Cancer Res. 2019, 25, 4592–4602. [Google Scholar] [CrossRef] [PubMed]
- Hellmann, M.D.; Nathanson, T.; Rizvi, H.; Creelan, B.C.; Sanchez-Vega, F.; Ahuja, A.; Ni, A.; Novik, J.B.; Mangarin, L.M.; Abu-Akeel, M. Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell 2018, 33, 843–852.e4. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.S.; Baas, P.; Kim, D.-W.; Felip, E.; Pérez-Gracia, J.L.; Han, J.-Y.; Molina, J.; Kim, J.-H.; Arvis, C.D.; Ahn, M.-J. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): A randomised controlled trial. Lancet 2016, 387, 1540–1550. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).