Cell-Free Fetal DNA Screening Analysis in Korean Pregnant Women: Six Years of Experience and a Retrospective Study of 9327 Patients Analyzed from 2017 to 2022
Abstract
:1. Introduction
2. Materials and Methods
2.1. Subject
2.2. NIPT Analysis
2.3. Additional Diagnostic Tests
2.4. Statistical Analysis
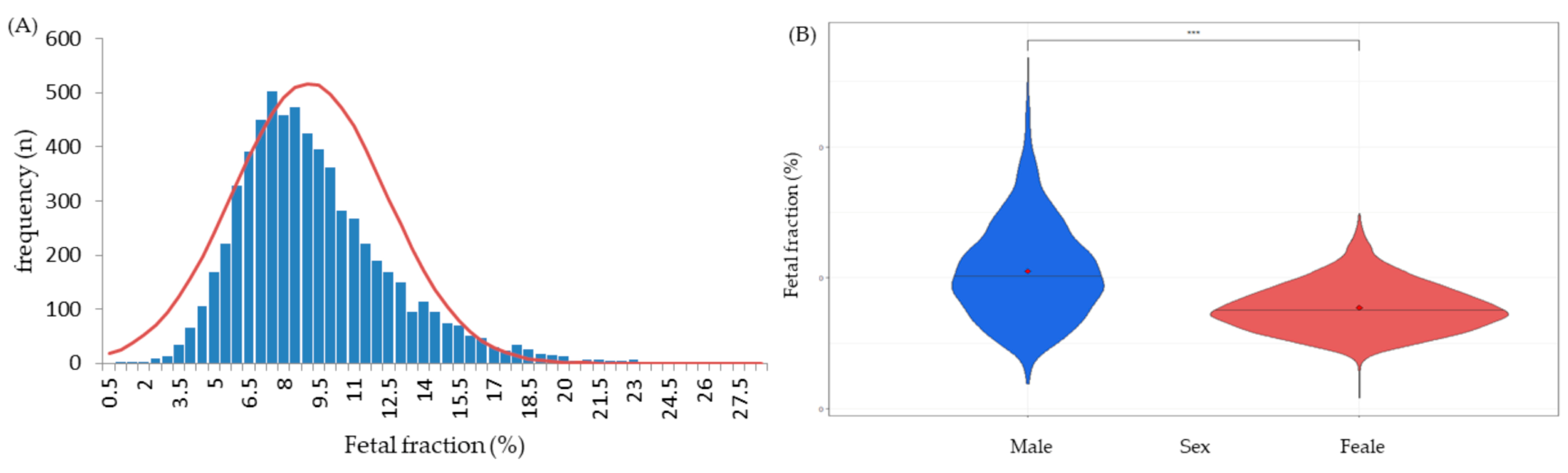
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- American College of Obstetricians and Gynecologists’ Committee on Practice Bulletins—Obstetrics. Committee on Genetics; Society for Maternal-Fetal Medicine. Screening for Fetal Chromosomal Abnormalities: ACOG Practice Bulletin, Number 226. Obstet. Gynecol. 2020, 136, e48–e69. [Google Scholar] [CrossRef] [PubMed]
- Institute of Health Economics. First and Second Trimester Prenatal Screening Update; Institute of Health Economics (IHE): Edmonton, AB, Canada, 2014. [Google Scholar]
- Nicolaides, K.H. Screening for fetal aneuploidies at 11 to 13 weeks. Prenat. Diagn. 2011, 31, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Alfirevic, Z.; Mujezinovic, F.; Sundberg, K.; Brigham, S. Amniocentesis and chorionic villus sampling for prenatal diagnosis. Cochrane Database Syst. Rev. 2003, 3, CD003252. [Google Scholar] [CrossRef]
- Tabor, A.; Alfirevic, Z. Update on procedure-related risks for prenatal diagnosis techniques. Fetal Diagn. Ther. 2010, 27, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.C.; Blumenfeld, Y.J.; Chitkara, U.; Hudgins, L.; Quake, S.R. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc. Natl. Acad. Sci. USA 2008, 105, 16266–16271. [Google Scholar] [CrossRef] [PubMed]
- Palomaki, G.E.; Kloza, E.M.; Lambert-Messerlian, G.M.; Haddow, J.E.; Neveux, L.M.; Ehrich, M.; van den Boom, D.; Bombard, A.T.; Deciu, C.; Grody, W.W.; et al. DNA sequencing of maternal plasma to detect Down syndrome: An international clinical validation study. Genet. Med. 2011, 13, 913–920. [Google Scholar] [CrossRef] [PubMed]
- Benn, P.; Cuckle, H.; Pergament, E. Non-invasive prenatal testing for aneuploidy: Current status and future prospects. Ultrasound Obstet. Gynecol. 2013, 42, 15–33. [Google Scholar] [CrossRef] [PubMed]
- Green, E.D.; Rubin, E.M.; Olson, M.V. Future DNA Sequencing. Nature 2017, 550, 179–181. [Google Scholar] [CrossRef]
- Bianchi, D.W.; Chiu, R.W.K. Sequencing of circulating cell-free DNA during pregnancy. N. Engl. J. Med. 2018, 379, 464–473. [Google Scholar] [CrossRef]
- Norton, M.E.; Jacobsson, B.; Swamy, G.K.; Laurent, L.C.; Ranzini, A.C.; Brar, H.; Tomlinson, M.W.; Pereira, L.; Spitz, J.L.; Hollemon, D.; et al. Cell-free DNA analysis for Noninvasive trisomy examination. N. Engl. J. Med. 2015, 372, 1589–1597. [Google Scholar] [CrossRef]
- Livergood, M.C.; LeChien, K.A.; Trudell, A.S. Obesity and cell-free DNA “no calls”: Is there an optimal gestational age at time of sampling? Am. J. Obstet. Gynecol. 2017, 216, 413.e411–413.e419. [Google Scholar] [CrossRef] [PubMed]
- Gil, M.M.; Accurti, V.; Santacruz, B.; Plana, M.N.; Nicolaides, K.H. Analysis of cell-free DNA in maternal blood during screening for aneuploidies: An updated meta-analysis. Ultrasound Obstet. Gynecol. 2017, 50, 302–314. [Google Scholar] [CrossRef] [PubMed]
- Gil, M.M.; Quezada, M.S.; Revello, R.; Akolekar, R.; Nicolaides, K.H. Analysis of cell-free DNA in maternal blood during screening for fetal aneuploidies: An updated meta-analysis. Ultrasound Obstet. Gynecol. 2015, 45, 249–266. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Gao, Y.; Jiang, F.; Fu, M.; Yuan, Y.; Guo, Y.; Zhu, Z.; Lin, M.; Liu, Q.; Tian, Z.; et al. Non-invasive prenatal testing for trisomies 21, 18 and 13: Clinical experience from 146,958 pregnancies. Ultrasound Obstet. Gynecol. 2015, 45, 530–538. [Google Scholar] [CrossRef] [PubMed]
- Bevilacqua, E.; Gil, M.M.; Nicolaides, K.H.; Ordonez, E.; Cirigliano, V.; Dierickx, H.; Willems, P.J.; Jani, J.C. Screening for aneuploidies using cell-free DNA analysis of maternal blood during twin pregnancies. Ultrasound Obstet. Gynecol. 2015, 45, 61–66. [Google Scholar] [CrossRef]
- Kinnings, S.L.; Geis, J.A.; Almasri, E.; Wang, H.; Guan, X.; McCullough, R.M.; Bombard, A.T.; Saldivar, J.S.; Oeth, P.; Deciu, C. Factors affecting levels of circulating cell-free fetal DNA in maternal plasma and their implications for noninvasive prenatal testing. Prenat. Diagn. 2015, 35, 816–822. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.M.; Kim, S.H.; Park, J.E.; Kim, H.J.; Jang, H.Y.; Go, M.; Yang, S.H.; Ryu, S.W.; Bae, S.M.; Cha, D.H.; et al. Inconsistency between non-invasive prenatal testing (NIPT) and conventional prenatal diagnosis due to confined placental and fetal mosaicism: Two case reports. Front. Med. 2022, 15, 1063480. [Google Scholar] [CrossRef]
- Kim, M.; Kim, J.H.; Kim, K.; Kim, S. Cost-effective and accurate method of measuring fetal fraction using SNP imputation. Bioinformatics 2018, 34, 1086–1091. [Google Scholar] [CrossRef]
- Deng, C.; Liu, S. Factors Affecting the Fetal Fraction in Noninvasive Prenatal Screening: A Review. Front. Pediatr. 2022, 10, 812781. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhu, Z.; Gao, Y.; Yuan, Y.; Guo, Y.; Zhou, L.; Liao, K.; Wang, J.; Du, B.; Hou, Y.; et al. Effects of Maternal and Fetal Characteristics on Cell-Free Fetal DNA Fraction in Maternal Plasma. Reprod. Sci. 2015, 22, 1429–1435. [Google Scholar] [CrossRef]
- Ashoor, G.; Syngelaki, A.; Poon, L.C.; Rezende, J.C.; Nicolaides, K.H. Fetal fraction in maternal plasma cell-free DNA at 11–13 weeks’ gestation: Relation to maternal and fetal characteristics. Ultrasound Obstet. Gynecol. 2013, 41, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Galeva, S.; Gil, M.M.; Konstantinidou, L.; Akolekar, R.; Nicolaides, K.H. First-trimester screening for trisomies by cfDNA testing of maternal blood in singleton and twin pregnancies: Factors affecting test failure. Ultrasound Obstet. Gynecol. 2019, 53, 804–809. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.J.; Rolnik, D.L.; Menezes, M.A.; McLennan, A.C.; da Silva Costa, F. Cell-free fetal DNA testing in singleton IVF conceptions. Hum. Reprod. 2018, 33, 572–578. [Google Scholar] [CrossRef] [PubMed]
- Baranova, E.E.; Sagaydak, O.V.; Galaktionova, A.M.; Kuznetsova, E.S.; Kaplanova, M.T.; Makarova, M.V.; Belenikin, M.S.; Olenev, A.S.; Songolova, E.N. Whole genome non-invasive prenatal testing in prenatal screening algorithm: Clinical experience from 12,700 pregnancies. BMC Pregnancy Childbirth 2022, 22, 633. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Lin, Y.; Qiao, F.; Li, H.; Wang, Y.; Zhang, J.; Liu, A.; Ji, X.; Ma, D.; Jiang, T.; et al. Perinatal outcomes following cell-free DNA screening in >32,000 women: Clinical follow-up data from a single tertiary center and has relative lower sensitivities and specificities for T18, T13 and SCAs. Prenat. Diagn. 2018, 38, 755–764. [Google Scholar] [CrossRef] [PubMed]
- Petersen, A.K.; Cheung, S.W.; Smith, J.L.; Bi, W.; Ward, P.A.; Peacock, S.; Braxton, A.; van Den Veyver, I.B.; Breman, A.M. Positive predictive value estimates for cell-free noninvasive prenatal screening from data of a large referral genetic diagnostic laboratory. Am. J. Obstet. Gynecol. 2017, 217, 691.e1–691.e6. [Google Scholar] [CrossRef] [PubMed]
- Samura, O.; Okamoto, A. Causes of aberrant non-invasive prenatal testing for aneuploidy: A systematic review. Taiwan. J. Obstet. Gyn 2020, 59, 16–20. [Google Scholar] [CrossRef]
- Bianchi, D.W.; Wilkins-Haug, L. Integration of noninvasive DNA testing for aneuploidy into prenatal care: What has happened since the rubber met the road? Clin. Chem. 2014, 60, 78–87. [Google Scholar] [CrossRef]
- Grati, F.R.; Bajaj, K.; Malvestiti, F.; Agrati, C.; Grimi, B.; Malvestiti, B.; Pompilii, E.; Maggi, F.; Gross, S.; Simoni, G.; et al. The type of feto-placental aneuploidy detected by cfDNA testing may influence the choice of confirmatory diagnostic procedure. Prenat. Diagn. 2015, 35, 994–998. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Y.; Tian, F.; Zhang, J.; Song, Z.; Wu, Y.; Han, X.; Hu, W.; Ma, D.; Cram, D.; et al. Maternal mosaicism is a significant contributor to discordant sex chromosomal aneuploidies associated with noninvasive prenatal testing. Clin. Chem. 2014, 60, 251–259. [Google Scholar] [CrossRef]
- Rolnik, D.L.; Yong, Y.; Lee, T.J.; Tse, C.; McLennan, A.C.; da Silva, C.F. Influence of body mass index on fetal fraction increase with gestation and cell-free DNA test failure. Obstet. Gynecol. 2018, 132, 436–443. [Google Scholar] [CrossRef]
- Caldwell, S.; Almasri, E.; Schmidt, L.; Xu, C.; Dyr, B.; Wardrop, J.; Cacheris, P. Not all low fetal fraction cell-free DNA screening failures increase the risk of aneuploidy. Prenat. Diagn. 2021, 41, 1372–1379. [Google Scholar] [CrossRef]

| Characteristic | Results |
|---|---|
| Sample size | 9327 |
| Maternal age (y), mean ± SD | 36.3 ± 3.63 (range 23 to 47 years) |
| Maternal weight (kg), mean ± SD (IQR) * | 58.8 ± 9.27 (52.4, 63.4) |
| Maternal height (cm), mean ± SD (IQR) | 162.5 ± 5.08 (159.0, 166.0) |
| Body mass index (kg/m2), mean ± SD (IQR) | 22.3 ± 3.33 (19.9, 23.8) |
| Method of conception (%) | |
| Spontaneous | 5672 (60.81) |
| In vitro fertilization | 3655 (39.19) |
| Gestational age (weeks and days), mean (IQR) | 12 weeks 3.7 day (11 w 3 d, 12 w 4 d) |
| Fetus gender (%) ** | |
| Male | 4704 (50.43) |
| Female | 4582 (49.13) |
| Inability to conclude on sex chromosomes | 41 (0.44) |
| Group | Results | Subjects (n) | Portion (%) | Fetal Fraction (Mean % ± SD, p Value *) | BMI (Mean % ± SD, p Value *) |
|---|---|---|---|---|---|
| Passed test | 1st test passed | 9013 | 96.63 | 9.31 ± 3.23 | 22.15 ± 3.24 |
| 2nd test passed (resample and retest) | 218 | 2.34 | 4.48 ± 1.86, <0.001 | 24.71 ± 3.70, <0.001 | |
| Hemolysis | 16 | 0.17 | 9.50 ± 5.60, 0.89 | 22.16 ± 3.19, 0.98 | |
| No-call | Replace other diagnosis | 42 | 0.45 | 3.26 ± 0.93, <0.001 | 28.03 ± 4.98, <0.001 |
| 2nd test fail (no-call) | 35 | 0.38 | 3.85 ± 1.02, <0.001 | 27.48 ± 4.99, <0.001 | |
| IUFD ** before results | 3 | 0.03 | 6.44 ± 3.96, 0.413 | 20.60 ± 1.56, 0.296 |
| BMI Group | Sample (n) | Fetal Fraction (Mean% ± SD) | p Value * | |||
|---|---|---|---|---|---|---|
| Total | Male | Female | Not Determined for Sex ** | |||
| Underweight | 714 | 10.45 ± 3.53 | 12.32 ±3.81 | 8.61 ± 3.81 | 8.95 ± 1.96 | <0.001 |
| Normal | 5400 | 9.71 ± 3.28 | 11.24 ± 3.61 | 8.08 ± 1.90 | 11.84 ± 4.90 | |
| Overweight | 1571 | 8.60 ± 2.82 | 9.66 ± 3.35 | 7.42 ± 1.72 | 9.26 ± 2.37 | <0.001 |
| Obese | 1330 | 7.76 ± 2.81 | 8.49 ± 3.44 | 6.68 ± 1.69 | 8.51 ± 2.56 | <0.001 |
| Highly obese | 312 | 6.37 ± 2.05 | 6.58 ± 2.49 | 5.97 ± 1.58 | - | <0.05 |
| Reason for No-Call | 1st Diagnosis Failed (n, %) | 2nd Diagnosis (n, % *) | ||
|---|---|---|---|---|
| Alternative Test | Resample | No-Call | ||
| Underweight, n = 714 | 10 (0.01) * | 9 (1.26) | 1 (0.14) | |
| Low FF | 4 (0.56) ** | 4 | 1 | |
| Fluctuated z-score | 4 (0.56) ** | 3 | ||
| Hemolysis | 2 (0.28) ** | 2 | ||
| Normal, n = 5400 | 95 (0.06) * | 12 (0.22) | 77 (1.43) | 6 (0.11) |
| Low FF | 63 (3.22) ** | 10 | 49 | 4 |
| Fluctuated z-score | 23 (8.82) ** | 2 | 19 | 2 |
| Hemolysis | 9 (1.26) ** | 9 | ||
| Overweight, n = 1571 | 52 (0.04) ** | 4 (0.25) | 44 (2.80) | 4 (0.25) |
| Low FF | 42 (5.88) ** | 4 | 34 | 4 |
| Fluctuated z-score | 10 (1.40) ** | 10 | ||
| Obese, n =1330 | 111 (0.14) * | 15 (1.13) | 83 (6.24) | 13 (0.98) |
| Low FF | 92 (12.89) ** | 14 | 67 | 11 |
| Fluctuated z-score | 14 (1.96) ** | 1 | 11 | 2 |
| Hemolysis | 5 (0.70) ** | 5 | ||
| Highly obese, n =312 | 46 (6.44) * | 14(4.49) | 21(6.73) | 11 (3.53) |
| Low FF | 42 (5.88) ** | 14 | 18 | 10 |
| Fluctuated z-score | 4 (0.56) ** | 3 | 1 | |
| Sum | 314 (3.37) * | 45(0.48) * | 234(2.51) * | 35(0.38) * |
| GA Group | Sample (n) | Fetal Fraction (%) (Mean ± SD) | p Value * |
|---|---|---|---|
| <10 | 52 | 9.02 ± 3.33 | 0.77 |
| 11–12 | 7912 | 9.15 ± 3.27 | |
| 13–14 | 828 | 8.97 ± 3.38 | 0.14 |
| 15–16 | 343 | 9.19 ± 3.71 | 0.80 |
| 17–18 | 155 | 9.51 ± 3.28 | 0.18 |
| 19–20 | 12 | 8.47 ± 2.52 | 0.47 |
| >21 | 25 | 17.16 ± 2.29 | <0.05 |
| Maternal Age (Years) | Samples (n) | Portion (%) | Fetal Fraction (Mean %, SD) | p Value * |
|---|---|---|---|---|
| 20–29 | 359 | 3.8 | 9.50 ± 3.31 | 0.13 |
| 30–39 | 7201 | 77.2 | 9.23 ± 3.34 | |
| 40–49 | 1767 | 18.9 | 8.74 ± 3.20 | <0.001 |
| Alternative Test | Cases | Follow-Up |
|---|---|---|
| Invasive test (Amniocentesis or CVS) | 37 | 36 of 37 were identified as normal diploid, but one fetus was identified as 46,00,der?(14) via amniocentesis. A patient with normal amniocentesis was diagnosed with acute myeloid leukemia (AML) in the third trimester of pregnancy. |
| Non-invasive test (Quadruple or integrated test) | 7 | 5 out of 7 women received a low-risk result in the serum test, but 2 patients obtained scores of 2.725 and 2.90, respectively in the neural tube defect risk test. These two patients were judged as low-risk group via the AFP test. |
| Follow-up loss | 33 | Thirty-three pregnant women either transferred to another institution for examination or refused further examination. |
| IUFD | 3 | Three pregnant women underwent IUFD at 11 w + 5 d, 12 w, and 12 w + 6 d, during the NIPT or after determining the no-call result. Three cases had a fetal fraction of 3.04%, 4.28%, and 12.0%, respectively. |
| High Risk | True Positive | False Positive | Follow-Up Loss | PPV (%) | |
|---|---|---|---|---|---|
| Trisomy 21 | 56 | 45 | 1 * | 10 | 97.8 |
| Trisomy 18 | 21 | 9 | 10 | 2 | 47.4 |
| Trisomy 13 | 8 | 1 | 6 | 1 | 14.3 |
| Autosomal abnormality | 20 | 1 | 15 | 4 | 6.3 |
| Sex chromosome abnormality | 41 | 15 | 19 | 7 | 44.1 |
| Abnormality or variation in maternal origin | 19 | 12 ** | 1 *** | 6 | 92.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, J.E.; Kang, K.M.; Kim, H.; Jang, H.Y.; Go, M.; Yang, S.H.; Jeong, D.; Jeong, H.; Kim, J.C.; Lim, S.Y.; et al. Cell-Free Fetal DNA Screening Analysis in Korean Pregnant Women: Six Years of Experience and a Retrospective Study of 9327 Patients Analyzed from 2017 to 2022. J. Pers. Med. 2023, 13, 1468. https://doi.org/10.3390/jpm13101468
Park JE, Kang KM, Kim H, Jang HY, Go M, Yang SH, Jeong D, Jeong H, Kim JC, Lim SY, et al. Cell-Free Fetal DNA Screening Analysis in Korean Pregnant Women: Six Years of Experience and a Retrospective Study of 9327 Patients Analyzed from 2017 to 2022. Journal of Personalized Medicine. 2023; 13(10):1468. https://doi.org/10.3390/jpm13101468
Chicago/Turabian StylePark, Ji Eun, Kyung Min Kang, Hyunjin Kim, Hee Yeon Jang, Minyeon Go, So Hyun Yang, Daeun Jeong, Hyeonmin Jeong, Jong Chul Kim, Seo Young Lim, and et al. 2023. "Cell-Free Fetal DNA Screening Analysis in Korean Pregnant Women: Six Years of Experience and a Retrospective Study of 9327 Patients Analyzed from 2017 to 2022" Journal of Personalized Medicine 13, no. 10: 1468. https://doi.org/10.3390/jpm13101468
APA StylePark, J. E., Kang, K. M., Kim, H., Jang, H. Y., Go, M., Yang, S. H., Jeong, D., Jeong, H., Kim, J. C., Lim, S. Y., Cha, D. H., & Shim, S. H. (2023). Cell-Free Fetal DNA Screening Analysis in Korean Pregnant Women: Six Years of Experience and a Retrospective Study of 9327 Patients Analyzed from 2017 to 2022. Journal of Personalized Medicine, 13(10), 1468. https://doi.org/10.3390/jpm13101468






