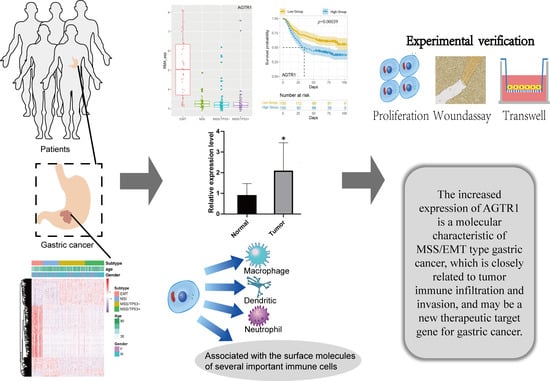
Screening of Differentially Expressed Genes Based on the ACRG Molecular Subtypes of Gastric Cancer and the Significance and Mechanism of AGTR1 Gene Expression
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microarray Data
Screening of Genes Related to the ACRG Subtypes of GC in the Database
2.2. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment Analysis
2.3. Kaplan-Meier Survival Analysis of Differential Gene Expression in Different GC Subtypes
2.4. Tissue Specimens and Immunohistochemical (IHC) Staining of Patient Samples
2.5. The Relationship between AGTR1 and the Genes and Pathways Related to Tumor Immune Infiltration in GC
2.6. The Relationship between AGTR1 and the Immune Cells in Tumor Immune Infiltration in GC
2.7. PPI Network Information
2.8. AGTR1-siRNA Synthesis and Transfection
2.9. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Analysis
2.10. Western Blot Analysis
2.11. Cell Counting Kit-8(CCK-8) Assay
2.12. Wound-Healing Assay
2.13. Transwell Invasion Assay
2.14. Statistical Analysis
3. Results
3.1. Screening of DEGs in GC with Different Molecular Subtypes Using Bioinformatics
3.2. GO and KEGG Enrichment Analysis of the Functions of DEGs
3.3. Analysis of the Expression Levels of DEGs in Different GC Subtypes
3.4. Kaplan-Meier Survival Analysis of the Expression of DEGs in Different GC Subtypes
3.5. AGTR1 Expression Verification
3.6. The Relationship between AGTR1 and Related Pathways Related to Tumor Immune Infiltration in GC
3.7. Gene Association Analysis of the AGTR1-Related Gene Network and Pathways Using the STRING Database
3.8. Relationship between AGTR1 and a Variety of Immune Cells
3.9. AGTR1 Expression in GC Cell Lines
3.10. Effect of AGTR1 on the Activity, Invasion and Migration of GC Cell Lines
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Pretzsch, E.; Bösch, F.; Todorova, R.; Nieß, H.; Jacob, S.; Guba, M.; Kirchner, T.; Werner, J.; Klauschen, F.; Angele, M.K.; et al. Molecular subtyping of gastric cancer according to ACRG using immunohistochemistry—Correlation with clinical parameters. Pathol. Res. Pract. 2022, 231, 153797. [Google Scholar] [CrossRef]
- Chen, X.; Yi, C.H.; Ya, K.G. Renin-angiotensin system inhibitor use and colorectal cancer risk and mortality: A dose-response meta analysis. J. Renin-Angiotensin-Aldosterone Syst. JRAAS 2020, 21, 1470320319895646. [Google Scholar] [CrossRef]
- Afsar, B.; Afsar, R.E.; Ertuglu, L.A.; Kuwabara, M.; Ortiz, A.; Covic, A.; Kanbay, M. Renin-angiotensin system and cancer: Epidemiology, cell signaling, genetics and epigenetics. Clin. Transl. Oncol. 2021, 23, 682–696. [Google Scholar] [CrossRef]
- Moore, A.R.; Rosenberg, S.C.; McCormick, F.; Malek, S. RAS-targeted therapies: Is the undruggable drugged? Nat. Rev. Drug Discov. 2020, 19, 533–552. [Google Scholar] [CrossRef]
- Sun, H.; Li, T.; Zhuang, R.; Cai, W.; Zheng, Y. Do renin-angiotensin system inhibitors influence the recurrence, metastasis, and survival in cancer patients?: Evidence from a meta-analysis including 55 studies. Medicine 2017, 96, e6394. [Google Scholar] [CrossRef]
- Rasha, F.; Ramalingam, L.; Menikdiwela, K.; Hernandez, A.; Moussa, H.; Gollahon, L.; Rahman, R.L.; Moustaid-Moussa, N. Renin angiotensin system inhibition attenuates adipocyte-breast cancer cell interactions. Exp. Cell Res. 2020, 394, 112114. [Google Scholar] [CrossRef]
- Khoshghamat, N.; Jafari, N.; Toloue-Pouya, V.; Azami, S.; Mirnourbakhsh, S.H.; Khazaei, M.; Ferns, G.A.; Rajabian, M.; Avan, A. The therapeutic potential of renin-angiotensin system inhibitors in the treatment of pancreatic cancer. Life Sci. 2021, 270, 119118. [Google Scholar] [CrossRef]
- Xiong, L.; Wei, Y.; Zhou, X.; Dai, P.; Cai, Y.; Zhou, X.; Xu, M.; Zhao, J.; Tang, H. AGTR1 Inhibits the Progression of Lung Adenocarcinoma. Cancer Manag. Res. 2021, 13, 8535–8550. [Google Scholar] [CrossRef]
- Singh, A.; Srivastava, N.; Amit, S.; Prasad, S.N.; Misra, M.P.; Ateeq, B. Association of AGTR1 (A1166C) and ACE (I/D) Polymorphisms with Breast Cancer Risk in North Indian Population. Transl. Oncol. 2018, 11, 233–242. [Google Scholar] [CrossRef]
- Delforce, S.; Lumbers, E.R.; De Meaultsart, C.C.; Wang, Y.; Proietto, A.; Otton, G.; Scurry, J.; Verrills, N.; Scott, R.J.; Pringle, K.G. Expression of renin-angiotensin system (RAS) components in endometrial cancer. Endocr. Connect. 2017, 6, 9–19. [Google Scholar] [CrossRef]
- Zhang, D.; Liu, H.; Yang, B.; Hu, J.; Cheng, Y. L-securinine inhibits cell growth and metastasis of human androgen-independent prostate cancer DU145 cells via regulating mitochondrial and AGTR1/MEK/ERK/STAT3/PAX2 apoptotic pathways. Biosci. Rep. 2019, 39, BSR20190469. [Google Scholar] [CrossRef]
- Cheng, H.; Wang, Y.; Liu, C.; Wu, T.; Chen, S.; Chen, M. Development and Verification of a Prostate Cancer Prognostic Signature Based on an Immunogenomic Landscape Analysis. Front. Oncol. 2021, 11, 711258. [Google Scholar] [CrossRef]
- Zhang, Q.; Yu, S.; Lam, M.M.T.; Poon, T.C.W.; Sun, L.; Jiao, Y.; Wong, A.S.T.; Lee, L.T.O. Angiotensin II promotes ovarian cancer spheroid formation and metastasis by upregulation of lipid desaturation and suppression of endoplasmic reticulum stress. J. Exp. Clin. Cancer Res. CR 2019, 38, 116. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Xia, Z.; Ye, C.; Lu, C.; Zhou, S.; Pan, J.; Liu, C.; Zhang, J.; Liu, T.; Hu, T.; et al. AGTR1 promotes lymph node metastasis in breast cancer by upregulating CXCR4/SDF-1α and inducing cell migration and invasion. Aging 2019, 11, 3969–3992. [Google Scholar] [CrossRef]
- Qiao, Z.W.; Jiang, Y.; Wang, L.; Wang, L.; Jiang, J.; Zhang, J.R.; Mu, P. LINC00852 promotes the proliferation and invasion of ovarian cancer cells by competitively binding with miR-140-3p to regulate AGTR1 expression. BMC Cancer 2021, 21, 1004. [Google Scholar] [CrossRef]
- Wang, Q.; Xie, Q.; Liu, Y.; Guo, H.; Ren, Y.; Li, J.; Zhao, Q. Clinical characteristics and prognostic significance of TCGA and ACRG classification in gastric cancer among the Chinese population. Mol. Med. Rep. 2020, 22, 828–840. [Google Scholar] [CrossRef]
- Pickup, M.W.; Mouw, J.K.; Weaver, V.M. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014, 15, 1243–1253. [Google Scholar] [CrossRef] [Green Version]
- Gelse, K.; Pöschl, E.; Aigner, T. Collagens—Structure, function, and biosynthesis. Adv. Drug Deliv. Rev. 2003, 55, 1531–1546. [Google Scholar] [CrossRef] [Green Version]
- Ricard-Blum, S. The collagen family. Cold Spring Harb. Perspect. Biol. 2011, 3, a004978. [Google Scholar] [CrossRef] [Green Version]
- Sjöblom, B.; Salmazo, A.; Djinović-Carugo, K. Alpha-actinin structure and regulation. Cell. Mol. Life Sci. CMLS 2008, 65, 2688–2701. [Google Scholar] [CrossRef]
- Izdebska, M.; Zielińska, W.; Hałas-Wiśniewska, M.; Grzanka, A. Involvement of Actin and Actin-Binding Proteins in Carcinogenesis. Cells 2020, 9, 2245. [Google Scholar] [CrossRef]
- Sadras, F.; Monteith, G.R.; Roberts-Thomson, S.J. An Emerging Role for Calcium Signaling in Cancer-Associated Fibroblasts. Int. J. Mol. Sci. 2021, 22, 11366. [Google Scholar] [CrossRef]
- Brown, N.H.; Gregory, S.L.; Rickoll, W.L.; Fessler, L.I.; Prout, M.; White, R.A.; Fristrom, J.W. Talin is essential for integrin function in Drosophila. Dev. Cell 2002, 3, 569–579. [Google Scholar] [CrossRef]
- Weber, M.; Hauschild, R.; Schwarz, J.; Moussion, C.; de Vries, I.; Legler, D.F.; Luther, S.A.; Bollenbach, T.; Sixt, M. Interstitial dendritic cell guidance by haptotactic chemokine gradients. Science 2013, 339, 328–332. [Google Scholar] [CrossRef] [Green Version]
- Hara, K.; Nakagawa, T.; Enomoto, H.; Suzuki, M.; Yamamoto, M.; Simons, B.D.; Yoshida, S. Mouse spermatogenic stem cells continually interconvert between equipotent singly isolated and syncytial states. Cell Stem Cell 2014, 14, 658–672. [Google Scholar] [CrossRef] [Green Version]
- Schaffer, A.E.; Breuss, M.W.; Caglayan, A.O.; Al-Sanaa, N.; Al-Abdulwahed, H.Y.; Kaymakçalan, H.; Yılmaz, C.; Zaki, M.S.; Rosti, R.O.; Copeland, B.; et al. Biallelic loss of human CTNNA2, encoding αN-catenin, leads to ARP2/3 complex overactivity and disordered cortical neuronal migration. Nat. Genet. 2018, 50, 1093–1101. [Google Scholar] [CrossRef]
- Cheung, K.J.; Gabrielson, E.; Werb, Z.; Ewald, A.J. Collective invasion in breast cancer requires a conserved basal epithelial program. Cell 2013, 155, 1639–1651. [Google Scholar] [CrossRef] [Green Version]
- Mok, S.; Tsoi, J.; Koya, R.C.; Hu-Lieskovan, S.; West, B.L.; Bollag, G.; Graeber, T.G.; Ribas, A. Inhibition of colony stimulating factor-1 receptor improves antitumor efficacy of BRAF inhibition. BMC Cancer 2015, 15, 356. [Google Scholar] [CrossRef] [Green Version]
- Hao, Q.; Vadgama, J.V.; Wang, P. CCL2/CCR2 signaling in cancer pathogenesis. Cell Commun. Signal. 2020, 18, 82. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Ge, Y.; Xiao, M.; Lopez-Coral, A.; Azuma, R.; Somasundaram, R.; Zhang, G.; Wei, Z.; Xu, X.; Rauscher, F.J.; et al. Melanoma-derived conditioned media efficiently induce the differentiation of monocytes to macrophages that display a highly invasive gene signature. Pigment Cell Melanoma Res. 2012, 25, 493–505. [Google Scholar] [CrossRef] [Green Version]
- Yoon, J.Y.; Sy, K.; Brezden-Masley, C.; Streutker, C.J. Histo- and immunohistochemistry-based estimation of the TCGA and ACRG molecular subtypes for gastric carcinoma and their prognostic significance: A single-institution study. PLoS ONE 2019, 14, e0224812. [Google Scholar] [CrossRef]
- Pinto, M.P.; Córdova-Delgado, M.; Retamal, I.N.; Muñoz-Medel, M.; Bravo, M.L.; Durán, D.; Villanueva, F.; Sanchez, C.; Acevedo, F.; Mondaca, S.; et al. A Molecular Stratification of Chilean Gastric Cancer Patients with Potential Clinical Applicability. Cancers 2020, 12, 1863. [Google Scholar] [CrossRef] [PubMed]
- Puliga, E.; Corso, S.; Pietrantonio, F.; Giordano, S. Microsatellite instability in Gastric Cancer: Between lights and shadows. Cancer Treat. Rev. 2021, 95, 102175. [Google Scholar] [CrossRef] [PubMed]
- Patel, T.H.; Cecchini, M. Targeted Therapies in Advanced Gastric Cancer. Curr. Treat. Options Oncol. 2020, 21, 70. [Google Scholar] [CrossRef] [PubMed]
- Pisanidou, V.; Apostolou, P.; Beis, G.; Hatzidaki, E.; Papasotiriou, I. Cancer Comprehensive Analysis in Gastric Carcinoma: Benefits and New Perspectives. Case Rep. Oncol. 2021, 14, 1682–1690. [Google Scholar] [CrossRef]
- Ekambaram, P.; Lee, J.L.; Hubel, N.E.; Hu, D.; Yerneni, S.; Campbell, P.G.; Pollock, N.; Klei, L.R.; Concel, V.J.; Delekta, P.C.; et al. The CARMA3-Bcl10-MALT1 Signalosome Drives NFκB Activation and Promotes Aggressiveness in Angiotensin II Receptor-Positive Breast Cancer. Cancer Res. 2018, 78, 1225–1240. [Google Scholar] [CrossRef] [Green Version]
- Moschetta-Pinheiro, M.G.; Colombo, J.; Godoy, B.L.V.; Balan, J.F.; Nascimento, B.C.; Zuccari, D. Modulation of Epithelial Mesenchymal Transition after AGTR-1 Gene Edition by Crispr/Cas9 and Losartan Treatment in Mammary Tumor Cell Line: A Comparative Study between Human and Canine Species. Life 2021, 11, 1427. [Google Scholar] [CrossRef]
- Exner, S.; Schuldt, C.; Sachindra, S.; Du, J.; Heing-Becker, I.; Licha, K.; Wiedenmann, B.; Grötzinger, C. AGTR1 Is Overexpressed in Neuroendocrine Neoplasms, Regulates Secretion and May Potentially Serve as a Target for Molecular Imaging and Therapy. Cancers 2020, 12, 3138. [Google Scholar] [CrossRef]
- Singh, A.; Srivastava, N.; Yadav, A.; Ateeq, B. Targeting AGTR1/NF-κB/CXCR4 axis by miR-155 attenuates oncogenesis in glioblastoma. Neoplasia 2020, 22, 497–510. [Google Scholar] [CrossRef] [PubMed]
- Renziehausen, A.; Wang, H.; Rao, B.; Weir, L.; Nigro, C.L.; Lattanzio, L.; Merlano, M.; Rioja, A.V.; Fernandez-Carranco, M.D.C.; Hajji, N.; et al. The renin angiotensin system (RAS) mediates bifunctional growth regulation in melanoma and is a novel target for therapeutic intervention. Oncogene 2019, 38, 2320–2336. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Zhang, Y.; Hao, M.; Zhu, R. LINC00665 functions as a competitive endogenous RNA to regulate AGTR1 expression by sponging miR-34a-5p in glioma. Oncol. Rep. 2021, 45, 1202–1212. [Google Scholar] [CrossRef] [PubMed]
- Panza, S.; Malivindi, R.; Caruso, A.; Russo, U.; Giordano, F.; Győrffy, B.; Gelsomino, L.; De Amicis, F.; Barone, I.; Conforti, F.L.; et al. Novel Insights into the Antagonistic Effects of Losartan against Angiotensin II/AGTR1 Signaling in Glioblastoma Cells. Cancers 2021, 13, 4555. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Zhen, S.; Ding, P.; Tan, B.; Wang, H.; Liu, W.; Tian, Y.; Zhao, Q. Screening of Differentially Expressed Genes Based on the ACRG Molecular Subtypes of Gastric Cancer and the Significance and Mechanism of AGTR1 Gene Expression. J. Pers. Med. 2023, 13, 560. https://doi.org/10.3390/jpm13030560
Zhang H, Zhen S, Ding P, Tan B, Wang H, Liu W, Tian Y, Zhao Q. Screening of Differentially Expressed Genes Based on the ACRG Molecular Subtypes of Gastric Cancer and the Significance and Mechanism of AGTR1 Gene Expression. Journal of Personalized Medicine. 2023; 13(3):560. https://doi.org/10.3390/jpm13030560
Chicago/Turabian StyleZhang, Haoran, Shuman Zhen, Pingan Ding, Bibo Tan, Hongyan Wang, Wenbo Liu, Yuan Tian, and Qun Zhao. 2023. "Screening of Differentially Expressed Genes Based on the ACRG Molecular Subtypes of Gastric Cancer and the Significance and Mechanism of AGTR1 Gene Expression" Journal of Personalized Medicine 13, no. 3: 560. https://doi.org/10.3390/jpm13030560
APA StyleZhang, H., Zhen, S., Ding, P., Tan, B., Wang, H., Liu, W., Tian, Y., & Zhao, Q. (2023). Screening of Differentially Expressed Genes Based on the ACRG Molecular Subtypes of Gastric Cancer and the Significance and Mechanism of AGTR1 Gene Expression. Journal of Personalized Medicine, 13(3), 560. https://doi.org/10.3390/jpm13030560