Contemporary Biomarkers for Renal Transplantation: A Narrative Overview
Abstract
:1. Introduction
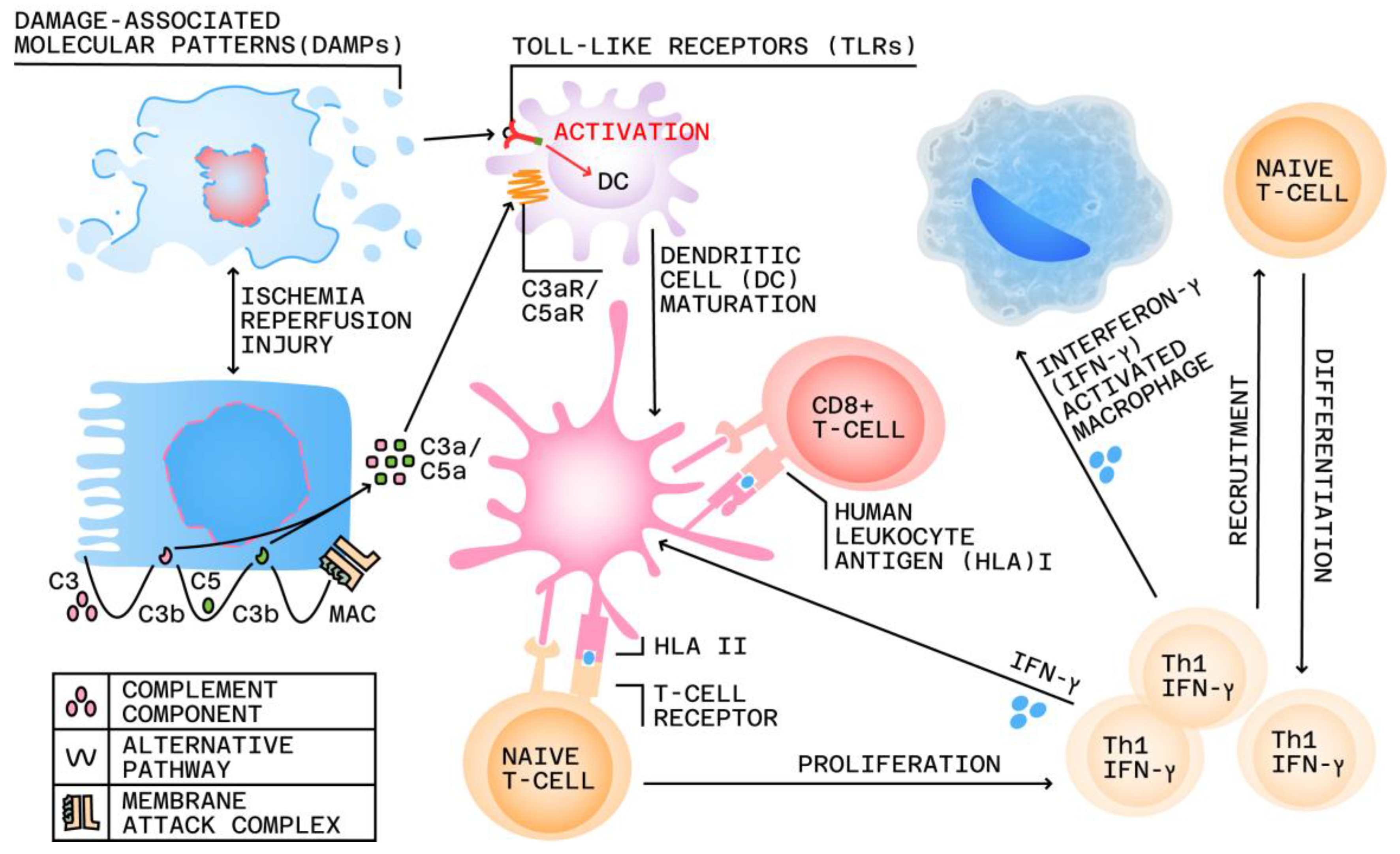
2. Immunopathology of Nephron Injury and Allograft Rejection
3. Glomerular vs. Tubular Biomarkers for Allograft Nephron Damage Assessment
4. Biomarkers for Non-Surgical Renal Allograft Complications
4.1. Acute Allograft Complications
4.1.1. Delayed Allograft Function
4.1.2. Acute Allograft Rejection
4.2. Chronic Allograft Rejection vs. Dysfunction
5. Immune Tolerance and Therapeutic Drug Monitoring
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, F.; Liao, M.; Wang, P.; Yang, Z.; Liu, Y. The Cost-Effectiveness of Kidney Replacement Therapy Modalities: A Systematic Review of Full Economic Evaluations. Appl. Health Econ. Health Policy 2021, 19, 163–180. [Google Scholar] [CrossRef] [PubMed]
- Peddi, V.R.; First, M.R. Recent Advances in Immunosuppressive Therapy for Renal Transplantation. Semin. Dial. 2001, 14, 218–222. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kriesche, H.-U.; Schold, J.D.; Srinivas, T.R.; Kaplan, B. Lack of Improvement in Renal Allograft Survival despite a Marked Decrease in Acute Rejection Rates over the Most Recent Era. Am. J. Transpl. 2004, 4, 378–383. [Google Scholar] [CrossRef]
- Salvadori, M.; Tsalouchos, A. Biomarkers in Renal Transplantation: An Updated Review. World J. Transpl. 2017, 7, 161–178. [Google Scholar] [CrossRef] [PubMed]
- Redfield, R.R.; McCune, K.R.; Rao, A.; Sadowski, E.; Hanson, M.; Kolterman, A.J.; Robbins, J.; Guite, K.; Mohamed, M.; Parajuli, S.; et al. Nature, Timing, and Severity of Complications from Ultrasound-guided Percutaneous Renal Transplant Biopsy. Transpl. Int. 2016, 29, 167–172. [Google Scholar] [CrossRef] [Green Version]
- Naesens, M.; Khatri, P.; Li, L.; Sigdel, T.K.; Vitalone, M.J.; Chen, R.; Butte, A.J.; Salvatierra, O.; Sarwal, M.M. Progressive Histological Damage in Renal Allografts Is Associated with Expression of Innate and Adaptive Immunity Genes. Kidney Int. 2011, 80, 1364–1376. [Google Scholar] [CrossRef] [Green Version]
- Biomarkers Definitions Working Group. Biomarkers and Surrogate Endpoints: Preferred Definitions and Conceptual Framework. Clin. Pharmacol. Ther. 2001, 69, 89–95. [Google Scholar] [CrossRef]
- Loupy, A.; Haas, M.; Solez, K.; Racusen, L.; Glotz, D.; Seron, D.; Nankivell, B.J.; Colvin, R.B.; Afrouzian, M.; Akalin, E.; et al. The Banff 2015 Kidney Meeting Report: Current Challenges in Rejection Classification and Prospects for Adopting Molecular Pathology. Am. J. Transplant. 2017, 17, 28–41. [Google Scholar] [CrossRef]
- Swanson, K.J.; Aziz, F.; Garg, N.; Mohamed, M.; Mandelbrot, D.; Djamali, A.; Parajuli, S. Role of Novel Biomarkers in Kidney Transplantation. World J. Transpl. 2020, 10, 230–255. [Google Scholar] [CrossRef]
- Kępka, A.; Waszkiewicz, N.; Chojnowska, S.; Zalewska-Szajda, B.; Ładny, J.R.; Wasilewska, A.; Zwierz, K.; Szajda, S.D.; Kępka, A.; Waszkiewicz, N.; et al. Utility of Urinary Biomarkers in Kidney Transplant Function Assessment. In Current Issues and Future Direction in Kidney Transplantation; IntechOpen: London, UK, 2013. [Google Scholar] [CrossRef] [Green Version]
- Mao, S.; Wang, C.; Dong, G. Evaluation of Inter-Laboratory and Cross-Platform Concordance of DNA Microarrays through Discriminating Genes and Classifier Transferability. J. Bioinform. Comput. Biol. 2009, 7, 157–173. [Google Scholar] [CrossRef]
- Sato, F.; Tsuchiya, S.; Terasawa, K.; Tsujimoto, G. Intra-Platform Repeatability and Inter-Platform Comparability of MicroRNA Microarray Technology. PLoS ONE 2009, 4, e5540. [Google Scholar] [CrossRef] [Green Version]
- Menon, M.C.; Murphy, B.; Heeger, P.S. Moving Biomarkers toward Clinical Implementation in Kidney Transplantation. J. Am. Soc. Nephrol. 2017, 28, 735. [Google Scholar] [CrossRef] [Green Version]
- Lo, D.J.; Kaplan, B.; Kirk, A.D. Biomarkers for Kidney Transplant Rejection. Nat. Rev. Nephrol. 2014, 10, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.; Terasaki, P.I. Significance of the Positive Crossmatch Test in Kidney Transplantation. N. Engl. J. Med. 1969, 280, 735–739. [Google Scholar] [CrossRef]
- Eikmans, M.; Gielis, E.M.; Ledeganck, K.J.; Yang, J.; Abramowicz, D.; Claas, F.F.J. Non-Invasive Biomarkers of Acute Rejection in Kidney Transplantation: Novel Targets and Strategies. Front. Med. 2019, 5, 358. [Google Scholar] [CrossRef]
- Naesens, M.; Kuypers, D.R.J.; De Vusser, K.; Evenepoel, P.; Claes, K.; Bammens, B.; Meijers, B.; Sprangers, B.; Pirenne, J.; Monbaliu, D.; et al. The Histology of Kidney Transplant Failure: A Long-Term Follow-Up Study. Transplantation 2014, 98, 427. [Google Scholar] [CrossRef] [PubMed]
- Chong, A.S. Mechanisms of Organ Transplant Injury Mediated by B Cells and Antibodies: Implications for Antibody-Mediated Rejection. Am. J. Transplant. 2020, 20, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Francis, J.; Gautam, A.; Pelletier, L.; Sanchorawala, V.; Quillen, K. Durable Renal Response after Combination of Bortezomib, Corticosteroids, Rituximab, and Plasmapheresis for Late Antibody-Mediated Renal Transplant Rejection. Clin. Nephrol. 2018, 89, 252–259. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, J.G.; Samaniego, M.; Barrio, M.C.; Potena, L.; Zeevi, A.; Djamali, A.; Cozzi, E. The Influence of Immunosuppressive Agents on the Risk of De Novo Donor-Specific HLA Antibody Production in Solid Organ Transplant Recipients. Transplantation 2016, 100, 39–53. [Google Scholar] [CrossRef] [Green Version]
- Filler, G.; Todorova, E.K.; Bax, K.; Alvarez-Elías, A.C.; Huang, S.-H.S.; Kobrzynski, M.C. Minimum Mycophenolic Acid Levels Are Associated with Donor-Specific Antibody Formation. Pediatr. Transplant. 2016, 20, 34–38. [Google Scholar] [CrossRef]
- Ginevri, F.; Nocera, A.; Comoli, P.; Innocente, A.; Cioni, M.; Parodi, A.; Fontana, I.; Magnasco, A.; Nocco, A.; Tagliamacco, A.; et al. Posttransplant De Novo Donor-Specific HLA Antibodies Identify Pediatric Kidney Recipients at Risk for Late Antibody-Mediated Rejection. Am. J. Transplant. 2012, 12, 3355–3362. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.J.; Balasubramanian, R.; Michaelides, G.; Wittenhagen, P.; Sebire, N.J.; Mamode, N.; Shaw, O.; Vaughan, R.; Marks, S.D. The Clinical Spectrum of De Novo Donor-Specific Antibodies in Pediatric Renal Transplant Recipients. Am. J. Transplant. 2014, 14, 2350–2358. [Google Scholar] [CrossRef] [PubMed]
- Sellarés, J.; de Freitas, D.G.; Mengel, M.; Reeve, J.; Einecke, G.; Sis, B.; Hidalgo, L.G.; Famulski, K.; Matas, A.; Halloran, P.F. Understanding the Causes of Kidney Transplant Failure: The Dominant Role of Antibody-Mediated Rejection and Nonadherence. Am. J. Transplant. 2012, 12, 388–399. [Google Scholar] [CrossRef] [PubMed]
- Loupy, A.; Lefaucheur, C.; Vernerey, D.; Prugger, C.; van Huyen, J.-P.D.; Mooney, N.; Suberbielle, C.; Frémeaux-Bacchi, V.; Méjean, A.; Desgrandchamps, F.; et al. Complement-Binding Anti-HLA Antibodies and Kidney-Allograft Survival. N. Engl. J. Med. 2013, 369, 1215–1226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macedo, C.; Orkis, E.A.; Popescu, I.; Elinoff, B.D.; Zeevi, A.; Shapiro, R.; Lakkis, F.G.; Metes, D. Contribution of Naïve and Memory T-Cell Populations to the Human Alloimmune Response. Am. J. Transplant. 2009, 9, 2057–2066. [Google Scholar] [CrossRef]
- Wood, K.J.; Goto, R. Mechanisms of Rejection: Current Perspectives. Transplantation 2012, 93, 1. [Google Scholar] [CrossRef] [Green Version]
- Nankivell, B.J.; Alexander, S.I. Rejection of the Kidney Allograft. N. Engl. J. Med. 2010, 363, 1451–1462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segerer, S.; Cui, Y.; Eitner, F.; Goodpaster, T.; Hudkins, K.L.; Mack, M.; Cartron, J.-P.; Colin, Y.; Schlondorff, D.; Alpers, C.E. Expression of Chemokines and Chemokine Receptors during Human Renal Transplant Rejection. Am. J. Kidney Dis. 2001, 37, 518–531. [Google Scholar] [CrossRef]
- Zuidwijk, K.; de Fijter, J.W.; Mallat, M.J.K.; Eikmans, M.; van Groningen, M.C.; Goemaere, N.N.; Bajema, I.M.; van Kooten, C. Increased Influx of Myeloid Dendritic Cells during Acute Rejection Is Associated with Interstitial Fibrosis and Tubular Atrophy and Predicts Poor Outcome. Kidney Int. 2012, 81, 64–75. [Google Scholar] [CrossRef] [Green Version]
- Requião-Moura, L.R.; de Durão, M.S.; de Matos, A.C.C.; Pacheco-Silva, A. Ischemia and Reperfusion Injury in Renal Transplantation: Hemodynamic and Immunological Paradigms. Einstein 2015, 13, 129–135. [Google Scholar] [CrossRef]
- Leemans, J.C.; Kors, L.; Anders, H.-J.; Florquin, S. Pattern Recognition Receptors and the Inflammasome in Kidney Disease. Nat. Rev. Nephrol. 2014, 10, 398–414. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, L.A.J.; Golenbock, D.; Bowie, A.G. The History of Toll-like Receptors—Redefining Innate Immunity. Nat. Rev. Immunol. 2013, 13, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Devarajan, P. Neutrophil Gelatinase-Associated Lipocalin: A Promising Biomarker for Human Acute Kidney Injury. Biomark. Med. 2010, 4, 265–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acute Kidney Injury (AKI)—KDIGO. Available online: https://kdigo.org/guidelines/acute-kidney-injury/ (accessed on 27 May 2023).
- Hull, R.P.; Goldsmith, D.J.A. Nephrotic Syndrome in Adults. BMJ 2008, 336, 1185–1189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grahammer, F.; Schell, C.; Huber, T.B. The Podocyte Slit Diaphragm—From a Thin Grey Line to a Complex Signalling Hub. Nat. Rev. Nephrol. 2013, 9, 587–598. [Google Scholar] [CrossRef]
- Ahmad, A.; Roderick, P.; Ward, M.; Steenkamp, R.; Burden, R.; O’Donoghue, D.; Ansell, D.; Feest, T. Current Chronic Kidney Disease Practice Patterns in the UK: A National Survey. QJM 2006, 99, 245–251. [Google Scholar] [CrossRef] [Green Version]
- Lisowska-Myjak, B. Serum and Urinary Biomarkers of Acute Kidney Injury. Blood Purif. 2010, 29, 357–365. [Google Scholar] [CrossRef]
- Alachkar, N.; Rabb, H.; Jaar, B.G. Urinary Biomarkers in Acute Kidney Transplant Dysfunction. Nephron Clin. Pract. 2011, 118, c173–c181; discussion c181. [Google Scholar] [CrossRef]
- Metzger, J.; Kirsch, T.; Schiffer, E.; Ulger, P.; Mentes, E.; Brand, K.; Weissinger, E.M.; Haubitz, M.; Mischak, H.; Herget-Rosenthal, S. Urinary Excretion of Twenty Peptides Forms an Early and Accurate Diagnostic Pattern of Acute Kidney Injury. Kidney Int. 2010, 78, 1252–1262. [Google Scholar] [CrossRef] [Green Version]
- Finney, H.; Newman, D.J.; Thakkar, H.; Fell, J.M.; Price, C.P. Reference Ranges for Plasma Cystatin C and Creatinine Measurements in Premature Infants, Neonates, and Older Children. Arch. Dis. Child. 2000, 82, 71–75. [Google Scholar] [CrossRef] [Green Version]
- Filler, G.; Bökenkamp, A.; Hofmann, W.; Le Bricon, T.; Martínez-Brú, C.; Grubb, A. Cystatin C as a Marker of GFR--History, Indications, and Future Research. Clin. Biochem. 2005, 38, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Campo, A.; Lanfranco, G.; Gramaglia, L.; Goia, F.; Cottino, R.; Giusto, V. Could Plasma Cystatin C Be Useful as a Marker of Hemodialysis Low Molecular Weight Proteins Removal? Nephron Clin. Pract. 2004, 98, c79–c82. [Google Scholar] [CrossRef] [PubMed]
- Herget-Rosenthal, S.; Feldkamp, T.; Volbracht, L.; Kribben, A. Measurement of Urinary Cystatin C by Particle-Enhanced Nephelometric Immunoassay: Precision, Interferences, Stability and Reference Range. Ann. Clin. Biochem. 2004, 41, 111–118. [Google Scholar] [CrossRef] [Green Version]
- Haraldsson, B.; Sörensson, J. Why Do We Not All Have Proteinuria? An Update of Our Current Understanding of the Glomerular Barrier. News Physiol. Sci. 2004, 19, 7–10. [Google Scholar] [CrossRef] [Green Version]
- Halbesma, N.; Kuiken, D.-S.; Brantsma, A.H.; Bakker, S.J.L.; Wetzels, J.F.M.; De Zeeuw, D.; De Jong, P.E.; Gansevoort, R.T. Macroalbuminuria Is a Better Risk Marker than Low Estimated GFR to Identify Individuals at Risk for Accelerated GFR Loss in Population Screening. J. Am. Soc. Nephrol. 2006, 17, 2582–2590. [Google Scholar] [CrossRef] [Green Version]
- Abbate, M.; Zoja, C.; Remuzzi, G. How Does Proteinuria Cause Progressive Renal Damage? J. Am. Soc. Nephrol. 2006, 17, 2974–2984. [Google Scholar] [CrossRef] [Green Version]
- Eddy, A.A. Proteinuria and Interstitial Injury. Nephrol. Dial. Transplant. 2004, 19, 277–281. [Google Scholar] [CrossRef] [Green Version]
- Tryggvason, K.; Pettersson, E. Causes and Consequences of Proteinuria: The Kidney Filtration Barrier and Progressive Renal Failure. J. Intern. Med. 2003, 254, 216–224. [Google Scholar] [CrossRef] [Green Version]
- Zoja, C.; Morigi, M.; Remuzzi, G. Proteinuria and Phenotypic Change of Proximal Tubular Cells. J. Am. Soc. Nephrol. 2003, 14 (Suppl. 1), S36–S41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ofstad, J.; Iversen, B.M. Glomerular and Tubular Damage in Normotensive and Hypertensive Rats. Am. J. Physiol.-Ren. Physiol. 2005, 288, F665–F672. [Google Scholar] [CrossRef]
- Kang, N.R.; Lee, J.E.; Huh, W.; Kim, S.J.; Kim, Y.-G.; Kim, D.J.; Oh, H.Y. Minimal Proteinuria One Year after Transplant Is a Risk Factor for Graft Survival in Kidney Transplantation. J. Korean Med. Sci. 2009, 24 (Suppl. S1), S129–S134. [Google Scholar] [CrossRef] [Green Version]
- Remuzzi, G.; Benigni, A.; Remuzzi, A. Mechanisms of Progression and Regression of Renal Lesions of Chronic Nephropathies and Diabetes. J. Clin. Investig. 2006, 116, 288–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramesh Prasad, G.v.; Bandukwala, F.; Huang, M.; Zaltzman, J.S. Microalbuminuria Post-Renal Transplantation: Relation to Cardiovascular Risk Factors and C-Reactive Protein. Clin. Transplant. 2009, 23, 313–320. [Google Scholar] [CrossRef]
- Erman, A.; Rahamimov, R.; Mashraki, T.; Levy-Drummer, R.S.; Winkler, J.; David, I.; Hirsh, Y.; Gafter, U.; Chagnac, A. The Urine Albumin-to-Creatinine Ratio: Assessment of Its Performance in the Renal Transplant Recipient Population. Clin. J. Am. Soc. Nephrol. 2011, 6, 892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nauta, F.L.; Bakker, S.J.L.; van Oeveren, W.; Navis, G.; van der Heide, J.J.H.; van Goor, H.; de Jong, P.E.; Gansevoort, R.T. Albuminuria, Proteinuria, and Novel Urine Biomarkers as Predictors of Long-Term Allograft Outcomes in Kidney Transplant Recipients. Am. J. Kidney Dis. 2011, 57, 733–743. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, M.; Eidelman, O.; Torosyan, Y.; Jozwik, C.; Mannon, R.B.; Pollard, H.B. Elevated Expression Levels of ANXA11, Integrins Β3 and A3, and TNF-α Contribute to a Candidate Proteomic Signature in Urine for Kidney Allograft Rejection. PROTEOMICS-Clin. Appl. 2011, 5, 311–321. [Google Scholar] [CrossRef] [Green Version]
- Reinhold, S.W.; Straub, R.H.; Krüger, B.; Kaess, B.; Bergler, T.; Weingart, C.; Banas, M.C.; Krämer, B.K.; Banas, B. Elevated Urinary SVCAM-1, IL6, SIL6R and TNFR1 Concentrations Indicate Acute Kidney Transplant Rejection in the First 2weeks after Transplantation. Cytokine 2012, 57, 379–388. [Google Scholar] [CrossRef]
- Teppo, A.M.; von Willebrand, E.; Honkanen, E.; Ahonen, J.; Grönhagen-Riska, C. Soluble Intercellular Adhesion Molecule-1 (SICAM-1) after Kidney Transplantation: The Origin and Role of Urinary SICAM-1? Transplantation 2001, 71, 1113–1119. [Google Scholar] [CrossRef]
- van Ree, R.M.; Oterdoom, L.H.; de Vries, A.P.J.; Homan van der Heide, J.J.; van Son, W.J.; Navis, G.; Gans, R.O.B.; Bakker, S.J.L. Circulating Markers of Endothelial Dysfunction Interact with Proteinuria in Predicting Mortality in Renal Transplant Recipients. Transplantation 2008, 86, 1713–1719. [Google Scholar] [CrossRef]
- Gwinner, W. Renal Transplant Rejection Markers. World J. Urol. 2007, 25, 445–455. [Google Scholar] [CrossRef]
- Guder, W.G.; Hofmann, W. Clinical Role of Urinary Low Molecular Weight Proteins: Their Diagnostic and Prognostic Implications. Scand. J. Clin. Lab. Investig. 2008, 68, 95–98. [Google Scholar] [CrossRef] [PubMed]
- Câmara, N.O.S.; Williams, W.W., Jr.; Pacheco-Silva, A. Proximal Tubular Dysfunction as an Indicator of Chronic Graft Dysfunction. Braz. J. Med. Biol. Res. 2009, 42, 229–236. [Google Scholar] [CrossRef] [Green Version]
- Kuźniar, J.; Marchewka, Z.; Krasnowski, R.; Boratyńska, M.; Długosz, A.; Klinger, M. Enzymuria and Low Molecular Weight Protein Excretion as the Differentiating Marker of Complications in the Early Post Kidney Transplantation Period. Int. Urol. Nephrol. 2006, 38, 753–758. [Google Scholar] [CrossRef] [PubMed]
- Uchida, K.; Gotoh, A. Measurement of Cystatin-C and Creatinine in Urine. Clin. Chim. Acta 2002, 323, 121–128. [Google Scholar] [CrossRef]
- Bagshaw, S.M.; Langenberg, C.; Haase, M.; Wan, L.; May, C.N.; Bellomo, R. Urinary Biomarkers in Septic Acute Kidney Injury. Intensive Care Med. 2007, 33, 1285–1296. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, J.H.; Richard, G.A. Urinary Adenosine Deaminase Binding Protein as a Predictor of Renal Transplant Rejection in Children. Transpl. Proc. 1994, 26, 75–76. [Google Scholar]
- Refaie, M.O.I.; Abo-Zaid, H.; Gomma, N.A.; Aboul-Enein, H.Y. Determination of Urinary and Serum β-Glucuronidase and Alkaline Phosphatase in Various Renal Disease and Kidney Rejection Transplanted Patients. Prep. Biochem. Biotechnol. 2000, 30, 93–106. [Google Scholar] [CrossRef]
- Santos, C.; Marcelino, P.; Carvalho, T.; Coelho, J.; Bispo, M.; Mourão, L.; Perdigoto, R.; Barroso, E. The Value of Tubular Enzymes for Early Detection of Acute Kidney Injury After Liver Transplantation: An Observational Study. Transplant. Proc. 2010, 42, 3639–3643. [Google Scholar] [CrossRef]
- Branten, A.J.W.; Mulder, T.P.J.; Peters, W.M.; Assmann, K.J.M.; Wetzels, J.F.M. Urinary Excretion of Glutathione S Transferases Alpha and Pi in Patients with Proteinuria: Reflection of the Site of Tubular Injury. Nephron 2000, 85, 120–126. [Google Scholar] [CrossRef]
- Westhuyzen, J.; Endre, Z.H.; Reece, G.; Reith, D.M.; Saltissi, D.; Morgan, T.J. Measurement of Tubular Enzymuria Facilitates Early Detection of Acute Renal Impairment in the Intensive Care Unit. Nephrol. Dial. Transplant. 2003, 18, 543–551. [Google Scholar] [CrossRef] [Green Version]
- Trof, R.J.; Di Maggio, F.; Leemreis, J.; Groeneveld, A.B.J. Biomarkers of acute renal injury and renal failure. Shock 2006, 26, 245. [Google Scholar] [CrossRef]
- Polak, W.P.; Kosieradzki, M.; Kwiatkowski, A.; Danielewicz, R.; Lisik, W.; Michalak, G.; Paczek, L.; Lao, M.; Wałaszewski, J.; Rowiński, W.A. Activity of Glutathione S-Transferases in the Urine of Kidney Transplant Recipients during the First Week after Transplantation. Ann. Transpl. 1999, 4, 42–45. [Google Scholar]
- Gautier, J.-C.; Riefke, B.; Walter, J.; Kurth, P.; Mylecraine, L.; Guilpin, V.; Barlow, N.; Gury, T.; Hoffman, D.; Ennulat, D.; et al. Evaluation of Novel Biomarkers of Nephrotoxicity in Two Strains of Rat Treated with Cisplatin. Toxicol. Pathol. 2010, 38, 943–956. [Google Scholar] [CrossRef] [Green Version]
- Liangos, O.; Perianayagam, M.C.; Vaidya, V.S.; Han, W.K.; Wald, R.; Tighiouart, H.; MacKinnon, R.W.; Li, L.; Balakrishnan, V.S.; Pereira, B.J.G.; et al. Urinary N-Acetyl-β-(D)-Glucosaminidase Activity and Kidney Injury Molecule-1 Level Are Associated with Adverse Outcomes in Acute Renal Failure. J. Am. Soc. Nephrol. 2007, 18, 904. [Google Scholar] [CrossRef] [Green Version]
- Holdt-Lehmann, B.; Lehmann, A.; Korten, G.; Nagel, H.-R.; Nizze, H.; Schuff-Werner, P. Diagnostic Value of Urinary Alanine Aminopeptidase and N-Acetyl-β-d-Glucosaminidase in Comparison to A1-Microglobulin as a Marker in Evaluating Tubular Dysfunction in Glomerulonephritis Patients. Clin. Chim. Acta 2000, 297, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Kotanko, P.; Keiler, R.; Knabl, L.; Aulitzky, W.; Margreiter, R.; Gstraunthaler, G.; Pfaller, W. Urinary Enzyme Analysis in Renal Allograft Transplantation. Clin. Chim. Acta 1986, 160, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Kotanko, P.; Margreiter, R.; Pfaller, W. Graft Ischemia Correlates with Urinary Excretion of the Proximal Marker Enzyme Fructose-1,6-Bisphosphatase in Human Kidney Transplantation. Nephron 2008, 77, 62–67. [Google Scholar] [CrossRef]
- Mazloum, M.; Jouffroy, J.; Brazier, F.; Legendre, C.; Neuraz, A.; Garcelon, N.; Prié, D.; Anglicheau, D.; Bienaimé, F. Osmoregulation Performance and Kidney Transplant Outcome. J. Am. Soc. Nephrol. 2019, 30, 1282–1293. [Google Scholar] [CrossRef]
- Kaden, J.; Groth, J.; May, G.; Liedvogel, B. Urinary Tamm-Horsfall Protein as a Marker of Renal Transplant Function. Urol. Res. 1994, 22, 131–136. [Google Scholar] [CrossRef]
- Thongboonkerd, V.; Malasit, P. Renal and Urinary Proteomics: Current Applications and Challenges. PROTEOMICS 2005, 5, 1033–1042. [Google Scholar] [CrossRef]
- Florek, M.; Bauer, N.; Janich, P.; Wilsch-Braeuninger, M.; Fargeas, C.A.; Marzesco, A.-M.; Ehninger, G.; Thiele, C.; Huttner, W.B.; Corbeil, D. Prominin-2 Is a Cholesterol-Binding Protein Associated with Apical and Basolateral Plasmalemmal Protrusions in Polarized Epithelial Cells and Released into Urine. Cell Tissue Res. 2007, 328, 31–47. [Google Scholar] [CrossRef]
- Jászai, J.; Farkas, L.M.; Fargeas, C.A.; Janich, P.; Haase, M.; Huttner, W.B.; Corbeil, D. Prominin-2 Is a Novel Marker of Distal Tubules and Collecting Ducts of the Human and Murine Kidney. Histochem. Cell Biol. 2010, 133, 527–539. [Google Scholar] [CrossRef] [PubMed]
- Tonomura, Y.; Tsuchiya, N.; Torii, M.; Uehara, T. Evaluation of the Usefulness of Urinary Biomarkers for Nephrotoxicity in Rats. Toxicology 2010, 273, 53–59. [Google Scholar] [CrossRef]
- Ting, Y.-T.; Coates, P.T.; Walker, R.J.; Mclellan, A.D. Urinary Tubular Biomarkers as Potential Early Predictors of Renal Allograft Rejection. Nephrology 2012, 17, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Szeto, C.-C.; Kwan, B.C.-H.; Lai, K.-B.; Lai, F.M.-M.; Chow, K.-M.; Wang, G.; Luk, C.C.-W.; Li, P.K.-T. Urinary Expression of Kidney Injury Markers in Renal Transplant Recipients. Clin. J. Am. Soc. Nephrol. 2010, 5, 2329. [Google Scholar] [CrossRef] [Green Version]
- Josephson, M.A. Monitoring and Managing Graft Health in the Kidney Transplant Recipient. Clin. J. Am. Soc. Nephrol. 2011, 6, 1774. [Google Scholar] [CrossRef] [Green Version]
- Colvin, R.B. Antibody-Mediated Renal Allograft Rejection: Diagnosis and Pathogenesis. J. Am. Soc. Nephrol. 2007, 18, 1046. [Google Scholar] [CrossRef] [Green Version]
- Nankivell, B.J.; Chapman, J.R. The Significance of Subclinical Rejection and the Value of Protocol Biopsies. Am. J. Transplant. 2006, 6, 2006–2012. [Google Scholar] [CrossRef]
- Schwarz, A.; Gwinner, W.; Hiss, M.; Radermacher, J.; Mengel, M.; Haller, H. Safety and Adequacy of Renal Transplant Protocol Biopsies. Am. J. Transplant. 2005, 5, 1992–1996. [Google Scholar] [CrossRef] [PubMed]
- Furness, P.N.; Taub, N. International Variation in the Interpretation of Renal Transplant Biopsies: Report of the CERTPAP Project1. Kidney Int. 2001, 60, 1998–2012. [Google Scholar] [CrossRef] [Green Version]
- Hernández, D.; Rufino, M.; Armas, S.; González, A.; Gutiérrez, P.; Barbero, P.; Vivancos, S.; Rodríguez, C.; de Vera, J.R.; Torres, A. Retrospective Analysis of Surgical Complications Following Cadaveric Kidney Transplantation in the Modern Transplant Era. Nephrol. Dial. Transplant. 2006, 21, 2908–2915. [Google Scholar] [CrossRef] [Green Version]
- Salvadori, M.; Rosso, G.; Bertoni, E. Update on Ischemia-Reperfusion Injury in Kidney Transplantation: Pathogenesis and Treatment. World J. Transpl. 2015, 5, 52–67. [Google Scholar] [CrossRef]
- Cheung, K.P.; Kasimsetty, S.G.; McKay, D.B. Innate Immunity in Donor Procurement. Curr. Opin. Organ. Transpl. 2013, 18, 154–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mueller, T.F.; Solez, K.; Mas, V. Assessment of Kidney Organ Quality and Prediction of Outcome at Time of Transplantation. Semin. Immunopathol. 2011, 33, 185–199. [Google Scholar] [CrossRef] [PubMed]
- Ojo, A.O.; Wolfe, R.A.; Held, P.J.; Port, F.K.; Schmouder, R.L. Delayed Graft Function: Risk Factors and Implications for Renal Allograft Survival. Transplantation 1997, 63, 968–974. [Google Scholar] [CrossRef] [PubMed]
- Hollmen, M.E.; Kyllönen, L.E.; Inkinen, K.A.; Lalla, M.L.T.; Merenmies, J.; Salmela, K.T. Deceased Donor Neutrophil Gelatinase-Associated Lipocalin and Delayed Graft Function after Kidney Transplantation: A Prospective Study. Crit. Care 2011, 15, R121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reese, P.P.; Hall, I.E.; Weng, F.L.; Schröppel, B.; Doshi, M.D.; Hasz, R.D.; Thiessen-Philbrook, H.; Ficek, J.; Rao, V.; Murray, P.; et al. Associations between Deceased-Donor Urine Injury Biomarkers and Kidney Transplant Outcomes. J. Am. Soc. Nephrol. 2016, 27, 1534–1543. [Google Scholar] [CrossRef] [Green Version]
- Koo, T.Y.; Jeong, J.C.; Lee, Y.; Ko, K.-P.; Lee, K.-B.; Lee, S.; Park, S.J.; Park, J.B.; Han, M.; Lim, H.J.; et al. Pre-Transplant Evaluation of Donor Urinary Biomarkers Can Predict Reduced Graft Function After Deceased Donor Kidney Transplantation. Medicine 2016, 95, e3076. [Google Scholar] [CrossRef]
- Sadeghi, M.; Daniel, V.; Naujokat, C.; Mehrabi, A.; Opelz, G. Association of High Pretransplant SIL-6R Plasma Levels with Acute Tubular Necrosis in Kidney Graft Recipients. Transplantation 2006, 81, 1716–1724. [Google Scholar] [CrossRef]
- Nguyen, M.-T.J.P.; Fryml, E.; Sahakian, S.K.; Liu, S.; Cantarovich, M.; Lipman, M.; Tchervenkov, J.I.; Paraskevas, S. Pretransplant Recipient Circulating CD4+CD127lo/- Tumor Necrosis Factor Receptor 2+ Regulatory T Cells: A Surrogate of Regulatory T Cell-Suppressive Function and Predictor of Delayed and Slow Graft Function After Kidney Transplantation. Transplantation 2016, 100, 314–324. [Google Scholar] [CrossRef]
- Haase, M.; Bellomo, R.; Devarajan, P.; Schlattmann, P.; Haase-Fielitz, A.; NGAL Meta-analysis Investigator Group. Accuracy of Neutrophil Gelatinase-Associated Lipocalin (NGAL) in Diagnosis and Prognosis in Acute Kidney Injury: A Systematic Review and Meta-Analysis. Am. J. Kidney Dis. 2009, 54, 1012–1024. [Google Scholar] [CrossRef] [Green Version]
- Siew, E.D.; Ware, L.B.; Ikizler, T.A. Biological Markers of Acute Kidney Injury. J. Am. Soc. Nephrol. 2011, 22, 810–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fonseca, I.; Oliveira, J.C.; Almeida, M.; Cruz, M.; Malho, A.; Martins, L.S.; Dias, L.; Pedroso, S.; Santos, J.; Lobato, L.; et al. Neutrophil Gelatinase-Associated Lipocalin in Kidney Transplantation Is an Early Marker of Graft Dysfunction and Is Associated with One-Year Renal Function. J. Transpl. 2013, 2013, 650123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishra, J.; Ma, Q.; Kelly, C.; Mitsnefes, M.; Mori, K.; Barasch, J.; Devarajan, P. Kidney NGAL Is a Novel Early Marker of Acute Injury Following Transplantation. Pediatr. Nephrol. 2006, 21, 856–863. [Google Scholar] [CrossRef]
- Sureshkumar, K.K.; Marcus, R.J. Urinary Biomarkers as Predictors of Long-Term Allograft Function after Renal Transplantation. Transplantation 2010, 90, 688–689. [Google Scholar] [CrossRef] [PubMed]
- Pajek, J.; Škoberne, A.; Šosterič, K.; Adlešič, B.; Leskošek, B.; Bučar Pajek, M.; Osredkar, J.; Lindič, J. Non-Inferiority of Creatinine Excretion Rate to Urinary L-FABP and NGAL as Predictors of Early Renal Allograft Function. BMC Nephrol. 2014, 15, 117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malyszko, J.; Koc-Zorawska, E.; Malyszko, J.S.; Mysliwiec, M. Kidney Injury Molecule-1 Correlates with Kidney Function in Renal Allograft Recipients. Transpl. Proc. 2010, 42, 3957–3959. [Google Scholar] [CrossRef]
- Lacquaniti, A.; Caccamo, C.; Salis, P.; Chirico, V.; Buemi, A.; Cernaro, V.; Noto, A.; Pettinato, G.; Santoro, D.; Bertani, T.; et al. Delayed Graft Function and Chronic Allograft Nephropathy: Diagnostic and Prognostic Role of Neutrophil Gelatinase-Associated Lipocalin. Biomarkers 2016, 21, 371–378. [Google Scholar] [CrossRef]
- Pianta, T.J.; Peake, P.W.; Pickering, J.W.; Kelleher, M.; Buckley, N.A.; Endre, Z.H. Clusterin in Kidney Transplantation: Novel Biomarkers versus Serum Creatinine for Early Prediction of Delayed Graft Function. Transplantation 2015, 99, 171–179. [Google Scholar] [CrossRef]
- Schwarz, C.; Regele, H.; Steininger, R.; Hansmann, C.; Mayer, G.; Oberbauer, R. The Contribution of Adhesion Molecule Expression in Donor Kidney Biopsies to Early Allograft Dysfunction. Transplantation 2001, 71, 1666–1670. [Google Scholar] [CrossRef] [Green Version]
- Schwarz, C.; Hauser, P.; Steininger, R.; Regele, H.; Heinze, G.; Mayer, G.; Oberbauer, R. Failure of BCL-2 up-Regulation in Proximal Tubular Epithelial Cells of Donor Kidney Biopsy Specimens Is Associated with Apoptosis and Delayed Graft Function. Lab. Investig. 2002, 82, 941–948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hauser, P.; Schwarz, C.; Mitterbauer, C.; Regele, H.M.; Mühlbacher, F.; Mayer, G.; Perco, P.; Mayer, B.; Meyer, T.W.; Oberbauer, R. Genome-Wide Gene-Expression Patterns of Donor Kidney Biopsies Distinguish Primary Allograft Function. Lab. Investig. 2004, 84, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Kamińska, D.; Kościelska-Kasprzak, K.; Drulis-Fajdasz, D.; Hałoń, A.; Polak, W.; Chudoba, P.; Jańczak, D.; Mazanowska, O.; Patrzałek, D.; Klinger, M. Kidney Ischemic Injury Genes Expressed after Donor Brain Death Are Predictive for the Outcome of Kidney Transplantation. Transpl. Proc. 2011, 43, 2891–2894. [Google Scholar] [CrossRef]
- McGuinness, D.; Leierer, J.; Shapter, O.; Mohammed, S.; Gingell-Littlejohn, M.; Kingsmore, D.B.; Little, A.-M.; Kerschbaum, J.; Schneeberger, S.; Maglione, M.; et al. Identification of Molecular Markers of Delayed Graft Function Based on the Regulation of Biological Ageing. PLoS ONE 2016, 11, e0146378. [Google Scholar] [CrossRef] [Green Version]
- Wilflingseder, J.; Sunzenauer, J.; Toronyi, E.; Heinzel, A.; Kainz, A.; Mayer, B.; Perco, P.; Telkes, G.; Langer, R.M.; Oberbauer, R. Molecular Pathogenesis of Post-Transplant Acute Kidney Injury: Assessment of Whole-Genome MRNA and MiRNA Profiles. PLoS ONE 2014, 9, e104164. [Google Scholar] [CrossRef]
- Del Prete, G.; De Carli, M.; Almerigogna, F.; Daniel, C.K.; D’Elios, M.M.; Zancuoghi, G.; Vinante, F.; Pizzolo, G.; Romagnani, S. Preferential Expression of CD30 by Human CD4+ T Cells Producing Th2-Type Cytokines. FASEB J. 1995, 9, 81–86. [Google Scholar] [CrossRef]
- Weimer, R.; Zipperle, S.; Daniel, V.; Carl, S.; Staehler, G.; Opelz, G. Pretransplant CD4 Helper Function and Interleukin 10 Response Predict Risk of Acute Kidney Graft Rejection. Transplantation 1996, 62, 1606–1614. [Google Scholar] [CrossRef] [PubMed]
- Rajakariar, R.; Jivanji, N.; Varagunam, M.; Rafiq, M.; Gupta, A.; Sheaff, M.; Sinnott, P.; Yaqoob, M.M. High Pre-Transplant Soluble CD30 Levels Are Predictive of the Grade of Rejection. Am. J. Transpl. 2005, 5, 1922–1925. [Google Scholar] [CrossRef] [PubMed]
- Cinti, P.; Pretagostini, R.; Arpino, A.; Tamburro, M.L.; Mengasini, S.; Lattanzi, R.; De Simone, P.; Berloco, P.; Molajoni, E.R. Evaluation of Pretransplant Immunologic Status in Kidney-Transplant Recipients by Panel Reactive Antibody and Soluble CD30 Determinations. Transplantation 2005, 79, 1154–1156. [Google Scholar] [CrossRef]
- Sengul, S.; Keven, K.; Gormez, U.; Kutlay, S.; Erturk, S.; Erbay, B. Identification of Patients at Risk of Acute Rejection by Pretransplantation and Posttransplantation Monitoring of Soluble CD30 Levels in Kidney Transplantation. Transplantation 2006, 81, 1216–1219. [Google Scholar] [CrossRef]
- Altermann, W.; Schlaf, G.; Rothhoff, A.; Seliger, B. High Variation of Individual Soluble Serum CD30 Levels of Pre-Transplantation Patients: SCD30 a Feasible Marker for Prediction of Kidney Allograft Rejection? Nephrol. Dial. Transpl. 2007, 22, 2795–2799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shooshtarizadeh, T.; Mohammadali, A.; Ossareh, S.; Ataipour, Y. Relation between Pretransplant Serum Levels of Soluble CD30 and Acute Rejection during the First 6 Months after a Kidney Transplant. Exp. Clin. Transpl. 2013, 11, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Augustine, J.J.; Siu, D.S.; Clemente, M.J.; Schulak, J.A.; Heeger, P.S.; Hricik, D.E. Pre-Transplant IFN-Gamma ELISPOTs Are Associated with Post-Transplant Renal Function in African American Renal Transplant Recipients. Am. J. Transpl. 2005, 5, 1971–1975. [Google Scholar] [CrossRef] [PubMed]
- Bendjelloul, F.; Desin, T.S.; Shoker, A.S. Donor Non-Specific IFN-γ Production by Primed Alloreactive Cells as a Potential Screening Test to Predict the Alloimmune Response. Transpl. Immunol. 2004, 12, 167–176. [Google Scholar] [CrossRef]
- Heeger, P.S.; Greenspan, N.S.; Kuhlenschmidt, S.; Dejelo, C.; Hricik, D.E.; Schulak, J.A.; Tary-Lehmann, M. Pretransplant Frequency of Donor-Specific, IFN-γ-Producing Lymphocytes Is a Manifestation of Immunologic Memory and Correlates with the Risk of Posttransplant Rejection Episodes. J. Immunol. 1999, 163, 2267–2275. [Google Scholar] [CrossRef]
- Bellisola, G.; Tridente, G.; Nacchia, F.; Fior, F.; Boschiero, L. Monitoring of Cellular Immunity by Interferon-Gamma Enzyme-Linked Immunosorbent Spot Assay in Kidney Allograft Recipients: Preliminary Results of a Longitudinal Study. Transpl. Proc. 2006, 38, 1014–1017. [Google Scholar] [CrossRef]
- Rotondi, M.; Rosati, A.; Buonamano, A.; Lasagni, L.; Lazzeri, E.; Pradella, F.; Fossombroni, V.; Cirami, C.; Liotta, F.; La Villa, G.; et al. High Pretransplant Serum Levels of CXCL10/IP-10 Are Related to Increased Risk of Renal Allograft Failure. Am. J. Transpl. 2004, 4, 1466–1474. [Google Scholar] [CrossRef]
- Lazzeri, E.; Rotondi, M.; Mazzinghi, B.; Lasagni, L.; Buonamano, A.; Rosati, A.; Pradella, F.; Fossombroni, V.; La Villa, G.; Gacci, M.; et al. High CXCL10 Expression in Rejected Kidneys and Predictive Role of Pretransplant Serum CXCL10 for Acute Rejection and Chronic Allograft Nephropathy. Transplantation 2005, 79, 1215–1220. [Google Scholar] [CrossRef]
- Hricik, D.E.; Nickerson, P.; Formica, R.N.; Poggio, E.D.; Rush, D.; Newell, K.A.; Goebel, J.; Gibson, I.W.; Fairchild, R.L.; Riggs, M.; et al. Multicenter Validation of Urinary CXCL9 as a Risk-Stratifying Biomarker for Kidney Transplant Injury. Am. J. Transpl. 2013, 13, 2634–2644. [Google Scholar] [CrossRef] [Green Version]
- Srinivas, T.R.; Kaplan, B. Urinary Biomarkers and Kidney Transplant Rejection: Fine-Tuning the Radar. Am. J. Transpl. 2013, 13, 2519–2521. [Google Scholar] [CrossRef]
- Kim, S.C.; Page, E.K.; Knechtle, S.J. Urine Proteomics in Kidney Transplantation. Transpl. Rev 2014, 28, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Hirt-Minkowski, P.; De Serres, S.A.; Ho, J. Developing Renal Allograft Surveillance Strategies—Urinary Biomarkers of Cellular Rejection. Can. J. Kidney Health Dis. 2015, 2, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Augustine, J.J.; Hricik, D.E. T-Cell Immune Monitoring by the ELISPOT Assay for Interferon Gamma. Clin. Chim. Acta 2012, 413, 1359–1363. [Google Scholar] [CrossRef]
- Bestard, O.; Crespo, E.; Stein, M.; Lúcia, M.; Roelen, D.L.; de Vaal, Y.J.; Hernandez-Fuentes, M.P.; Chatenoud, L.; Wood, K.J.; Claas, F.H.; et al. Cross-Validation of IFN-γ Elispot Assay for Measuring Alloreactive Memory/Effector T Cell Responses in Renal Transplant Recipients. Am. J. Transpl. 2013, 13, 1880–1890. [Google Scholar] [CrossRef] [PubMed]
- Gielis, E.M.; Ledeganck, K.J.; De Winter, B.Y.; Del Favero, J.; Bosmans, J.-L.; Claas, F.H.J.; Abramowicz, D.; Eikmans, M. Cell-Free DNA: An Upcoming Biomarker in Transplantation. Am. J. Transpl. 2015, 15, 2541–2551. [Google Scholar] [CrossRef] [Green Version]
- García Moreira, V.; Prieto García, B.; Baltar Martín, J.M.; Ortega Suárez, F.; Alvarez, F.V. Cell-Free DNA as a Noninvasive Acute Rejection Marker in Renal Transplantation. Clin. Chem. 2009, 55, 1958–1966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freue, G.V.C.; Sasaki, M.; Meredith, A.; Günther, O.P.; Bergman, A.; Takhar, M.; Mui, A.; Balshaw, R.F.; Ng, R.T.; Opushneva, N.; et al. Proteomic Signatures in Plasma during Early Acute Renal Allograft Rejection. Mol. Cell Proteom. 2010, 9, 1954–1967. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; Zhu, D.; Xu, M.; Rong, R.; Tang, Q.; Wang, X.; Zhu, T. Analysis of Transcriptional Factors and Regulation Networks in Patients with Acute Renal Allograft Rejection. J. Proteome Res. 2011, 10, 175–181. [Google Scholar] [CrossRef]
- Perez, J.D.; Sakata, M.M.; Colucci, J.A.; Spinelli, G.A.; Felipe, C.R.; Carvalho, V.M.; Cardozo, K.H.M.; Medina-Pestana, J.O.; Tedesco-Silva, H.; Schor, N.; et al. Plasma Proteomics for the Assessment of Acute Renal Transplant Rejection. Life Sci. 2016, 158, 111–120. [Google Scholar] [CrossRef]
- Sigdel, T.K.; Kaushal, A.; Gritsenko, M.; Norbeck, A.D.; Qian, W.-J.; Xiao, W.; Camp, D.G.; Smith, R.D.; Sarwal, M.M. Shotgun Proteomics Identifies Proteins Specific for Acute Renal Transplant Rejection. Proteom. Clin. Appl. 2010, 4, 32–47. [Google Scholar] [CrossRef] [Green Version]
- Loftheim, H.; Midtvedt, K.; Hartmann, A.; Reisæter, A.V.; Falck, P.; Holdaas, H.; Jenssen, T.; Reubsaet, L.; Åsberg, A. Urinary Proteomic Shotgun Approach for Identification of Potential Acute Rejection Biomarkers in Renal Transplant Recipients. Transpl. Res. 2012, 1, 9. [Google Scholar] [CrossRef] [Green Version]
- Sigdel, T.K.; Salomonis, N.; Nicora, C.D.; Ryu, S.; He, J.; Dinh, V.; Orton, D.J.; Moore, R.J.; Hsieh, S.-C.; Dai, H.; et al. The Identification of Novel Potential Injury Mechanisms and Candidate Biomarkers in Renal Allograft Rejection by Quantitative Proteomics. Mol. Cell Proteom. 2014, 13, 621–631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasconcellos, L.M.; Schachter, A.D.; Zheng, X.X.; Vasconcellos, L.H.; Shapiro, M.; Harmon, W.E.; Strom, T.B. Cytotoxic Lymphocyte Gene Expression in Peripheral Blood Leukocytes Correlates with Rejecting Renal Allografts. Transplantation 1998, 66, 562–566. [Google Scholar] [CrossRef]
- Li, B.; Hartono, C.; Ding, R.; Sharma, V.K.; Ramaswamy, R.; Qian, B.; Serur, D.; Mouradian, J.; Schwartz, J.E.; Suthanthiran, M. Noninvasive Diagnosis of Renal-Allograft Rejection by Measurement of Messenger RNA for Perforin and Granzyme B in Urine. N. Engl. J. Med. 2001, 344, 947–954. [Google Scholar] [CrossRef]
- Afaneh, C.; Muthukumar, T.; Lubetzky, M.; Ding, R.; Snopkowski, C.; Sharma, V.K.; Seshan, S.; Dadhania, D.; Schwartz, J.E.; Suthanthiran, M. Urinary Cell Levels of MRNA for OX40, OX40L, PD-1, PD-L1 or PD-L2 and Acute Rejection of Human Renal Allografts. Transplantation 2010, 90, 1381–1387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muthukumar, T.; Dadhania, D.; Ding, R.; Snopkowski, C.; Naqvi, R.; Lee, J.B.; Hartono, C.; Li, B.; Sharma, V.K.; Seshan, S.V.; et al. Messenger RNA for FOXP3 in the Urine of Renal-Allograft Recipients. N. Engl. J. Med. 2005, 353, 2342–2351. [Google Scholar] [CrossRef] [PubMed]
- Suthanthiran, M.; Schwartz, J.E.; Ding, R.; Abecassis, M.; Dadhania, D.; Samstein, B.; Knechtle, S.J.; Friedewald, J.; Becker, Y.T.; Sharma, V.K.; et al. Urinary-Cell MRNA Profile and Acute Cellular Rejection in Kidney Allografts. N. Engl. J. Med. 2013, 369, 20–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sui, W.; Yang, M.; Li, F.; Chen, H.; Chen, J.; Ou, M.; Zhang, Y.; Lin, H.; Xue, W.; Dai, Y. Serum MicroRNAs as New Diagnostic Biomarkers for Pre- and Post-Kidney Transplantation. Transpl. Proc. 2014, 46, 3358–3362. [Google Scholar] [CrossRef]
- Lorenzen, J.M.; Volkmann, I.; Fiedler, J.; Schmidt, M.; Scheffner, I.; Haller, H.; Gwinner, W.; Thum, T. Urinary MiR-210 as a Mediator of Acute T-Cell Mediated Rejection in Renal Allograft Recipients. Am. J. Transpl. 2011, 11, 2221–2227. [Google Scholar] [CrossRef]
- Betts, G.; Shankar, S.; Sherston, S.; Friend, P.; Wood, K.J. Examination of Serum MiRNA Levels in Kidney Transplant Recipients with Acute Rejection. Transplantation 2014, 97, e28–e30. [Google Scholar] [CrossRef]
- Grigoryev, Y.A.; Kurian, S.M.; Hart, T.; Nakorchevsky, A.A.; Chen, C.; Campbell, D.; Head, S.R.; Yates, J.R.; Salomon, D.R. MicroRNA Regulation of Molecular Networks Mapped by Global MicroRNA, MRNA, and Protein Expression in Activated T Lymphocytes. J. Immunol. 2011, 187, 2233–2243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarwal, M.M.; Ettenger, R.B.; Dharnidharka, V.; Benfield, M.; Mathias, R.; Portale, A.; McDonald, R.; Harmon, W.; Kershaw, D.; Vehaskari, V.M.; et al. Complete Steroid Avoidance Is Effective and Safe in Children with Renal Transplants: A Multicenter Randomized Trial with Three-Year Follow-Up. Am. J. Transpl. 2012, 12, 2719–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Khatri, P.; Sigdel, T.K.; Tran, T.; Ying, L.; Vitalone, M.; Chen, A.; Hsieh, S.; Dai, H.; Zhang, M.; et al. A Five-Gene Peripheral Blood Diagnostic Test for Acute Rejection in Renal Transplantation. Am. J. Transpl. 2012, 12, 2710–2718. [Google Scholar] [CrossRef] [Green Version]
- Allison, S.J. Biomarkers in Peripheral Blood Detect Acute Rejection. Nat. Rev. Nephrol. 2012, 8, 681. [Google Scholar] [CrossRef] [PubMed]
- Roedder, S.; Sigdel, T.; Salomonis, N.; Hsieh, S.; Dai, H.; Bestard, O.; Metes, D.; Zeevi, A.; Gritsch, A.; Cheeseman, J.; et al. The KSORT Assay to Detect Renal Transplant Patients at High Risk for Acute Rejection: Results of the Multicenter AART Study. PLoS Med. 2014, 11, e1001759. [Google Scholar] [CrossRef] [Green Version]
- Shen-Orr, S.S.; Tibshirani, R.; Khatri, P.; Bodian, D.L.; Staedtler, F.; Perry, N.M.; Hastie, T.; Sarwal, M.M.; Davis, M.M.; Butte, A.J. Cell Type-Specific Gene Expression Differences in Complex Tissues. Nat. Methods 2010, 7, 287–289. [Google Scholar] [CrossRef]
- Li, L.; Khush, K.; Hsieh, S.-C.; Ying, L.; Luikart, H.; Sigdel, T.; Roedder, S.; Yang, A.; Valantine, H.; Sarwal, M.M. Identification of Common Blood Gene Signatures for the Diagnosis of Renal and Cardiac Acute Allograft Rejection. PLoS ONE 2013, 8, e82153. [Google Scholar] [CrossRef]
- Crespo, E.; Roedder, S.; Sigdel, T.; Hsieh, S.-C.; Luque, S.; Cruzado, J.M.; Tran, T.Q.; Grinyó, J.M.; Sarwal, M.M.; Bestard, O. Molecular and Functional Noninvasive Immune Monitoring in the ESCAPE Study for Prediction of Subclinical Renal Allograft Rejection. Transplantation 2017, 101, 1400–1409. [Google Scholar] [CrossRef]
- Khatri, P.; Roedder, S.; Kimura, N.; De Vusser, K.; Morgan, A.A.; Gong, Y.; Fischbein, M.P.; Robbins, R.C.; Naesens, M.; Butte, A.J.; et al. A Common Rejection Module (CRM) for Acute Rejection across Multiple Organs Identifies Novel Therapeutics for Organ Transplantation. J. Exp. Med. 2013, 210, 2205–2221. [Google Scholar] [CrossRef]
- Sigdel, T.K.; Bestard, O.; Tran, T.Q.; Hsieh, S.-C.; Roedder, S.; Damm, I.; Vincenti, F.; Sarwal, M.M. A Computational Gene Expression Score for Predicting Immune Injury in Renal Allografts. PLoS ONE 2015, 10, e0138133. [Google Scholar] [CrossRef] [Green Version]
- The Urine Common Rejection Module (uCRM) Is a Sentinal Assay for Graft Rejection. ATC Abstracts. Available online: https://atcmeetingabstracts.com/abstract/the-urine-common-rejection-module-ucrm-is-a-sentinal-assay-for-graft-rejection/ (accessed on 6 May 2023).
- Novacescu, D.; Feciche, B.O.; Cumpanas, A.A.; Bardan, R.; Rusmir, A.V.; Bitar, Y.A.; Barbos, V.I.; Cut, T.G.; Raica, M.; Latcu, S.C. Contemporary Clinical Definitions, Differential Diagnosis, and Novel Predictive Tools for Renal Cell Carcinoma. Biomedicines 2022, 10, 2926. [Google Scholar] [CrossRef] [PubMed]
- Novacescu, D.; Cut, T.G.; Cumpanas, A.A.; Latcu, S.C.; Bardan, R.; Ferician, O.; Secasan, C.-C.; Rusmir, A.; Raica, M. Evaluating Established Roles, Future Perspectives and Methodological Heterogeneity for Wilms’ Tumor 1 (WT1) Antigen Detection in Adult Renal Cell Carcinoma, Using a Novel N-Terminus Targeted Antibody (Clone WT49). Biomedicines 2022, 10, 912. [Google Scholar] [CrossRef] [PubMed]
- Novacescu, D.; Cut, T.G.; Cumpanas, A.A.; Bratosin, F.; Ceausu, R.A.; Raica, M. Novel Expression of Thymine Dimers in Renal Cell Carcinoma, Demonstrated through Immunohistochemistry. Biomedicines 2022, 10, 2673. [Google Scholar] [CrossRef] [PubMed]
- Radu-Cosnita, A.D.; Nesiu, A.; Berzava, P.L.; Cerbu, S.; Cosma, A.; Comsa, S.; Sarb, S.; Ferician, A.M.; Ferician, O.C.; Cimpean, A.M. Anti-Chloride Intracellular Channel Protein 1 (CLIC1) Antibodies Induce Tumour Necrosis and Angiogenesis Inhibition on In Vivo Experimental Models of Human Renal Cancer. Anticancer Res. 2022, 42, 1313–1325. [Google Scholar] [CrossRef] [PubMed]
- Ferician, A.M.; Ferician, O.C.; Cumpanas, A.D.; Berzava, P.L.; Nesiu, A.; Barmayoun, A.; Cimpean, A.M. Heterogeneity of Platelet Derived Growth Factor Pathway Gene Expression Profile Defines Three Distinct Subgroups of Renal Cell Carcinomas. Cancer Genom. Proteom. 2022, 19, 477–489. [Google Scholar] [CrossRef]
- Ferician, A.M.; Ferician, O.C.; Nesiu, A.; Cosma, A.A.; Caplar, B.D.; Melnic, E.; Cimpean, A.M. The Mutually Mediated Chloride Intracellular Channel Protein 1 (CLIC1) Relationship between Malignant Cells and Tumor Blood Vessel Endothelium Exhibits a Significant Impact on Tumor Angiogenesis, Progression, and Metastasis in Clear Cell Renal Cell Carcinoma (CcRCC). Cancers 2022, 14, 5981. [Google Scholar] [CrossRef]
- Sigdel, T.K.; Vitalone, M.J.; Tran, T.Q.; Dai, H.; Hsieh, S.-C.; Salvatierra, O.; Sarwal, M.M. A Rapid Noninvasive Assay for the Detection of Renal Transplant Injury. Transplantation 2013, 96, 97–101. [Google Scholar] [CrossRef] [Green Version]
- Halloran, P.F.; Madill-Thomsen, K.S.; Reeve, J. The Molecular Phenotype of Kidney Transplants: Insights From the MMDx Project. Transplantation 2023. [Google Scholar] [CrossRef]
- Nakorchevsky, A.; Hewel, J.A.; Kurian, S.M.; Mondala, T.S.; Campbell, D.; Head, S.R.; Marsh, C.L.; Yates, J.R.; Salomon, D.R. Molecular Mechanisms of Chronic Kidney Transplant Rejection via Large-Scale Proteogenomic Analysis of Tissue Biopsies. J. Am. Soc. Nephrol. 2010, 21, 362–373. [Google Scholar] [CrossRef] [Green Version]
- Sigdel, T.K.; Gao, Y.; He, J.; Wang, A.; Nicora, C.D.; Fillmore, T.L.; Shi, T.; Webb-Robertson, B.-J.; Smith, R.D.; Qian, W.-J.; et al. Mining the Human Urine Proteome for Monitoring Renal Transplant Injury. Kidney Int. 2016, 89, 1244–1252. [Google Scholar] [CrossRef] [Green Version]
- Solez, K.; Colvin, R.B.; Racusen, L.C.; Sis, B.; Halloran, P.F.; Birk, P.E.; Campbell, P.M.; Cascalho, M.; Collins, A.B.; Demetris, A.J.; et al. Banff ’05 Meeting Report: Differential Diagnosis of Chronic Allograft Injury and Elimination of Chronic Allograft Nephropathy (‘CAN’). Am. J. Transpl. 2007, 7, 518–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colvin, R.B. Chronic Allograft Nephropathy. N. Engl. J. Med. 2003, 349, 2288–2290. [Google Scholar] [CrossRef] [PubMed]
- Nankivell, B.J.; Borrows, R.J.; Fung, C.L.-S.; O’Connell, P.J.; Allen, R.D.M.; Chapman, J.R. The Natural History of Chronic Allograft Nephropathy. N. Engl. J. Med. 2003, 349, 2326–2333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bosmans, J.-L.; Ysebaert, D.K.; Verpooten, G.A. Chronic Allograft Nephropathy: What Have We Learned from Protocol Biopsies? Transplantation 2008, 85, S38–S41. [Google Scholar] [CrossRef] [PubMed]
- Cosio, F.G.; Grande, J.P.; Larson, T.S.; Gloor, J.M.; Velosa, J.A.; Textor, S.C.; Griffin, M.D.; Stegall, M.D. Kidney Allograft Fibrosis and Atrophy Early after Living Donor Transplantation. Am. J. Transpl. 2005, 5, 1130–1136. [Google Scholar] [CrossRef] [PubMed]
- Serón, D.; Moreso, F. Protocol Biopsies in Renal Transplantation: Prognostic Value of Structural Monitoring. Kidney Int. 2007, 72, 690–697. [Google Scholar] [CrossRef] [Green Version]
- Quintana, L.F.; Solé-Gonzalez, A.; Kalko, S.G.; Bañon-Maneus, E.; Solé, M.; Diekmann, F.; Gutierrez-Dalmau, A.; Abian, J.; Campistol, J.M. Urine Proteomics to Detect Biomarkers for Chronic Allograft Dysfunction. J. Am. Soc. Nephrol. 2009, 20, 428–435. [Google Scholar] [CrossRef] [Green Version]
- Quintana, L.F.; Campistol, J.M.; Alcolea, M.P.; Bañon-Maneus, E.; Sol-González, A.; Cutillas, P.R. Application of Label-Free Quantitative Peptidomics for the Identification of Urinary Biomarkers of Kidney Chronic Allograft Dysfunction. Mol. Cell Proteom. 2009, 8, 1658–1673. [Google Scholar] [CrossRef] [Green Version]
- Johnston, O.; Cassidy, H.; O’Connell, S.; O’Riordan, A.; Gallagher, W.; Maguire, P.B.; Wynne, K.; Cagney, G.; Ryan, M.P.; Conlon, P.J.; et al. Identification of Β2-Microglobulin as a Urinary Biomarker for Chronic Allograft Nephropathy Using Proteomic Methods. Proteom. Clin. Appl. 2011, 5, 422–431. [Google Scholar] [CrossRef]
- Bañón-Maneus, E.; Diekmann, F.; Carrascal, M.; Quintana, L.F.; Moya-Rull, D.; Bescós, M.; Ramírez-Bajo, M.J.; Rovira, J.; Gutierrez-Dalmau, A.; Solé-González, A.; et al. Two-Dimensional Difference Gel Electrophoresis Urinary Proteomic Profile in the Search of Nonimmune Chronic Allograft Dysfunction Biomarkers. Transplantation 2010, 89, 548–558. [Google Scholar] [CrossRef]
- Jung, H.-Y.; Lee, C.-H.; Choi, J.-Y.; Cho, J.-H.; Park, S.-H.; Kim, Y.-L.; Moon, P.-G.; Baek, M.-C.; Berm Park, J.; Hoon Kim, Y.; et al. Potential Urinary Extracellular Vesicle Protein Biomarkers of Chronic Active Antibody-Mediated Rejection in Kidney Transplant Recipients. J. Chromatogr. B 2020, 1138, 121958. [Google Scholar] [CrossRef]
- Kurian, S.M.; Heilman, R.; Mondala, T.S.; Nakorchevsky, A.; Hewel, J.A.; Campbell, D.; Robison, E.H.; Wang, L.; Lin, W.; Gaber, L.; et al. Biomarkers for Early and Late Stage Chronic Allograft Nephropathy by Proteogenomic Profiling of Peripheral Blood. PLoS ONE 2009, 4, e6212. [Google Scholar] [CrossRef] [Green Version]
- Scian, M.J.; Maluf, D.G.; David, K.G.; Archer, K.J.; Suh, J.L.; Wolen, A.R.; Mba, M.U.; Massey, H.D.; King, A.L.; Gehr, T.; et al. MicroRNA Profiles in Allograft Tissues and Paired Urines Associate with Chronic Allograft Dysfunction with IF/TA. Am. J. Transpl. 2011, 11, 2110–2122. [Google Scholar] [CrossRef] [Green Version]
- Maluf, D.G.; Dumur, C.I.; Suh, J.L.; Scian, M.J.; King, A.L.; Cathro, H.; Lee, J.K.; Gehrau, R.C.; Brayman, K.L.; Gallon, L.; et al. The Urine MicroRNA Profile May Help Monitor Post-Transplant Renal Graft Function. Kidney Int. 2014, 85, 439–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zununi Vahed, S.; Omidi, Y.; Ardalan, M.; Samadi, N. Dysregulation of Urinary MiR-21 and MiR-200b Associated with Interstitial Fibrosis and Tubular Atrophy (IFTA) in Renal Transplant Recipients. Clin. Biochem. 2017, 50, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Soltaninejad, E.; Nicknam, M.H.; Nafar, M.; Sharbafi, M.H.; Keshavarz Shahbaz, S.; Barabadi, M.; Yekaninejad, M.S.; Bahrami, T.; Ahmadpoor, P.; Amirzargar, A. Altered Expression of MicroRNAs Following Chronic Allograft Dysfunction with Interstitial Fibrosis and Tubular Atrophy. Iran. J. Allergy Asthma Immunol. 2015, 14, 615–623. [Google Scholar]
- Iwasaki, K.; Yamamoto, T.; Inanaga, Y.; Hiramitsu, T.; Miwa, Y.; Murotani, K.; Narumi, S.; Watarai, Y.; Katayama, A.; Uchida, K.; et al. MiR-142-5p and MiR-486-5p as Biomarkers for Early Detection of Chronic Antibody-Mediated Rejection in Kidney Transplantation. Biomarkers 2017, 22, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Mas, V.; Maluf, D.; Archer, K.; Yanek, K.; Mas, L.; King, A.; Gibney, E.; Massey, D.; Cotterell, A.; Fisher, R.; et al. Establishing the Molecular Pathways Involved in Chronic Allograft Nephropathy for Testing New Noninvasive Diagnostic Markers. Transplantation 2007, 83, 448. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.R.; Muthukumar, T.; Dadhania, D.; Ding, R.; Sharma, V.K.; Schwartz, J.E.; Suthanthiran, M. Urinary Cell MRNA Profiles Predictive of Human Kidney Allograft Status. Immunol. Rev. 2014, 258, 218–240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Connell, P.J.; Zhang, W.; Menon, M.C.; Yi, Z.; Schröppel, B.; Gallon, L.; Luan, Y.; Rosales, I.A.; Ge, Y.; Losic, B.; et al. Biopsy Transcriptome Expression Profiling to Identify Kidney Transplants at Risk of Chronic Injury: A Multicentre, Prospective Study. Lancet 2016, 388, 983–993. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Greene, I.; Readhead, B.; Menon, M.C.; Kidd, B.A.; Uzilov, A.V.; Wei, C.; Philippe, N.; Schroppel, B.; He, J.C.; et al. Novel Therapeutics Identification for Fibrosis in Renal Allograft Using Integrative Informatics Approach. Sci. Rep. 2017, 7, 39487. [Google Scholar] [CrossRef] [Green Version]
- Peruzzi, L.; Deaglio, S. Rejection Markers in Kidney Transplantation: Do New Technologies Help Children? Pediatr. Nephrol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Bergan, S.; Brunet, M.; Hesselink, D.A.; Johnson-Davis, K.L.; Kunicki, P.K.; Lemaitre, F.; Marquet, P.; Molinaro, M.; Noceti, O.; Pattanaik, S.; et al. Personalized Therapy for Mycophenolate: Consensus Report by the International Association of Therapeutic Drug Monitoring and Clinical Toxicology. Ther. Drug Monit. 2021, 43, 150. [Google Scholar] [CrossRef] [PubMed]
- Kuypers, D.R.J. Intrapatient Variability of Tacrolimus Exposure in Solid Organ Transplantation: A Novel Marker for Clinical Outcome. Clin. Pharmacol. Ther. 2020, 107, 347–358. [Google Scholar] [CrossRef]
- Eid, L.; Tuchman, S.; Moudgil, A. Late Acute Rejection: Incidence, Risk Factors, and Effect on Graft Survival and Function. Pediatr. Transplant. 2014, 18, 155–162. [Google Scholar] [CrossRef]
- Pollock-BarZiv, S.M.; Finkelstein, Y.; Manlhiot, C.; Dipchand, A.I.; Hebert, D.; Ng, V.L.; Solomon, M.; McCrindle, B.W.; Grant, D. Variability in Tacrolimus Blood Levels Increases the Risk of Late Rejection and Graft Loss after Solid Organ Transplantation in Older Children. Pediatr. Transplant. 2010, 14, 968–975. [Google Scholar] [CrossRef]
- Marquet, P.; Cros, F.; Micallef, L.; Jacqz-Aigrain, E.; Woillard, J.-B.; Monchaud, C.; Saint-Marcoux, F.; Debord, J. Tacrolimus Bayesian Dose Adjustment in Pediatric Renal Transplant Recipients. Ther. Drug Monit. 2021, 43, 472. [Google Scholar] [CrossRef]
- Davis, S.; Gralla, J.; Klem, P.; Tong, S.; Wedermyer, G.; Freed, B.; Wiseman, A.; Cooper, J.E. Lower Tacrolimus Exposure and Time in Therapeutic Range Increase the Risk of de Novo Donor-Specific Antibodies in the First Year of Kidney Transplantation. Am. J. Transplant. 2018, 18, 907–915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirk, A.D.; Hale, D.A.; Mannon, R.B.; Kleiner, D.E.; Hoffmann, S.C.; Kampen, R.L.; Cendales, L.K.; Tadaki, D.K.; Harlan, D.M.; Swanson, S.J. Results from a Human Renal Allograft Tolerance Trial Evaluating the Humanized CD52-Specific Monoclonal Antibody Alemtuzumab (CAMPATH-1H). Transplantation 2003, 76, 120–129. [Google Scholar] [CrossRef]
- Kowalski, R.J.; Post, D.R.; Mannon, R.B.; Sebastian, A.; Wright, H.I.; Sigle, G.; Burdick, J.; Elmagd, K.A.; Zeevi, A.; Lopez-Cepero, M.; et al. Assessing Relative Risks of Infection and Rejection: A Meta-Analysis Using an Immune Function Assay. Transplantation 2006, 82, 663–668. [Google Scholar] [CrossRef]
- Egli, A.; Humar, A.; Kumar, D. State-of-the-Art Monitoring of Cytomegalovirus-Specific Cell-Mediated Immunity after Organ Transplant: A Primer for the Clinician. Clin. Infect. Dis. 2012, 55, 1678–1689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ginevri, F.; Azzi, A.; Hirsch, H.H.; Basso, S.; Fontana, I.; Cioni, M.; Bodaghi, S.; Salotti, V.; Rinieri, A.; Botti, G.; et al. Prospective Monitoring of Polyomavirus BK Replication and Impact of Pre-Emptive Intervention in Pediatric Kidney Recipients. Am. J. Transpl. 2007, 7, 2727–2735. [Google Scholar] [CrossRef] [PubMed]
- Rittà, M.; Costa, C.; Sinesi, F.; Sidoti, F.; Di Nauta, A.; Mantovani, S.; Piceghello, A.; Simeone, S.; Ricci, D.; Boffini, M.; et al. Evaluation of Epstein-Barr Virus-Specific Immunologic Response in Solid Organ Transplant Recipients with an Enzyme-Linked ImmunoSpot Assay. Transpl. Proc. 2013, 45, 2754–2757. [Google Scholar] [CrossRef] [PubMed]
- Abate, D.; Saldan, A.; Mengoli, C.; Fiscon, M.; Silvestre, C.; Fallico, L.; Peracchi, M.; Furian, L.; Cusinato, R.; Bonfante, L.; et al. Comparison of Cytomegalovirus (CMV) Enzyme-Linked Immunosorbent Spot and CMV Quantiferon Gamma Interferon-Releasing Assays in Assessing Risk of CMV Infection in Kidney Transplant Recipients. J. Clin. Microbiol. 2013, 51, 2501–2507. [Google Scholar] [CrossRef] [Green Version]
- Walker, S.; Fazou, C.; Crough, T.; Holdsworth, R.; Kiely, P.; Veale, M.; Bell, S.; Gailbraith, A.; McNeil, K.; Jones, S.; et al. Ex Vivo Monitoring of Human Cytomegalovirus-Specific CD8+ T-Cell Responses Using QuantiFERON-CMV. Transpl. Infect. Dis. 2007, 9, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Mella, A.; Mariano, F.; Dolla, C.; Gallo, E.; Manzione, A.M.; Di Vico, M.C.; Cavallo, R.; De Rosa, F.G.; Costa, C.; Biancone, L. Bacterial and Viral Infection and Sepsis in Kidney Transplanted Patients. Biomedicines 2022, 10, 701. [Google Scholar] [CrossRef]
- Mafi, S.; Essig, M.; Rerolle, J.-P.; Lagathu, G.; Crochette, R.; Brodard, V.; Schvartz, B.; Gouarin, S.; Bouvier, N.; Engelmann, I.; et al. Torque Teno Virus Viremia and QuantiFERON®-CMV Assay in Prediction of Cytomegalovirus Reactivation in R+ Kidney Transplant Recipients. Front. Med. 2023, 10, 1180769. [Google Scholar] [CrossRef]
- Newell, K.A.; Asare, A.; Kirk, A.D.; Gisler, T.D.; Bourcier, K.; Suthanthiran, M.; Burlingham, W.J.; Marks, W.H.; Sanz, I.; Lechler, R.I.; et al. Identification of a B Cell Signature Associated with Renal Transplant Tolerance in Humans. J. Clin. Investig. 2010, 120, 1836–1847. [Google Scholar] [CrossRef] [Green Version]
- Sagoo, P.; Perucha, E.; Sawitzki, B.; Tomiuk, S.; Stephens, D.A.; Miqueu, P.; Chapman, S.; Craciun, L.; Sergeant, R.; Brouard, S.; et al. Development of a Cross-Platform Biomarker Signature to Detect Renal Transplant Tolerance in Humans. J. Clin. Investig. 2010, 120, 1848–1861. [Google Scholar] [CrossRef]
- Brouard, S.; Le Bars, A.; Dufay, A.; Gosselin, M.; Foucher, Y.; Guillet, M.; Cesbron-Gautier, A.; Thervet, E.; Legendre, C.; Dugast, E.; et al. Identification of a Gene Expression Profile Associated with Operational Tolerance among a Selected Group of Stable Kidney Transplant Patients. Transpl. Int. 2011, 24, 536–547. [Google Scholar] [CrossRef]
- Orlando, G.; Hematti, P.; Stratta, R.J.; Burke, G.W.; Di Cocco, P.; Pisani, F.; Soker, S.; Wood, K. Clinical Operational Tolerance after Renal Transplantation: Current Status and Future Challenges. Ann. Surg. 2010, 252, 915–928. [Google Scholar] [CrossRef] [Green Version]
- Lozano, J.J.; Pallier, A.; Martinez-Llordella, M.; Danger, R.; López, M.; Giral, M.; Londoño, M.C.; Rimola, A.; Soulillou, J.P.; Brouard, S.; et al. Comparison of Transcriptional and Blood Cell-Phenotypic Markers between Operationally Tolerant Liver and Kidney Recipients. Am. J. Transpl. 2011, 11, 1916–1926. [Google Scholar] [CrossRef]
- Danger, R.; Pallier, A.; Giral, M.; Martínez-Llordella, M.; Lozano, J.J.; Degauque, N.; Sanchez-Fueyo, A.; Soulillou, J.-P.; Brouard, S. Upregulation of MiR-142-3p in Peripheral Blood Mononuclear Cells of Operationally Tolerant Patients with a Renal Transplant. J. Am. Soc. Nephrol. 2012, 23, 597–606. [Google Scholar] [CrossRef] [Green Version]
- Viklicky, O.; Krystufkova, E.; Brabcova, I.; Sekerkova, A.; Wohlfahrt, P.; Hribova, P.; Wohlfahrtova, M.; Sawitzki, B.; Slatinska, J.; Striz, I.; et al. B-Cell-Related Biomarkers of Tolerance Are up-Regulated in Rejection-Free Kidney Transplant Recipients. Transplantation 2013, 95, 148–154. [Google Scholar] [CrossRef]
- Rebollo-Mesa, I.; Nova-Lamperti, E.; Mobillo, P.; Runglall, M.; Christakoudi, S.; Norris, S.; Smallcombe, N.; Kamra, Y.; Hilton, R.; Indices of Tolerance EU Consortium; et al. Biomarkers of Tolerance in Kidney Transplantation: Are We Predicting Tolerance or Response to Immunosuppressive Treatment? Am. J. Transpl. 2016, 16, 3443–3457. [Google Scholar] [CrossRef] [Green Version]
- Roedder, S.; Li, L.; Alonso, M.N.; Hsieh, S.-C.; Vu, M.T.; Dai, H.; Sigdel, T.K.; Bostock, I.; Macedo, C.; Metes, D.; et al. A Three-Gene Assay for Monitoring Immune Quiescence in Kidney Transplantation. J. Am. Soc. Nephrol. 2015, 26, 2042–2053. [Google Scholar] [CrossRef] [Green Version]
- Bohne, F.; Martínez-Llordella, M.; Lozano, J.-J.; Miquel, R.; Benítez, C.; Londoño, M.-C.; Manzia, T.-M.; Angelico, R.; Swinkels, D.W.; Tjalsma, H.; et al. Intra-Graft Expression of Genes Involved in Iron Homeostasis Predicts the Development of Operational Tolerance in Human Liver Transplantation. J. Clin. Investig. 2012, 122, 368–382. [Google Scholar] [CrossRef]

| Biomarker | Clinical Evidence | |
|---|---|---|
| Renal Corpuscle | Urea/ Creatinine | The oldest biomarkers of glomerular injury [42], urea (protein metabolism) and creatinine (muscular metabolism) are both byproducts filtered by the glomeruli and excreted in urine. ↑ serum urea/creatinine, i.e., nitrogen retention, are indicative of ↓ GFR and renal dysfunction. |
| Cystatin C (CYC) | CYC = Low-molecular-weight cysteine protease inhibitor [43], secreted by all nucleated cells at a constant rate, freely filtered by the glomerulus and completely reabsorbed by proximal tubule cells, i.e., negligible amounts in final urine [44]. Normal range of 0.05–10.47 mg/L [45]. ↑ urinary levels may indicate proximal tubule dysfunction and tubule-interstitial disease [39]. | |
| Proteinuria | Proteinuria is caused by ↑ filtration of plasma proteins and ↓ proximal tubular reabsorption [46,47]. Levels ≥ 0.5 g/24 h = independent risk factor for progressive tubular-interstitial fibrosis and a strong predictor of ESRD [48,49,50,51]. It may indicate established renal damage associated with ↓ GFR [52]. Post-RT, signals poor renal graft function and potential graft failure [53]. | |
| Albuminuria | Urinary albumin (the main plasmatic protein) is a more sensitive marker of GFR than proteinuria [54]. Micro-albuminuria (<200 μg/min) is a better indicator of kidney transplant condition than proteinuria [55]. It predicts renal graft loss and offers early detection of allograft changes [56], but also reflects both glomerular and chronic allograft damage and may be indicative of interstitial inflammation [57]. Thus, it is also considered a predictor of long-term allograft outcomes in RT recipients. Urine albumin-to-creatinine ratios (UACRs) are also recommended for post-RT monitoring [57]. | |
| Adhesion Molecules (between Podocytes & Basal Membranes) | Integrins = transmembranary glycoproteins with two subunits, α and β, which promote cellular attachment/migration/invasion, along the surrounding extracellular matrix (ECM), and are necessary to maintain cellular survival and functions. The β1 subtypes = major class of cell substrate receptors, specifically binding collagens, laminins, and fibronectins [58]. Integrin α3/β3 are particularly recommended for monitoring the function of allografts, both in the early and long-term setting, after RT [58]. | |
| Vascular cell adhesion molecule-1 (VCAM-1), Soluble vascular cell adhesion molecule 1 (sVCAM-1)/(CD106), and Anti-intercellular adhesion molecule-1 (ICAM-1), as members of the immunoglobulin (Ig) superfamily, are the chief endothelial cell proteins recognized by white cell integrins [10]. ↑ urinary sVCAM-1, interleukin (IL)6, sIL6R, and tumor necrosis factor (TNF)R1 levels in the first 2 weeks post RT indicate AR [59]. ↑ urinary sICAM-1 levels have also been reported in AR patients [60]. RT patients with proteinuria showed ↑ sVCAM/sICAM urinary levels [61]. Currently, the determination of cell adhesion molecules is recommended as a non-invasive monitoring tool for AR post RT [62]. | ||
| Proximal Tubules | α1-Microglobulin (α1M) | α1M = 27 kDa glycoprotein from the lipocalin family, structurally related to retinol binding protein (RBP), synthesized by liver cells, with various functions, i.e., immunoregulation by binding to T and B lymphocytes, and involvement in heme-complex catabolism [39,62,63]. Stability in acidic urine makes it a sensitive indicator of proximal tubule renal damage, i.e., ↑ urinary levels may be a consequence of GF deterioration. An ↑ α1M/creatinine ratio = an early and sensitive indicator of poor allograft function/prognosis/long-term survival after RT [64], i.e., 6 months post RT, 32% had microalbuminuria. |
| β2-Microglobulin (β2M) | β2M = 11.8 kDa protein, part of the major histocompatibility complex (MHC) class I molecules, found on the surface of all nucleated cells [63]. It undergoes GF and then is reabsorbed and catabolized in the proximal tubules. β2M excretion is typically used to evaluate nephrotoxic damage, such as that caused by aminoglycoside antibiotics or heavy metal salts. Urinary β2M can be helpful in evaluating the state of a transplanted kidney, yet the interpretation of results should be done with caution due to the variety of factors that can influence β2M plasmatic/urinary concentration, renal filtration ability, and tubular function, i.e., drugs, ischemia–reperfusion complications, or true renal graft rejection [64]. | |
| Retinol binding protein (RBP) | RBP = 21 kDa protein belonging to the lipocalin family, primarily synthesized in the liver, it mainly transports retinol (vitamin A) from the liver to peripheral tissues [63,64,65,66]. RBP is filtered by the glomerulus, and then reabsorbed and catabolized in proximal tubules. ↑ urinary RBP can be a result of impaired GF and/or reabsorption in the renal proximal tubules. Due to its greater stability in acidic urine, RBP is considered a better biomarker for proximal tubule damage than β2M [10]. | |
| Brush Border Tubular Enzymes (↑urinary Excretion = Tubular Brush Border Membrane Damage/Microvillus Loss [29]) | Adenosine deaminase binding protein (ABP) = 120 kDa glycoprotein found in various tissues such as lungs, liver, placenta, and brush border of renal proximal tubules. It is involved in the regulation of adenosine levels and has been implicated in several physiological and pathological processes [67]. ↑ urinary ABP is considered an early indicator of AKI and has been reported in various clinical situations, such as ischemia without sepsis, RT, toxic renal tubular damage, and neonatal sepsis. Some researchers have suggested that ABP may be the best marker of acute renal damage, even better than β2M or α1M [67]. Due to higher ABP excretion in RT recipients compared to those with normal renal function, ABP is a good indicator for detecting graft failure [68]. | |
| Alkaline phosphatase (AP) = 140 kDa membrane-bound glycoprotein found in various tissues, including renal proximal tubular structures. AP is involved in the metabolism of organic phosphates [69]. One common reason for declining function in allografts post-RT is the nephrotoxicity of chronic immunosuppressive therapy. ↑ urinary AP levels can be a sign of renal damage due to the use of immunosuppressive drugs, i.e., usually Cyclosporine A [69]. | ||
| γ-glutamyl-transferase (GGT) = ubiquitous enzyme found in the cell membranes of numerous tissues such as kidneys, bile duct, pancreas, gallbladder, spleen, heart, brain, and seminal vesicles. It plays an integral role in amino acid transport across the cell membrane and in the metabolism of leukotrienes. Notably, GGT is also involved in maintaining the balance of oxidative stress within the cell by participating in glutathione metabolism. ↑ urinary GGT provides reliable evidence of nephrotoxicity, such as that caused by prolonged use of anti-rejection drugs in RT patients. An absence of GGT/enzymes in urine suggests a return to normal function of the renal tubules [65]. | ||
| Alanyl-aminopeptidase (AAP), an enzyme that degrades oligopeptides, when ↑ in urine, is associated with severe conditions such as acute renal tubular necrosis, rejection of renal graft, or the toxic effects of immunosuppressive drugs [39,65,70]. | ||
| Cytosolic/Lysosomal Tubular Enzymes | α-/π-Glutathione-S-transferase (α-/π-GST) = a specific cytosolic enzyme of tubular epithelial cells, which consists of two main isoenzymes: α-GST that thrives in alkaline pH, and π-GST which prefers an acidic pH. The α-GST is found in the epithelium of proximal tubular cells, and the π-GST in distal tubules [71]. The determination of α-GST and π-GST in urine is utilized for diagnosing acute renal graft rejection with acute tubular necrosis [65]. A differentiated increase in the urinary excretion of α-GST vs. π-GST may indicate the location of nephron damage [45,71,72,73,74,75]. | |
| N-acetyl-β-D-hexosaminidase (HEX) = a lysosomal renal enzyme and one of the most commonly determined urinary markers for tubular damage, i.e., HEX activity increases early on, prior to the onset of disturbances in renal excretion. Mainly found in proximal tubular cells, HEX is thus specific, i.e., ↑ molecular weight (>130 kDa) prevents glomerular filtration [76,77]. During active kidney disease, HEX activity consistently rises. ↑ urinary activity of HEX/its isoenzyme HEXB indicates damage to renal tubular cellularity. Thus, urinary HEX and, particularly, HEXB, may serve as specific markers for proximal tubular damage post-RT [76,77]. | ||
| Fructose-1,6-bisphosphatase (FBP-1,6) = primarily localized in the convoluted and to a lesser extent in the straight portion of proximal renal tubules. Similar to HEX and GST, it indicates the precise location of allograft nephron damage [73,78]. ↑urinary FBP-1,6 was observed post-RT. Urinary FBP-1,6 excretion was significantly lower in patients with a median cold ischemia time of <22 h, compared to those with >22 h. Even in the absence of graft dysfunction, if the cold ischemia period is extended, urinary excretion of FBP-1,6 correlates with the extent of damage to the renal tubules [79] | ||
| Distal Tubules | Urinary Osmolality | Urine osmolality refers to the concentration of solutes in urine, and is regulated by the activity of antidiuretic hormone (ADH) in the distal nephron [80]. An important parameter for evaluating the function of distal renal tubules. ↓ urinary osmolality suggests the presence of distal tubular dysfunction [80]. |
| Tamm-Horsfall Glycoprotein (THP) | THP, i.e., uromodulin = protein synthesized by renal tubular cells in the thick ascending limb of Henle’s loop and the distal convoluted tubule. THP is the most abundant protein in normal urine, and its concentration is directly proportional to the number of functioning nephrons [81]. A ↓ THP excretion is a sensitive indicator of tubular dysfunction in patients with CKD [81]. | |
| Renal Kallikrein | Renal kallikrein = an enzyme that regulates blood pressure and sodium excretion in the kidney [82]. Urinary kallikrein is considered a sensitive marker of distal tubular dysfunction, and its levels have been shown to decrease in various types of renal disease [82]. | |
| Annexin A11 (ANX11) | ANX11 = a calcium-binding protein that is found in high quantities in distal tubular cells and glomerular epithelium. ANX11 has been identified as a useful marker of acute and chronic renal graft rejection [58]. | |
| Renal Papillary Antigen (RPA)-1 | RPA-1 = a sensitive and specific antigen of renal papillary cells, i.e., a useful marker of damage to renal collecting tubules. RPA-1 has been shown to be a sensitive and specific urinary marker of renal papillary cell injury in both animal models and humans [75]. | |
| Prominin-2 (PROM-2) | PROM-2 = a cellular membrane glycoprotein (112 kDa), with peak expression in epithelial cells of fully developed kidneys, i.e., a cholesterol-binding protein, associated with apical and basolateral plasmalemma protrusions in polarized renal epithelial cells that is released into urine [83]. PROM-2 has been identified as a novel biomarker, specific for distal tubules and collecting ducts, in human and murine kidneys, useful biomarker for the functional assessment of distal renal tubules [84]. | |
| μ-Glutathione-S-Transferase (μ-GST) | μ-GST = a conjugating glutathione present in tubular epithelial cells, i.e., mainly the ascending part of Henle’s loop [75], alongside π-GST. It represents a nephrotoxicity-specific biomarker. μ-GST is an early biomarker for Henle’s loop and distal tubule damage, and has been shown to be more specific than albuminuria for assessing nephrotoxicity [85]. ↑ urinary μ-GST levels can be observed in response to treatment with nephrotoxic drugs, such as cisplatin. |
| Pre-RT Applications | Post-RT Applications | ||
|---|---|---|---|
| Ischemia–reperfusoin Injury (IRI)/Delayed Graft Function (DGF) | Proteomic Data | Donor urinary biomarkers:
Recipient cytokines:
Recipient circulating regulatory T-cells:
| Recipient urinary biomarkers |
| Genomic/Transcriptomic Data | Predictive of DGF on pre-RT allograft biopsy samples:
| MicroRNAs (miRNAs) = short endogenous non-coding RNAs that inhibit gene expression; miR-182-5p and mi-21-3p, have been found to play a role in the pathogenesis of DGF [117]. Secretory Leukocyte Peptidase Inhibitor (SLPI): ↑ urine and serum transcript expression levels were reported in post-RT AKI cases [117]. | |
| Postoperative Biomarkers Specific for Acute Renal Allograft Rejection | |
|---|---|
| Proteomic Evidence | Plasmatic samples:
|
Urinary samples:
| |
| Transcriptomic Evidence | Messenger (m)RNAs:
|
MicroRNAs (miRNAs):
| |
| Genomic Evidence | Gene signatures (array technology on multicenter graft biopsies and paired peripheral blood samples):
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Novacescu, D.; Latcu, S.C.; Bardan, R.; Daminescu, L.; Cumpanas, A.A. Contemporary Biomarkers for Renal Transplantation: A Narrative Overview. J. Pers. Med. 2023, 13, 1216. https://doi.org/10.3390/jpm13081216
Novacescu D, Latcu SC, Bardan R, Daminescu L, Cumpanas AA. Contemporary Biomarkers for Renal Transplantation: A Narrative Overview. Journal of Personalized Medicine. 2023; 13(8):1216. https://doi.org/10.3390/jpm13081216
Chicago/Turabian StyleNovacescu, Dorin, Silviu Constantin Latcu, Razvan Bardan, Liviu Daminescu, and Alin Adrian Cumpanas. 2023. "Contemporary Biomarkers for Renal Transplantation: A Narrative Overview" Journal of Personalized Medicine 13, no. 8: 1216. https://doi.org/10.3390/jpm13081216
APA StyleNovacescu, D., Latcu, S. C., Bardan, R., Daminescu, L., & Cumpanas, A. A. (2023). Contemporary Biomarkers for Renal Transplantation: A Narrative Overview. Journal of Personalized Medicine, 13(8), 1216. https://doi.org/10.3390/jpm13081216







