Mismatch Repair Protein Expression in Endometrial Cancer: Assessing Concordance and Unveiling Pitfalls in Two Different Immunohistochemistry Assays
Abstract
:1. Introduction
2. Materials and Methods
2.1. Case Selection
2.2. IHC Analysis and Interpretation
2.3. Statistical Analysis
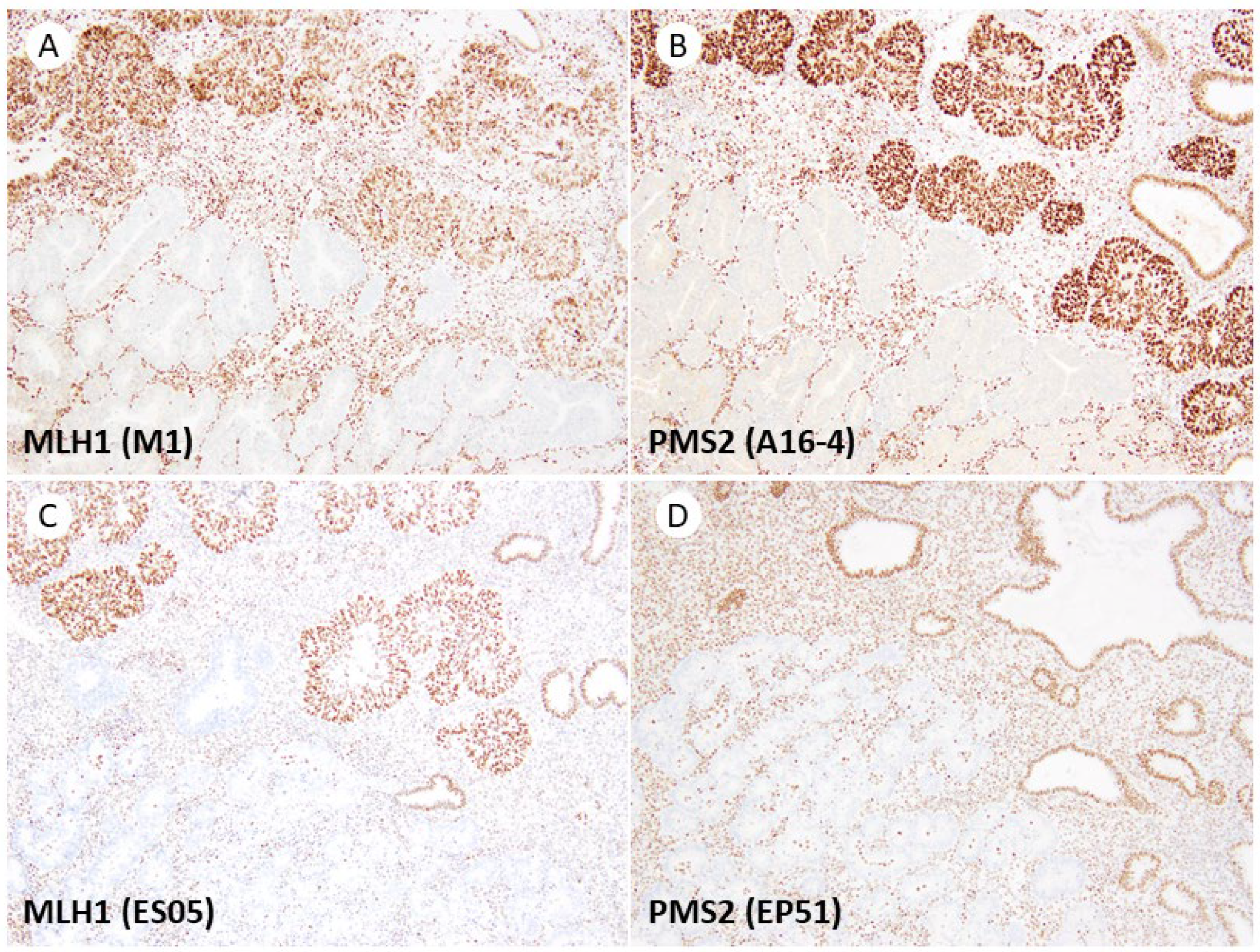
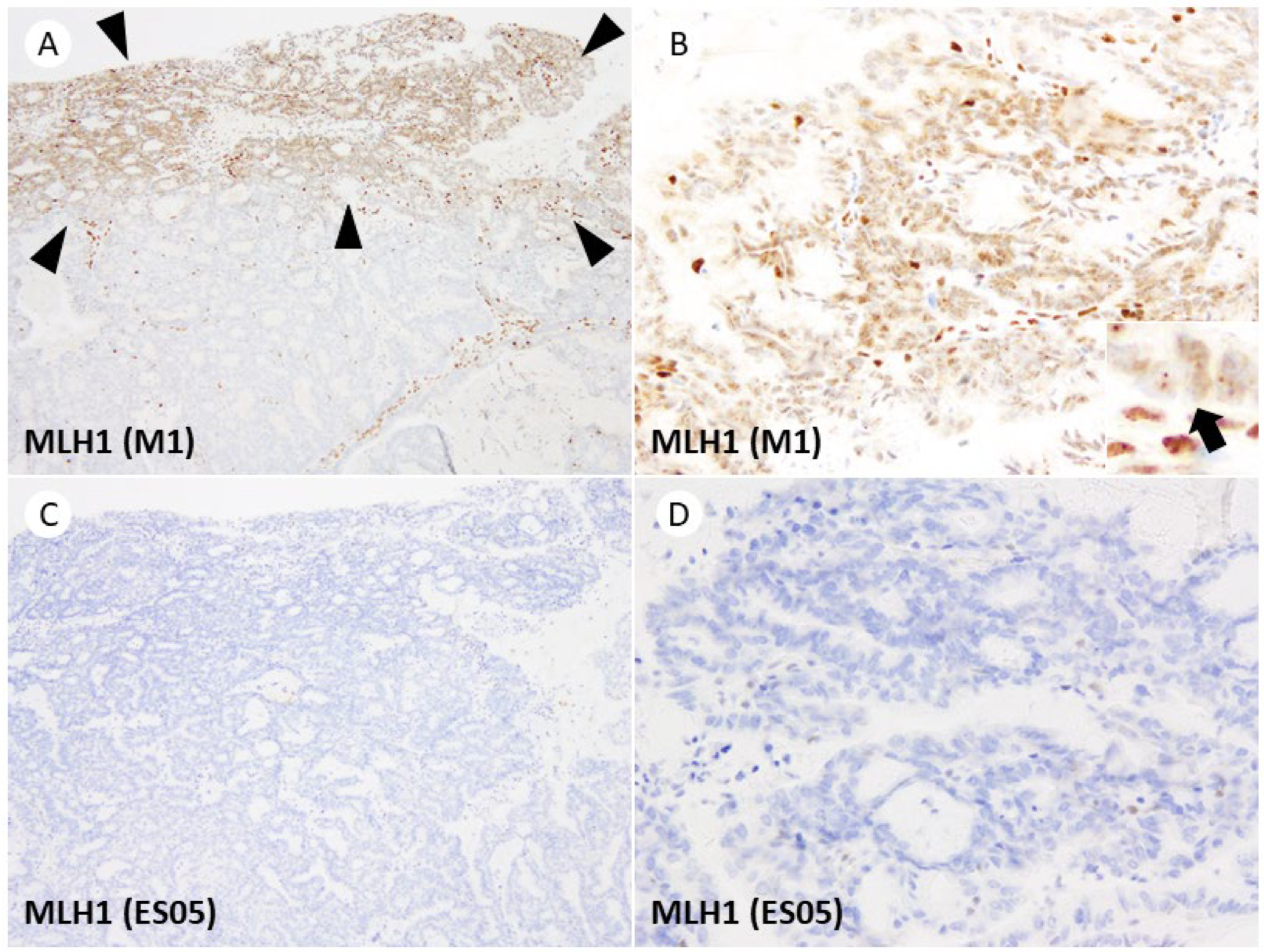
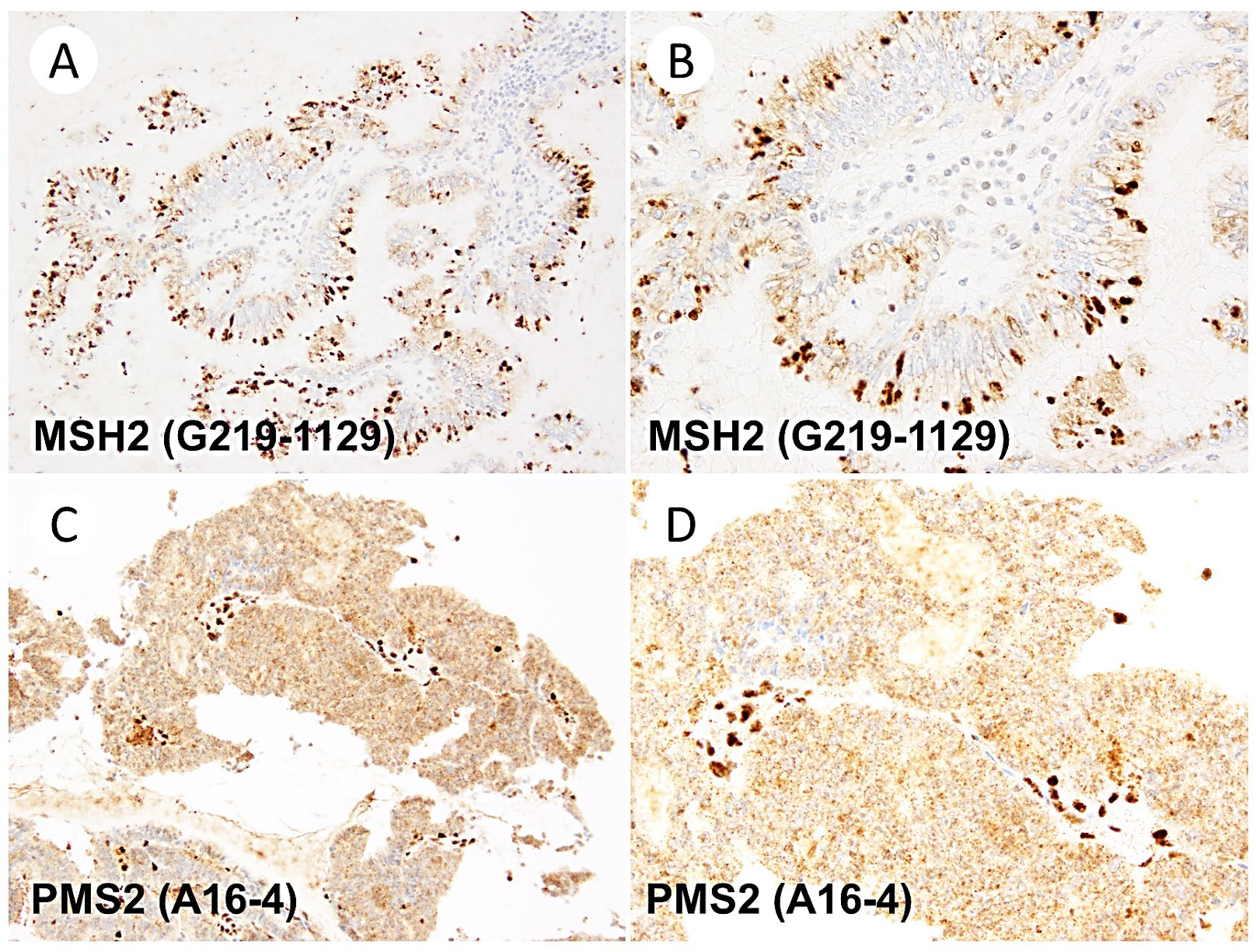
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Levine, D.A.; Getz, G.; Gabriel, S.B.; Cibulskis, K.; Lander, E.; Sivachenko, A.; Sougnez, C.; Lawrence, M.; Kandoth, C.; Dooling, D.; et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73. [Google Scholar] [CrossRef] [Green Version]
- Lu, K.H.; Broaddus, R.R. Endometrial Cancer. N. Engl. J. Med. 2020, 383, 2053–2064. [Google Scholar] [CrossRef]
- WHO Classification of Tumours Editorial Board. Female Genital Tumours, WHO Classification of Tumours, 5th ed.; International Agency for Research on Cancer: Lyon, France, 2020; Volume 4. [Google Scholar]
- Cerretelli, G.; Ager, A.; Arends, M.J.; Frayling, I.M. Molecular pathology of Lynch syndrome. J. Pathol. 2020, 250, 518–531. [Google Scholar] [CrossRef] [Green Version]
- Deshpande, M.; Romanski, P.A.; Rosenwaks, Z.; Gerhardt, J. Gynecological Cancers Caused by Deficient Mismatch Repair and Microsatellite Instability. Cancers 2020, 12, 3319. [Google Scholar] [CrossRef]
- Bartley, A.N.; Mills, A.M.; Konnick, E.; Overman, M.; Ventura, C.B.; Souter, L.; Colasacco, C.; Stadler, Z.K.; Kerr, S.; Howitt, B.E.; et al. Mismatch Repair and Microsatellite Instability Testing for Immune Checkpoint Inhibitor Therapy: Guideline from the College of American Pathologists in Collaboration With the Association for Molecular Pathology and Fight Colorectal Cancer. Arch. Pathol. Lab. Med. 2022, 146, 1194–1210. [Google Scholar] [CrossRef]
- Berek, J.S.; Matias-Guiu, X.; Creutzberg, C.; Fotopoulou, C.; Gaffney, D.; Kehoe, S.; Lindemann, K.; Mutch, D.; Concin, N.; Endometrial Cancer Staging Subcommittee, FIGO Women’s Cancer Committee. FIGO staging of endometrial cancer: 2023. Int. J. Gynaecol. Obstet. 2023, 162, 383–394. [Google Scholar] [CrossRef]
- NCCN Guidelines Version 2.2023 Uterine Neoplasm. 2023. Available online: https://www.nccn.org/professionals/physician_gls/pdf/uterine.pdf (accessed on 15 June 2023).
- Concin, N.; Creutzberg, C.L.; Vergote, I.; Cibula, D.; Mirza, M.R.; Marnitz, S.; Ledermann, J.A.; Bosse, T.; Chargari, C.; Fagotti, A.; et al. ESGO/ESTRO/ESP Guidelines for the management of patients with endometrial carcinoma. Virchows Arch. 2021, 478, 153–190. [Google Scholar] [CrossRef] [PubMed]
- Oaknin, A.; Bosse, T.J.; Creutzberg, C.L.; Giornelli, G.; Harter, P.; Joly, F.; Lorusso, D.; Marth, C.; Makker, V.; Mirza, M.R.; et al. Endometrial cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol. 2022, 33, 860–877. [Google Scholar] [CrossRef] [PubMed]
- Marabelle, A.; Le, D.T.; Ascierto, P.A.; Di Giacomo, A.M.; De Jesus-Acosta, A.; Delord, J.P.; Geva, R.; Gottfried, M.; Penel, N.; Hansen, A.R.; et al. Efficacy of Pembrolizumab in Patients with Noncolorectal High Microsatellite Instability/Mismatch Repair-Deficient Cancer: Results From the Phase II KEYNOTE-158 Study. J. Clin. Oncol. 2020, 38, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Parry, S.; Dodson, A. Mismatch Repair Protein antibodies and their performance in the UK National External Quality Assessment Scheme for Immunocytochemistry and In-situ Hybridisation. In Proceedings of the 31st European Congress of Pathology, Nice, France, 7–11 September 2019. PS-20-018 2019. [Google Scholar]
- MSH2 Run 57, Nordic Immunohistochemical Quality Control. 2019. Available online: https://www.nordiqc.org/downloads/assessments/122_82.pdf (accessed on 1 June 2023).
- MSH6 Run 61, Nordic Immunohistochemical Quality Control. 2021. Available online: https://www.nordiqc.org/downloads/assessments/145_83.pdf (accessed on 1 June 2023).
- PMS2 Run 62, Nordic Immunohistochemical Quality Control. 2021. Available online: https://www.nordiqc.org/downloads/assessments/146_84.pdf (accessed on 1 June 2023).
- MLH1 Run 67, Nordic Immunohistochemical Quality Control. 2023. Available online: https://www.nordiqc.org/downloads/assessments/171_81.pdf (accessed on 25 May 2023).
- Summary Report–Run 118 MMR (MLH1, PMS2, MSH2, MSH6); Canadian Pathology Quality Assurance: Richmond, BC, Canada, 2020.
- Hampel, H.; Pearlman, R.; de la Chapelle, A.; Pritchard, C.C.; Zhao, W.; Jones, D.; Yilmaz, A.; Chen, W.; Frankel, W.L.; Suarez, A.A.; et al. Double somatic mismatch repair gene pathogenic variants as common as Lynch syndrome among endometrial cancer patients. Gynecol. Oncol. 2021, 160, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Simpkins, S.B.; Bocker, T.; Swisher, E.M.; Mutch, D.G.; Gersell, D.J.; Kovatich, A.J.; Palazzo, J.P.; Fishel, R.; Goodfellow, P.J. MLH1 promoter methylation and gene silencing is the primary cause of microsatellite instability in sporadic endometrial cancers. Hum. Mol. Genet. 1999, 8, 661–666. [Google Scholar] [CrossRef] [Green Version]
- Casey, L.; Singh, N. POLE, MMR, and MSI Testing in Endometrial Cancer: Proceedings of the ISGyP Companion Society Session at the USCAP 2020 Annual Meeting. Int. J. Gynecol. Pathol. 2021, 40, 5–16. [Google Scholar] [CrossRef]
- Naveena Singh, R.W.; Tchrakian, N.; Allen, S.-G.; Clarke, B.; Gilks, C.B. Interpretation and Reporting Terminology for Mismatch Repair Protein Immunohistochemistry in Endometrial Cancer, BAGP Guidance Document: MMR Immunohistochemistry Interpretation and Terminology; Version 1.1; The British Association of Gynaecological Pathologists: London, UK, 2020. [Google Scholar]
- Dasgupta, S.; Ewing-Graham, P.C.; Groenendijk, F.H.; Stam, O.; Biermann, K.E.; Doukas, M.; Dubbink, H.J.; van Velthuysen, M.F.; Dinjens, W.N.M.; Van Bockstal, M.R. Granular dot-like staining with MLH1 immunohistochemistry is a clone-dependent artefact. Pathol. Res. Pract. 2020, 216, 152581. [Google Scholar] [CrossRef]
- Niu, B.T.; Hammond, R.F.L.; Leen, S.L.S.; Faruqi, A.Z.; Trevisan, G.; Gilks, C.B.; Singh, N. Artefactual punctate MLH1 staining can lead to erroneous reporting of isolated PMS2 loss. Histopathology 2018, 73, 703–705. [Google Scholar] [CrossRef] [PubMed]
- Asami, Y.; Kobayashi Kato, M.; Hiranuma, K.; Matsuda, M.; Shimada, Y.; Ishikawa, M.; Koyama, T.; Komatsu, M.; Hamamoto, R.; Nagashima, M.; et al. Utility of molecular subtypes and genetic alterations for evaluating clinical outcomes in 1029 patients with endometrial cancer. Br. J. Cancer 2023, 128, 1582–1591. [Google Scholar] [CrossRef]
- Stelloo, E.; Jansen, A.M.L.; Osse, E.M.; Nout, R.A.; Creutzberg, C.L.; Ruano, D.; Church, D.N.; Morreau, H.; Smit, V.; van Wezel, T.; et al. Practical guidance for mismatch repair-deficiency testing in endometrial cancer. Ann. Oncol. 2017, 28, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Scheiderer, A.; Riedinger, C.; Kimball, K.; Kilgore, L.; Orucevic, A. Reporting Subclonal Immunohistochemical Staining of Mismatch Repair Proteins in Endometrial Carcinoma in the Times of Ever-Changing Guidelines. Arch. Pathol. Lab. Med. 2022, 146, 1114–1121. [Google Scholar] [CrossRef]
- Dillon, J.L.; Gonzalez, J.L.; DeMars, L.; Bloch, K.J.; Tafe, L.J. Universal screening for Lynch syndrome in endometrial cancers: Frequency of germline mutations and identification of patients with Lynch-like syndrome. Hum. Pathol. 2017, 70, 121–128. [Google Scholar] [CrossRef]
- Watkins, J.C.; Nucci, M.R.; Ritterhouse, L.L.; Howitt, B.E.; Sholl, L.M. Unusual Mismatch Repair Immunohistochemical Patterns in Endometrial Carcinoma. Am. J. Surg. Pathol. 2016, 40, 909–916. [Google Scholar] [CrossRef]
- Markow, M.; Chen, W.; Frankel, W.L. Immunohistochemical Pitfalls: Common Mistakes in the Evaluation of Lynch Syndrome. Surg. Pathol. Clin. 2017, 10, 977–1007. [Google Scholar] [CrossRef] [PubMed]
- Loughrey, M.B.; Dunne, P.D.; Coleman, H.G.; McQuaid, S.; James, J.A. Punctate MLH1 mismatch repair immunostaining in colorectal cancer. Histopathology 2019, 74, 795–797. [Google Scholar] [CrossRef] [Green Version]
- Kommoss, S.; McConechy, M.K.; Kommoss, F.; Leung, S.; Bunz, A.; Magrill, J.; Britton, H.; Kommoss, F.; Grevenkamp, F.; Karnezis, A.; et al. Final validation of the ProMisE molecular classifier for endometrial carcinoma in a large population-based case series. Ann. Oncol. 2018, 29, 1180–1188. [Google Scholar] [CrossRef]
- Stelloo, E.; Nout, R.A.; Osse, E.M.; Jurgenliemk-Schulz, I.J.; Jobsen, J.J.; Lutgens, L.C.; van der Steen-Banasik, E.M.; Nijman, H.W.; Putter, H.; Bosse, T.; et al. Improved Risk Assessment by Integrating Molecular and Clinicopathological Factors in Early-stage Endometrial Cancer-Combined Analysis of the PORTEC Cohorts. Clin. Cancer Res. 2016, 22, 4215–4224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talhouk, A.; McConechy, M.K.; Leung, S.; Li-Chang, H.H.; Kwon, J.S.; Melnyk, N.; Yang, W.; Senz, J.; Boyd, N.; Karnezis, A.N.; et al. A clinically applicable molecular-based classification for endometrial cancers. Br. J. Cancer 2015, 113, 299–310. [Google Scholar] [CrossRef] [Green Version]
- Talhouk, A.; McConechy, M.K.; Leung, S.; Yang, W.; Lum, A.; Senz, J.; Boyd, N.; Pike, J.; Anglesio, M.; Kwon, J.S.; et al. Confirmation of ProMisE: A simple, genomics-based clinical classifier for endometrial cancer. Cancer 2017, 123, 802–813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Committee on Practice Bulletins-Gynecology, Society of Gynecologic Oncology. ACOG Practice Bulletin No. 147: Lynch syndrome. Obstet. Gynecol. 2014, 124, 1042–1054. [Google Scholar] [CrossRef]
- Chen, L.; Han, X. Anti-PD-1/PD-L1 therapy of human cancer: Past, present, and future. J. Clin. Investig. 2015, 125, 3384–3391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaguchi, H.; Hsu, J.M.; Yang, W.H.; Hung, M.C. Mechanisms regulating PD-L1 expression in cancers and associated opportunities for novel small-molecule therapeutics. Nat. Rev. Clin. Oncol. 2022, 19, 287–305. [Google Scholar] [CrossRef]
- Chan, T.A.; Yarchoan, M.; Jaffee, E.; Swanton, C.; Quezada, S.A.; Stenzinger, A.; Peters, S. Development of tumor mutation burden as an immunotherapy biomarker: Utility for the oncology clinic. Ann. Oncol. 2019, 30, 44–56. [Google Scholar] [CrossRef]
- Le, D.T.; Durham, J.N.; Smith, K.N.; Wang, H.; Bartlett, B.R.; Aulakh, L.K.; Lu, S.; Kemberling, H.; Wilt, C.; Luber, B.S.; et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017, 357, 409–413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandal, R.; Samstein, R.M.; Lee, K.W.; Havel, J.J.; Wang, H.; Krishna, C.; Sabio, E.Y.; Makarov, V.; Kuo, F.; Blecua, P.; et al. Genetic diversity of tumors with mismatch repair deficiency influences anti-PD-1 immunotherapy response. Science 2019, 364, 485–491. [Google Scholar] [CrossRef]
- Llosa, N.J.; Cruise, M.; Tam, A.; Wicks, E.C.; Hechenbleikner, E.M.; Taube, J.M.; Blosser, R.L.; Fan, H.; Wang, H.; Luber, B.S.; et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015, 5, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Malley, D.M.; Bariani, G.M.; Cassier, P.A.; Marabelle, A.; Hansen, A.R.; De Jesus Acosta, A.; Miller, W.H., Jr.; Safra, T.; Italiano, A.; Mileshkin, L.; et al. Pembrolizumab in Patients With Microsatellite Instability-High Advanced Endometrial Cancer: Results From the KEYNOTE-158 Study. J. Clin. Oncol. 2022, 40, 752–761. [Google Scholar] [CrossRef]

| Antibody | Clone | Manufacturer | Dilution | Antigen Retrieval | Incubation | Detection System | Auto Stainer |
|---|---|---|---|---|---|---|---|
| MLH1 | ES05 | Dako/Agilent | 1:200 | Dako PT Link * | 60 min RT | Envision | Link48 |
| M1 | Ventana/Roche | Prediluted | CC1 64 min | 24 min 37 °C | OptiView | Benchmark XT | |
| MSH2 | FE11 | Dako/Agilent | 1:200 | Dako PT Link * | 60 min RT | Envision + linker | Link48 |
| G219-1129 | Ventana/Roche | Prediluted | CC1 40 min | 12 min 37 °C | OptiView | Benchmark XT | |
| MSH6 | EP49 | Dako/Agilent | 1:400 | Dako PT Link * | 60 min RT | Envision | Link48 |
| SP93 | Ventana/Roche | Prediluted | CC1 64 min | 12 min 37 °C | OptiView | Benchmark XT | |
| PMS2 | EP51 | Dako/Agilent | 1:200 | Dako PT Link * | 60 min RT | Envision | Link48 |
| A16-4 | Ventana/Roche | Prediluted | CC1 92 min | 44 min 37 °C | OptiView + Amp | Benchmark XT |
| MLH1 | ES05 (Dako/Agilent) | ||
| Retained | Lost | ||
| M1 (Ventana/Roche) | Retained | 24 | 0 |
| Lost | 0 | 26 | |
| MSH2 | FE11 (Dako/Agilent) | ||
| Retained | Lost | ||
| G219-1129 (Ventana/Roche) | Retained | 38 | 0 |
| Lost | 0 | 12 | |
| MSH6 | EP49 (Dako/Agilent) | ||
| Retained | Lost | ||
| SP93 (Ventana/Roche) | Retained | 27 | 0 |
| Lost | 0 | 23 | |
| PMS2 | EP51 (Dako/Agilent) | ||
| Retained | Lost | ||
| A16-4 (Ventana/Roche) | Retained | 22 | 0 |
| Lost | 0 | 28 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoshida, H.; Takigawa, W.; Kobayashi-Kato, M.; Nishikawa, T.; Shiraishi, K.; Ishikawa, M. Mismatch Repair Protein Expression in Endometrial Cancer: Assessing Concordance and Unveiling Pitfalls in Two Different Immunohistochemistry Assays. J. Pers. Med. 2023, 13, 1260. https://doi.org/10.3390/jpm13081260
Yoshida H, Takigawa W, Kobayashi-Kato M, Nishikawa T, Shiraishi K, Ishikawa M. Mismatch Repair Protein Expression in Endometrial Cancer: Assessing Concordance and Unveiling Pitfalls in Two Different Immunohistochemistry Assays. Journal of Personalized Medicine. 2023; 13(8):1260. https://doi.org/10.3390/jpm13081260
Chicago/Turabian StyleYoshida, Hiroshi, Waku Takigawa, Mayumi Kobayashi-Kato, Tadaaki Nishikawa, Kouya Shiraishi, and Mitsuya Ishikawa. 2023. "Mismatch Repair Protein Expression in Endometrial Cancer: Assessing Concordance and Unveiling Pitfalls in Two Different Immunohistochemistry Assays" Journal of Personalized Medicine 13, no. 8: 1260. https://doi.org/10.3390/jpm13081260
APA StyleYoshida, H., Takigawa, W., Kobayashi-Kato, M., Nishikawa, T., Shiraishi, K., & Ishikawa, M. (2023). Mismatch Repair Protein Expression in Endometrial Cancer: Assessing Concordance and Unveiling Pitfalls in Two Different Immunohistochemistry Assays. Journal of Personalized Medicine, 13(8), 1260. https://doi.org/10.3390/jpm13081260






