Comprehensive Molecular Evaluation of HNF-1 Alpha, miR-27a, and miR-146 Gene Variants and Their Link with Predisposition and Progression in Type 2 Diabetes Patients
Abstract
:1. Introduction

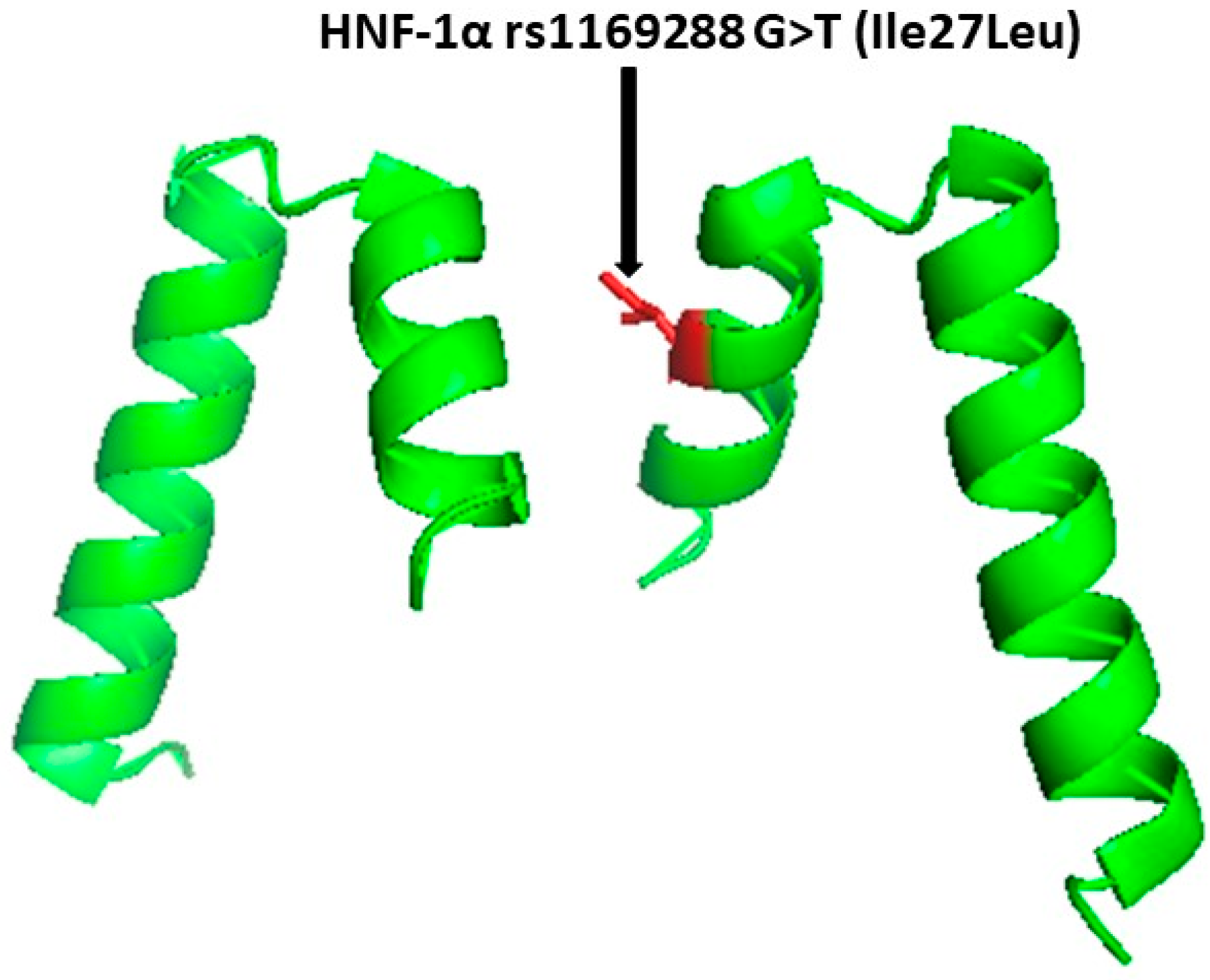
2. Methodology
2.1. Subjects of the Study
2.1.1. Inclusion Exclusion Criteria for T2D Cases
2.1.2. Inclusion Exclusion Criteria for Controls
2.2. Sample Collection and Extraction of Genomic DNA
2.3. Genotyping of miR-27a, miR-146, and HNF1alpha (rs1169288) SNVs
2.4. Genotyping of the HNF1alpha (rs1169288) A>C (I27L), miR-27a rs895819 A>G, and miR-146a-rs2910164 C>G
2.4.1. PCR Programming
2.4.2. Visualization of the PCR Product and Gel Electrophoresis
2.4.3. Amplification of microRNA-27a rs895819 A>G SNP
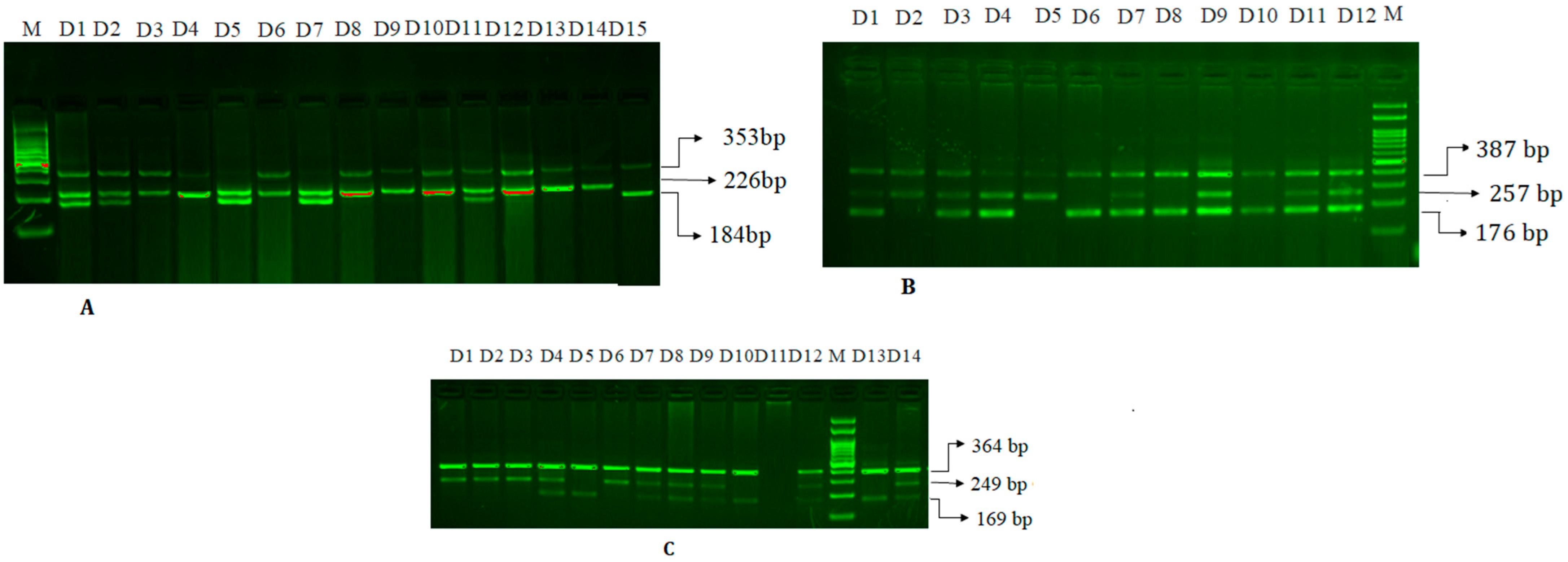
2.4.4. Amplification of HNF1A (rs1169288) A>C (I27L) SNP
2.4.5. Amplification of miR-146a rs2910164 C>G SNP
2.5. Statistical Analyses
3. Results
3.1. Demographic Characteristics of T2D Patients
3.2. Biochemical Characterization
3.3. Statistical Comparisons of T2D Patients and Controls for the HNF-1 rs1169288 G>T, miR-27a rs895819 A>G, and miR-146 rs2910164 C>G Genotypes
3.3.1. Association of HNF-1α rs1169288 G>T (Ile27Leu) Genotypes with T2D
3.3.2. Relationship between miR-27a rs895819 A>G Genotypes and T2D
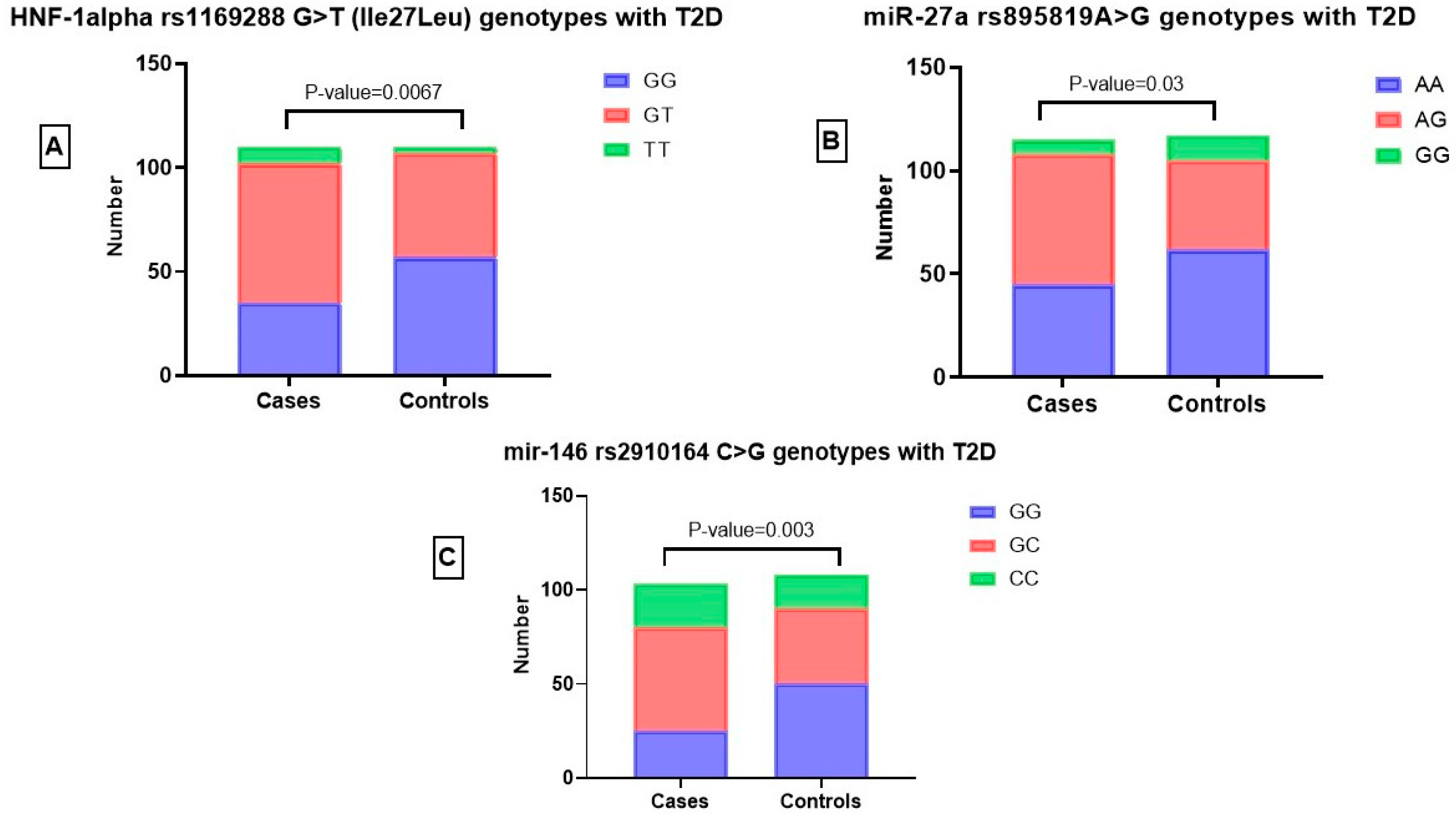
3.3.3. Relationship between miR-146 rs2910164 C>G Genotypes and T2D
3.4. Logistic Regression Analysis to Determine Association between HNF-1alpha rs1169288 G>T (Ile27Leu) Genotypes and Susceptibility to T2D
3.5. Association between HNF-1α rs1169288 G>T (Ile27Leu) Genotypes and the Clinicopathological Characteristics of the T2D Patients
3.6. Multivariate and Ordinal Regression Risk Factor Analysis for T2D with miR-27a rs895819 A>G Genotypes
3.7. Association between miR-27a-rs895819 G>A Genotypes and the Clinicopathological Characteristics of the T2D Patients
3.8. Logistic Regression Analysis of miR-146 rs2910164 C>G Genotypes to Predict Risk of T2D
3.9. Association between miR-146 rs2910164 C>G Genotypes and the Clinicopathological Characteristics of the T2D Patients
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Beckman, J.A.; Creager, M.A. Vascular Complications of Diabetes. Circ. Res. 2016, 118, 1771–1785. [Google Scholar] [CrossRef] [PubMed]
- American Diabetes Association. Economic Costs of Diabetes in the U.S. in 2017. Diabetes Care 2018, 41, 917–928. [Google Scholar] [CrossRef] [PubMed]
- Alotaibi, A.; Perry, L.; Gholizadeh, L.; Al-Ganmi, A. Incidence and prevalence rates of diabetes mellitus in Saudi Arabia: An overview. J. Epidemiol. Glob. Health 2017, 7, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Reed, J.; Bain, S.; Kanamarlapudi, V. A Review of Current Trends with Type 2 Diabetes Epidemiology, Aetiology, Pathogenesis, Treatments and Future Perspectives. Diabetes Metab. Syndr. Obes. 2021, 14, 3567–3602. [Google Scholar] [CrossRef]
- Jarrar, M.; Abusalah, M.A.H.; Albaker, W.; Al-Bsheish, M.; Alsyouf, A.; Al-Mugheed, K.; Issa, M.R.; Alumran, A. Prevalence of Type 2 Diabetes Mellitus in the General Population of Saudi Arabia, 2000–2020: A Systematic Review and Meta-Analysis of Observational Studies. Saudi J. Med. Med. Sci. 2023, 11, 1–10. [Google Scholar] [CrossRef]
- Galicia-Garcia, U.; Benito-Vicente, A.; Jebari, S.; Larrea-Sebal, A.; Siddiqi, H.; Uribe, K.B.; Ostolaza, H.; Martin, C. Pathophysiology of Type 2 Diabetes Mellitus. Int. J. Mol. Sci. 2020, 21, 6275. [Google Scholar] [CrossRef]
- Plows, J.F.; Stanley, J.L.; Baker, P.N.; Reynolds, C.M.; Vickers, M.H. The Pathophysiology of Gestational Diabetes Mellitus. Int. J. Mol. Sci. 2018, 19, 3342. [Google Scholar] [CrossRef]
- Kyrou, I.; Tsigos, C.; Mavrogianni, C.; Cardon, G.; Van Stappen, V.; Latomme, J.; Kivela, J.; Wikstrom, K.; Tsochev, K.; Nanasi, A.; et al. Sociodemographic and lifestyle-related risk factors for identifying vulnerable groups for type 2 diabetes: A narrative review with emphasis on data from Europe. BMC Endocr. Disord. 2020, 20, 134. [Google Scholar] [CrossRef]
- Elfaki, I.; Mir, R.; Mir, M.M.; AbuDuhier, F.M.; Babakr, A.T.; Barnawi, J. Potential Impact of MicroRNA Gene Polymorphisms in the Pathogenesis of Diabetes and Atherosclerotic Cardiovascular Disease. J. Pers. Med. 2019, 9, 51. [Google Scholar] [CrossRef]
- Elfaki, I.; Mir, R.; Abu-Duhier, F.; Khan, R.; Sakran, M. Phosphatidylinositol 3-kinase Glu545Lys and His1047Tyr Mutations are not Associated with T2D. Curr. Diabetes Rev. 2019, 15, 881–888. [Google Scholar] [CrossRef]
- Elfaki, I.; Mir, R.; Abu-Duhier, F.M.; Jha, C.K.; Ahmad Al-Alawy, A.I.; Babakr, A.T.; Habib, S.A.E. Analysis of the Potential Association of Drug-Metabolizing Enzymes CYP2C9*3 and CYP2C19*3 Gene Variations with Type 2 Diabetes: A Case-Control Study. Curr. Drug Metab. 2020, 21, 1152–1160. [Google Scholar] [CrossRef] [PubMed]
- Elfaki, I.; Mir, R.; Almutairi, F.M.; Duhier, F.M.A. Cytochrome P450: Polymorphisms and Roles in Cancer, Diabetes and Atherosclerosis. Asian Pac. J. Cancer Prev. 2018, 19, 2057–2070. [Google Scholar] [PubMed]
- Elfaki, I.; Mir, R.; Abu-Duhier, F.; Alotaibi, M.; Alalawy, A.; Barnawi, J.; Babakr, A.; Mir, M.; Mirghani, H. Clinical Implications of MiR128, Angiotensin I Converting Enzyme and Vascular Endothelial Growth Factor Gene Abnormalities and Their Association with T2D. Curr. Issues Mol. Biol. 2021, 43, 1859–1875. [Google Scholar] [CrossRef]
- Jha, C.K.; Mir, R.; Elfaki, I.; Javid, J.; Babakr, A.T.; Banu, S.; Chahal, S.M.S. Evaluation of the Association of Omentin 1 rs2274907 A>T and rs2274908 G>A Gene Polymorphisms with Coronary Artery Disease in Indian Population: A Case Control Study. J. Pers. Med. 2019, 9, 30. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S.J.; Fulton-Howard, B.; Goate, A. Interpretation of risk loci from genome-wide association studies of Alzheimer’s disease. Lancet Neurol. 2020, 19, 326–335. [Google Scholar] [CrossRef]
- Cano-Gamez, E.; Trynka, G. From GWAS to Function: Using Functional Genomics to Identify the Mechanisms Underlying Complex Diseases. Front. Genet. 2020, 11, 424. [Google Scholar] [CrossRef]
- Alzahrani, O.R.; Mir, R.; Alatwi, H.E.; Hawsawi, Y.M.; Alharbi, A.A.; Alessa, A.H.; Albalawi, E.S.; Elfaki, I.; Alalawi, Y.; Moharam, L.; et al. Potential Impact of PI3K-AKT Signaling Pathway Genes, KLF-14, MDM4, miRNAs 27a, miRNA-196a Genetic Alterations in the Predisposition and Progression of Breast Cancer Patients. Cancers 2023, 15, 1281. [Google Scholar] [CrossRef]
- Jalal, M.M.; Mir, R.; Hamadi, A.; Altayar, M.A.; Elfaki, I.; Barnawi, J.; Alkayyal, A.A.; Amr, M.; Hadeel, J.; Moawadh, M.S.; et al. Association of Genetic and Allelic Variants of Von Willebrand Factor (VWF), Glutathione S-Transferase and Tumor Necrosis Factor Alpha with Ischemic Stroke Susceptibility and Progression in the Saudi Population. Life 2023, 13, 1200. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Cione, E.; Cannataro, R.; Gallelli, L.; De Sarro, G.; Caroleo, M.C. Exosome microRNAs in Metabolic Syndrome as Tools for the Early Monitoring of Diabetes and Possible Therapeutic Options. Pharmaceuticals 2021, 14, 1257. [Google Scholar] [CrossRef]
- Chen, T.; Zhang, Y.; Liu, Y.; Zhu, D.; Yu, J.; Li, G.; Sun, Z.; Wang, W.; Jiang, H.; Hong, Z. MiR-27a promotes insulin resistance and mediates glucose metabolism by targeting PPAR-gamma-mediated PI3K/AKT signaling. Aging 2019, 11, 7510–7524. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.Y.; Cheng, J.; Zhou, M.Y.; Liang, L.L.; Lian, S.M.; Xie, X.S.; Xu, S.; Liu, X.; Xiong, X.D. The association between pre-miR-27a rs895819 polymorphism and myocardial infarction risk in a Chinese Han population. Lipids Health Dis. 2018, 17, 7. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Chen, Z.; Xu, J.; Li, W.; Zhao, J. Pre-mir-27a rs895819 polymorphism and cancer risk: A meta-analysis. Mol. Biol. Rep. 2013, 40, 3181–3186. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Gui, Y.F.; Liao, W.Y.; Zhang, Y.Q.; Zhang, X.B.; Huang, Y.P.; Wu, F.M.; Huang, Z.; Lu, Y.F. Association between miR-27a rs895819 polymorphism and breast cancer susceptibility: Evidence based on 6118 cases and 7042 controls. Medicine 2021, 100, e23834. [Google Scholar] [CrossRef] [PubMed]
- Roos, J.; Dahlhaus, M.; Funcke, J.B.; Kustermann, M.; Strauss, G.; Halbgebauer, D.; Boldrin, E.; Holzmann, K.; Moller, P.; Trojanowski, B.M.; et al. miR-146a regulates insulin sensitivity via NPR3. Cell Mol. Life Sci. 2021, 78, 2987–3003. [Google Scholar] [CrossRef]
- Mehanna, E.T.; Ghattas, M.H.; Mesbah, N.M.; Saleh, S.M.; Abo-Elmatty, D.M. Association of MicroRNA-146a rs2910164 Gene Polymorphism with Metabolic Syndrome. Folia Biol. 2015, 61, 43–48. [Google Scholar]
- Miyachi, Y.; Miyazawa, T.; Ogawa, Y. HNF1A Mutations and Beta Cell Dysfunction in Diabetes. Int. J. Mol. Sci. 2022, 23, 3222. [Google Scholar] [CrossRef]
- Beysel, S.; Eyerci, N.; Pinarli, F.A.; Kizilgul, M.; Ozcelik, O.; Caliskan, M.; Cakal, E. HNF1A gene p.I27L is associated with early-onset, maturity-onset diabetes of the young-like diabetes in Turkey. BMC Endocr. Disord. 2019, 19, 51. [Google Scholar] [CrossRef]
- Li, L.M.; Jiang, B.G.; Sun, L.L. HNF1A: From Monogenic Diabetes to Type 2 Diabetes and Gestational Diabetes Mellitus. Front. Endocrinol. 2022, 13, 829565. [Google Scholar] [CrossRef]
- Laakso, M. Biomarkers for type 2 diabetes. Mol. Metab. 2019, 27S, S139–S146. [Google Scholar] [CrossRef]
- Christodoulou, M.I.; Avgeris, M.; Kokkinopoulou, I.; Maratou, E.; Mitrou, P.; Kontos, C.K.; Pappas, E.; Boutati, E.; Scorilas, A.; Fragoulis, E.G. Blood-based analysis of type-2 diabetes mellitus susceptibility genes identifies specific transcript variants with deregulated expression and association with disease risk. Sci. Rep. 2019, 9, 1512. [Google Scholar] [CrossRef]
- World Health Organization. Diagnosis and Management of Type 2 Diabetes (HEARTS-D); World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Medrano, R.F.; de Oliveira, C.A. Guidelines for the tetra-primer ARMS-PCR technique development. Mol. Biotechnol. 2014, 56, 599–608. [Google Scholar] [CrossRef]
- Ahlawat, S.; Sharma, R.; Maitra, A.; Roy, M.; Tantia, M.S. Designing, optimization and validation of tetra-primer ARMS PCR protocol for genotyping mutations in caprine Fec genes. Meta Gene 2014, 2, 439–449. [Google Scholar] [CrossRef]
- Mashayekhi, S.; Saeidi Saedi, H.; Salehi, Z.; Soltanipour, S.; Mirzajani, E. Effects of miR-27a, miR-196a2 and miR-146a polymorphisms on the risk of breast cancer. Br. J. Biomed. Sci. 2018, 75, 76–81. [Google Scholar] [CrossRef]
- Mir, R.; Elfaki, I.; Jha, C.; Javid, D.J.; Rehman, S.; Banu, S.; Mir, M.; Babakr, A.; Chahal, S.M. Molecular Evaluation of MicroRNA-146 Gene Variability (rs2910164 C>G) and its Association with Increased Susceptibility to Coronary Artery Disease. MicroRNA 2020, 9, 363–372. [Google Scholar] [CrossRef]
- Al-Lawati, J.A. Diabetes Mellitus: A Local and Global Public Health Emergency! Oman Med. J. 2017, 32, 177–179. [Google Scholar] [CrossRef]
- Gardner, D.S.; Tai, E.S. Clinical features and treatment of maturity onset diabetes of the young (MODY). Diabetes Metab. Syndr. Obes. 2012, 5, 101–108. [Google Scholar] [CrossRef]
- Morita, K.; Saruwatari, J.; Tanaka, T.; Oniki, K.; Kajiwara, A.; Otake, K.; Ogata, Y.; Nakagawa, K. Associations between the common HNF1A gene variant p.I27L (rs1169288) and risk of type 2 diabetes mellitus are influenced by weight. Diabetes Metab. 2015, 41, 91–94. [Google Scholar] [CrossRef]
- Low, B.S.J.; Lim, C.S.; Ding, S.S.L.; Tan, Y.S.; Ng, N.H.J.; Krishnan, V.G.; Ang, S.F.; Neo, C.W.Y.; Verma, C.S.; Hoon, S.; et al. Decreased GLUT2 and glucose uptake contribute to insulin secretion defects in MODY3/HNF1A hiPSC-derived mutant beta cells. Nat. Commun. 2021, 12, 3133. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhang, Y.; Bai, R.; Yang, R.; Shan, Z.; Ma, C.; Yang, J.; Sun, D. Association of Genetic Polymorphisms in MicroRNAs With Type 2 Diabetes Mellitus in a Chinese Population. Front. Endocrinol. 2020, 11, 587561. [Google Scholar] [CrossRef]
- Nunez Lopez, Y.O.; Garufi, G.; Seyhan, A.A. Altered levels of circulating cytokines and microRNAs in lean and obese individuals with prediabetes and type 2 diabetes. Mol. Biosyst. 2016, 13, 106–121. [Google Scholar] [CrossRef]
- Ma, X.; Wang, D.; Zhao, W.; Xu, L. Deciphering the Roles of PPARgamma in Adipocytes via Dynamic Change of Transcription Complex. Front. Endocrinol. 2018, 9, 473. [Google Scholar] [CrossRef]
- Hou, N.; Mai, Y.; Qiu, X.; Yuan, W.; Li, Y.; Luo, C.; Liu, Y.; Zhang, G.; Zhao, G.; Luo, J.D. Carvacrol Attenuates Diabetic Cardiomyopathy by Modulating the PI3K/AKT/GLUT4 Pathway in Diabetic Mice. Front. Pharmacol. 2019, 10, 998. [Google Scholar] [CrossRef]
- Khan, A.A.; Agarwal, H.; Reddy, S.S.; Arige, V.; Natarajan, B.; Gupta, V.; Kalyani, A.; Barthwal, M.K.; Mahapatra, N.R. MicroRNA 27a Is a Key Modulator of Cholesterol Biosynthesis. Mol. Cell Biol. 2020, 40, e00470-19. [Google Scholar] [CrossRef]
- Alipoor, B.; Ghaedi, H.; Meshkani, R.; Torkamandi, S.; Saffari, S.; Iranpour, M.; Omrani, M.D. Association of MiR-146a Expression and Type 2 Diabetes Mellitus: A Meta-Analysis. Int. J. Mol. Cell Med. 2017, 6, 156–163. [Google Scholar]
- Iguchi, T.; Nambara, S.; Masuda, T.; Komatsu, H.; Ueda, M.; Kidogami, S.; Ogawa, Y.; Hu, Q.; Sato, K.; Saito, T.; et al. miR-146a Polymorphism (rs2910164) Predicts Colorectal Cancer Patients’ Susceptibility to Liver Metastasis. PLoS ONE 2016, 11, e0165912. [Google Scholar] [CrossRef]
- Saba, R.; Sorensen, D.L.; Booth, S.A. MicroRNA-146a: A Dominant, Negative Regulator of the Innate Immune Response. Front. Immunol. 2014, 5, 578. [Google Scholar] [CrossRef]
- Sindhu, S.; Akhter, N.; Kochumon, S.; Thomas, R.; Wilson, A.; Shenouda, S.; Tuomilehto, J.; Ahmad, R. Increased Expression of the Innate Immune Receptor TLR10 in Obesity and Type-2 Diabetes: Association with ROS-Mediated Oxidative Stress. Cell Physiol. Biochem. 2018, 45, 572–590. [Google Scholar] [CrossRef]
- Sun, X.J.; Kim, S.P.; Zhang, D.; Sun, H.; Cao, Q.; Lu, X.; Ying, Z.; Li, L.; Henry, R.R.; Ciaraldi, T.P.; et al. Deletion of interleukin 1 receptor-associated kinase 1 (Irak1) improves glucose tolerance primarily by increasing insulin sensitivity in skeletal muscle. J. Biol. Chem. 2017, 292, 12339–12350. [Google Scholar] [CrossRef]
- Chatzigeorgiou, A.; Seijkens, T.; Zarzycka, B.; Engel, D.; Poggi, M.; van den Berg, S.; van den Berg, S.; Soehnlein, O.; Winkels, H.; Beckers, L.; et al. Blocking CD40-TRAF6 signaling is a therapeutic target in obesity-associated insulin resistance. Proc. Natl. Acad. Sci. USA 2014, 111, 2686–2691. [Google Scholar] [CrossRef]
- Li, K.; Zhao, B.; Wei, D.; Wang, W.; Cui, Y.; Qian, L.; Liu, G. miR146a improves hepatic lipid and glucose metabolism by targeting MED1. Int. J. Mol. Med. 2020, 45, 543–555. [Google Scholar]
- Zhou, J.; Singh, B.K.; Ho, J.P.; Lim, A.; Bruinstroop, E.; Ohba, K.; Sinha, R.A.; Yen, P.M. MED1 mediator subunit is a key regulator of hepatic autophagy and lipid metabolism. Autophagy 2021, 17, 4043–4061. [Google Scholar] [CrossRef]
- Jia, Y.; Viswakarma, N.; Reddy, J.K. Med1 subunit of the mediator complex in nuclear receptor-regulated energy metabolism, liver regeneration, and hepatocarcinogenesis. Gene Expr. 2014, 16, 63–75. [Google Scholar] [CrossRef]
- Zhang, T.; Li, L.; Shang, Q.; Lv, C.; Wang, C.; Su, B. Circulating miR-126 is a potential biomarker to predict the onset of type 2 diabetes mellitus in susceptible individuals. Biochem. Biophys. Res. Commun. 2015, 463, 60–63. [Google Scholar] [CrossRef]
- Cui, X.; You, L.; Zhu, L.; Wang, X.; Zhou, Y.; Li, Y.; Wen, J.; Xia, Y.; Wang, X.; Ji, C.; et al. Change in circulating microRNA profile of obese children indicates future risk of adult diabetes. Metabolism 2018, 78, 95–105. [Google Scholar] [CrossRef]
- Karolina, D.S.; Tavintharan, S.; Armugam, A.; Sepramaniam, S.; Pek, S.L.; Wong, M.T.; Lim, S.C.; Sum, C.F.; Jeyaseelan, K. Circulating miRNA profiles in patients with metabolic syndrome. J. Clin. Endocrinol. Metab. 2012, 97, E2271–E2276. [Google Scholar] [CrossRef]
- Yang, Z.; Chen, H.; Si, H.; Li, X.; Ding, X.; Sheng, Q.; Chen, P.; Zhang, H. Serum miR-23a, a potential biomarker for diagnosis of pre-diabetes and type 2 diabetes. Acta Diabetol. 2014, 51, 823–831. [Google Scholar] [CrossRef]
- Shubrook, J.H.; Chen, W.; Lim, A. Evidence for the Prevention of Type 2 Diabetes Mellitus. J. Am. Osteopath. Assoc. 2018, 118, 730–737. [Google Scholar] [CrossRef]
- Sakane, N.; Sato, J.; Tsushita, K.; Tsujii, S.; Kotani, K.; Tsuzaki, K.; Tominaga, M.; Kawazu, S.; Sato, Y.; Usui, T.; et al. Prevention of type 2 diabetes in a primary healthcare setting: Three-year results of lifestyle intervention in Japanese subjects with impaired glucose tolerance. BMC Public Health 2011, 11, 40. [Google Scholar] [CrossRef]
- Kirwan, J.P.; Sacks, J.; Nieuwoudt, S. The essential role of exercise in the management of type 2 diabetes. Cleve Clin. J. Med. 2017, 84 (Suppl. S1), S15–S21. [Google Scholar] [CrossRef]
- Bekele, H.; Asefa, A.; Getachew, B.; Belete, A.M. Barriers and Strategies to Lifestyle and Dietary Pattern Interventions for Prevention and Management of TYPE-2 Diabetes in Africa, Systematic Review. J. Diabetes Res. 2020, 2020, 7948712. [Google Scholar] [CrossRef]
| Amplification Refractory Mutation System PCR Primers HNF1A (rs1169288) A>C (I27L) SNP | ||||
| HNF1A-Fo | 5′-GTGCCCACAGGGCTTGGCTAG-3′ | 387 bp | 62 °C | |
| HNF1A-Ro | 5′-CCATCGTCGTCCGTCTCGTCCTCG-3′ | |||
| HNF1A-FI | (G allele) | 5′-GGGCTGAGCAAAGAGGCACCG-3′ | 176 bp | |
| HNF1A-RI | (A allele) | 5′-CCCGGCTCACCCAGTGCCTGAAT-3′ | 257 bp | |
| ARMS primers for miR27a G>A gene variation | ||||
| miR-27a-Fo | 5′-GGC TTG ACC CCT GTT CCT GCT GAA CT-3′ | 353 bp | 63.5 °C | |
| miR-27a-Ro | 5′-TTG CTT CCT GTC ACA AAT CAC ATT GCC A-3′ | |||
| miR-27a-FI | (G allele) | 5′-GGA ACT TAG CCA CTG TGA ACA CGA CTT TGC-3′ | 184 bp | |
| miR-27a-RI | (A allele) | 5′-CTT AGC TGC TTG TGA GCA GGG TCC CCA-3′ | 226 bp | |
| Amplification Refractory Mutation System PCR primers for miR146a-rs2910164 C>G SNP | ||||
| miR146a Fo | 5′-GGC CTG GTC TCC TCC AGA TGT TTA T-3′ | 364 bp | 61.5 °C | |
| miR146a Ro | 5′-ATA CCT TCA GAG CCT GAG ACT CTG CC-3′ | |||
| miR146a FI | (C allele) | 5′-ATG GGT TGT GTC AGT GTC AGA CCT C-3′ | 169 bp | |
| miR146a RI | (G allele) | 5′-GAT ATC CCA GCT GAA GAA CTG AAT TTC AC-3′ | 249 bp | |
| Clinical Features | N= | % |
|---|---|---|
| 115 | ||
| Male | 82 | 71.30% |
| Female | 33 | 28.70% |
| Age > 40 | 91 | 79.13% |
| Age < 40 | 24 | 20.87% |
| FBG < 100 mg/dL | 24 | 20.87% |
| FBG > 100 mg/dL | 91 | 79.13% |
| HBA1c > 6% | 90 | 78.26% |
| HBA1c < 6% | 25 | 21.74% |
| Triglycerides mg/dL < 200 | 39 | 33.91% |
| Triglycerides mg/dL > 200 | 76 | 66.09% |
| Cholesterol mg/dL < 200 | 70 | 60.87% |
| Cholesterol mg/dL > 200 | 45 | 39.13% |
| LDL-C mg/dL < 100 | 57 | 49.57% |
| LDL-C mg/dL > 100 | 58 | 50.43% |
| HDL-L mg/dL < 55 | 48 | 41.74% |
| HDL-L mg/dL > 55 | 67 | 58.26% |
| Genotypes | Healthy Controls (N = 110) | T2D Cases (N = 110) | Odd Ratio OR (95% CI) | Risk Ratio RR (95% CI) | p-Value |
|---|---|---|---|---|---|
| Codominant Inheritance model | |||||
| HNF-1α-GG | 57 | 35 | Ref | Ref | |
| HNF-1α-GT | 50 | 67 | 2.18(1.2491 to 3.8127) | 1.44(1.1135 to 1.8876) | 0.0061 |
| HNF-1α-TT | 03 | 08 | 4.34(1.0794 to 17.4722) | 2.27(0.8541 to 6.0423) | 0.038 |
| Dominant Inheritance model | |||||
| HNF-1α-GG | 57 | 35 | Ref | Ref | |
| HNF-1α-(GT+TT) | 53 | 75 | 2.30(1.3316 to 3.9885) | 1.49(1.1526 to 1.9425) | 0.0029 |
| Recessive Inheritance model | |||||
| HNF-1α-(GT+GG) | 107 | 102 | Ref | Ref | |
| HNF-1α-TT | 03 | 08 | 2.79(0.7220 to 10.8379) | 1.87(0.7087 to 4.9721) | 0.136 |
| Allele | |||||
| HNF-1α-G | 164 | 137 | Ref | Ref | |
| HNF-1α-T | 56 | 83 | 1.77(1.1800 to 2.6677) | 1.35(1.0775 to 1.6974) | 0.0059 |
| Over dominant Inheritance model | |||||
| HNF-1α-(GG+TT) | 60 | 43 | Ref | Ref | |
| HNF-1α-(GT) | 50 | 67 | 1.86(1.0937 to 3.1964) | 1.36(1.0448 to 1.7784) | 0.022 |
| Clinical Feature | N= | GG | GA | AA | X2 | DF | p-Value |
|---|---|---|---|---|---|---|---|
| Association of HNF-1α SNV with Gender | |||||||
| Male | 80 | 20(25%) | 55(68.75%) | 05(6.25%) | 7.67 | 2 | 0.021 |
| Female | 30 | 15(50%) | 12(40%) | 3(10%) | |||
| Association of HNF-1α SNV with Age | |||||||
| >40 | 78 | 23(29.48%) | 49(62.82%) | 06(7.69%) | 0.68 | 2 | 0.711 |
| <40 | 32 | 12(37.5%) | 18(56.25%) | 02(6.25%) | |||
| Association of HNF-1 alpha SNV with Fasting glucose mg/dL | |||||||
| <100 | 21 | 13(61.90%) | 5(23.80%) | 03(14.28%) | 15 | 2 | 0.0006 |
| >100 | 89 | 22(24.71%) | 62(69.66%) | 5(5.61%) | |||
| Association of HNF-1alpha SNV with HbA1c % | |||||||
| >6 | 88 | 22(25%) | 60(68.18%) | 5(5.68%) | 11.4 | 2 | 0.003 |
| <6 | 22 | 13(59.09%) | 7(31.81%) | 3(13.63%) | |||
| Association of HNF-1alpha SNV with Triglycerides mg/dL | |||||||
| <200 | 77 | 22(28.57%) | 50(64.93%) | 05(6.49%) | 1.75 | 2 | 0.41 |
| >200 | 33 | 13(39.39%) | 17(51.51%) | 03(9.09%) | |||
| Association of HNF-1alpha SNV with Cholesterol mg/dL | |||||||
| <200 | 70 | 19(27.14%) | 48(68.57%) | 03(4.28%) | 5.54 | 2 | 0.062 |
| >200 | 40 | 16(40%) | 19(47.5%) | 05(12.5%) | |||
| Association of HNF-1alpha SNV with LDL-C mg/dL | |||||||
| <100 | 57 | 16(28.07%) | 56(98.24%) | 05(8.77%) | 0.99 | 2 | 0.609 |
| >100 | 53 | 19(35.84%) | 31(58.49%) | 03(5.66%) | |||
| Association of HNF-1alpha SNV with HDL-L mg/dL | |||||||
| <55 | 44 | 10(22.72%) | 33(75%) | 01(2.27%) | 6.82 | 2 | 0.033 |
| >55 | 66 | 25(37.87%) | 34(51.51%) | 07(10.60%) | |||
| Genotypes | Healthy Controls (N = 117) | T2D Cases N = 115 | OR (95% CI) | Risk Ratio (RR) | p-Value |
|---|---|---|---|---|---|
| Codominant model | |||||
| miR-27a-AA | 62 | 45 | 1 Ref | 1 Ref | |
| miR-27a-AG | 43 | 63 | 2.01(1.169 to 3.483) | 1.42(1.0706 to 1.899) | 0.011 |
| miR-27a-GG | 12 | 07 | 0.80(0.2935 to 2.206) | 0.91(0.6279 to 1.347) | 0.67 |
| Dominant model | |||||
| miR-27a-AA | 62 | 45 | 1 Ref | 1 Ref | |
| miR-27a-(GG+GA) | 55 | 68 | 1.70(1.009 to 2.870) | 1.29(1.009 to 1.674) | 0.046 |
| Recessive model | |||||
| miR-27a-(GA+AA) | 105 | 108 | 1 Ref | 1 Ref | |
| miR-27a-GG | 12 | 07 | 0.56(0.215 to 1.493) | 0.78(0.539 to 1.1294) | 0.25 |
| Allele | |||||
| miR-27a-A | 167 | 153 | 1 Ref | 1 Ref | |
| miR-27a-G | 67 | 75 | 1.22(0.822 to 1.817) | 1.10(0.902 to 1.355) | 0.32 |
| Variable | N= | AA | AG | GG | X2 | DF | p-Value |
|---|---|---|---|---|---|---|---|
| 115 | 45 | 63 | 07 | ||||
| Association of miR-27a SNP with Gender | |||||||
| Male | 82 | 28(34.14%) | 50(60.97%) | 4(4.87%) | 4.5 | 2 | 0.101 |
| Female | 33 | 17(51.51%) | 13(39.39%) | 3(9%) | |||
| Association of miR-27a SNP with Age | |||||||
| Age > 40 | 91 | 33(36.16%) | 52(57.14%) | 6(6.59%) | 1.54 | 2 | 0.46 |
| Age < 40 | 24 | 12(42.85%) | 11(45.83%) | 1(4.16%) | |||
| Association of miR-27a SNP with FBG mg/dL | |||||||
| FBG < 100 mg/dL | 24 | 10(41.66%) | 10(41.66%) | 4(16.66%) | 6.53 | 2 | 0.037 |
| FBG > 100 mg/dL | 91 | 35(38.46%) | 53(58.24%) | 3(3.29%) | |||
| Association of miR-27a SNP with HBA1c% | |||||||
| HBA1c > 6% | 90 | 28(31.11%) | 57(63.33%) | 05(5.55%) | 15.52 | 2 | 0.009 |
| HBA1c < 6% | 25 | 17(68%) | 6(24%) | 2(8%) | |||
| Association of miR-27a SNP with Triglycerides mg/dL | |||||||
| Triglycerides mg/dL < 200 | 39 | 23(58.97%) | 13(33.33%) | 3(7.69%) | 11.14 | 2 | 0.003 |
| Triglycerides mg/dL > 200 | 76 | 22(28.94%) | 50(65.78%) | 4(5.26%) | |||
| Association of miR-27a SNP with Cholesterol mg/dL | |||||||
| Cholesterol mg/dL < 200 | 70 | 24(34.28%) | 43(61.42%) | 3(4.28%) | 3.47 | 2 | 0.176 |
| Cholesterol mg/dL > 200 | 45 | 21(46.66%) | 20(44.44%) | 4(8.88%) | |||
| Association of miR-27a SNP with LDL mg/dL | |||||||
| LDL mg/dL < 100 | 57 | 15(26.31%) | 39(68.42%) | 3(5.26%) | 8.71 | 2 | 0.012 |
| LDL mg/dL > 100 | 58 | 30(51.72%) | 24 (41.37%) | 4(6.89%) | |||
| Association of miR-27a SNP with HDL-L mg/dL | |||||||
| HDL-L mg/dL < 55 | 48 | 12(25%) | 33(68.75%) | 3(6.25%) | 7.14 | 2 | 0.02 |
| HDL-L mg/dL > 55 | 67 | 33(49.25%) | 30(44.77%) | 4(5.97%) | |||
| Genotypes | Healthy Controls | T2D Cases | OR (95% CI) | Risk Ratio (RR) | p-Value |
|---|---|---|---|---|---|
| (N = 108) | (N = 103) | ||||
| Codominant | |||||
| miR146-GG | 50 | 25 | 1(reference) | 1(reference) | |
| miR146-GC | 40 | 55 | 2.75(1.465 to 5.161) | 1.58(1.190 to 2.105) | 0.0016 |
| miR146-CC | 18 | 23 | 2.55(1.169 to 5.584) | 1.53(1.037 to 2.223) | 0.0186 |
| Dominant | |||||
| miR-46-GG | 50 | 25 | 1(reference) | 1(reference) | |
| miR-146-(GC+CC) | 58 | 78 | 2.68(1.493 to 4.843) | 1.56(1.214 to 2.011) | 0.0005 |
| Recessive | |||||
| miR-146-(GG+GC) | 90 | 80 | 1(reference) | 1(reference) | |
| miR-146-CC | 18 | 23 | 1.43(0.72 to 2.855) | 1.20(0.82 to 1.75) | 0.300 |
| Allele | |||||
| miR-146-G | 140 | 105 | 1(reference) | 1(reference) | |
| miR-146-C | 76 | 101 | 1.77(1.198 to 2.618) | 1.33(1.088 to 1.627) | 0.004 |
| Clinical Feature | N= | AA | AG | GG | X2 | DF | p-Value |
|---|---|---|---|---|---|---|---|
| Association of miR-146 SNV with Gender | |||||||
| Male | 70 | 20(28.57%) | 36(51.42%) | 14(20%) | 2.35 | 2 | 0.301 |
| Female | 33 | 5(15.15%) | 19(57.57%) | 9(27.27%) | |||
| Association of miR-146 SNV with Age | |||||||
| >40 | 79 | 20(25.31%) | 40(50.63%) | 19(24.05%) | 1.09 | 2 | 0.579 |
| <40 | 24 | 5(20.83%) | 15(62.5%) | 4(16.66%) | |||
| Association of miR-146 SNV with Fasting blood glucose (FBG) mg/dL | |||||||
| <100 | 24 | 3(12.5%) | 10(41.66%) | 11(45.83%) | 10.33 | 2 | 0.005 |
| >100 | 79 | 22(27.84%) | 45(56.96%) | 12(15.18%) | |||
| Association of miR-146 SNV with HBA1c% | |||||||
| >6 | 78 | 19(24.35%) | 48(61.53%) | 11(14.10%) | 13.73 | 2 | 0.001 |
| <6 | 25 | 06(24%) | 07(28%) | 12(48%) | |||
| Association of miR-146 SNV with Triglycerides mg/dL | |||||||
| <200 | 39 | 12(30.76%) | 15(28.46%) | 12(30.76%) | 5.72 | 2 | 0.057 |
| >200 | 64 | 13(20.31%) | 40(62.5%) | 11(17.18%) | |||
| Association of miR-146 SNV with Cholesterol mg/dL | |||||||
| <200 | 58 | 15(25.86%) | 27(46.55%) | 16(27.58%) | 2.95 | 2 | 0.22 |
| >200 | 45 | 10(22.22%) | 28(62.22%) | 7(15.55%) | |||
| Association of miR-146 SNV with LDL-C mg/dL | |||||||
| <100 | 47 | 13(27.65%) | 22(46.80%) | 12(25.53%) | 1.51 | 2 | 0.47 |
| >100 | 56 | 12(21.42%) | 33(58.92%) | 11(19.64%) | |||
| Association of miR-146 SNV with HDL-L mg/dL | |||||||
| <55 | 48 | 11(22.91%) | 27(56.25%) | 10(20.83%) | 0.3 | 2 | 0.86 |
| >55 | 55 | 14(25.45%) | 28(50.90%) | 13(23.63%) | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mir, R.; Elfaki, I.; Elangeeb, M.E.; Moawadh, M.S.; Tayeb, F.J.; Barnawi, J.; Albalawi, I.A.; Alharbi, A.A.; Alhelali, M.H.; Alsaedi, B.S.O. Comprehensive Molecular Evaluation of HNF-1 Alpha, miR-27a, and miR-146 Gene Variants and Their Link with Predisposition and Progression in Type 2 Diabetes Patients. J. Pers. Med. 2023, 13, 1270. https://doi.org/10.3390/jpm13081270
Mir R, Elfaki I, Elangeeb ME, Moawadh MS, Tayeb FJ, Barnawi J, Albalawi IA, Alharbi AA, Alhelali MH, Alsaedi BSO. Comprehensive Molecular Evaluation of HNF-1 Alpha, miR-27a, and miR-146 Gene Variants and Their Link with Predisposition and Progression in Type 2 Diabetes Patients. Journal of Personalized Medicine. 2023; 13(8):1270. https://doi.org/10.3390/jpm13081270
Chicago/Turabian StyleMir, Rashid, Imadeldin Elfaki, M. E. Elangeeb, Mamdoh S. Moawadh, Faris Jamal Tayeb, Jameel Barnawi, Ibrahim Altedlawi Albalawi, Amnah A. Alharbi, Marwan H. Alhelali, and Basim S. O. Alsaedi. 2023. "Comprehensive Molecular Evaluation of HNF-1 Alpha, miR-27a, and miR-146 Gene Variants and Their Link with Predisposition and Progression in Type 2 Diabetes Patients" Journal of Personalized Medicine 13, no. 8: 1270. https://doi.org/10.3390/jpm13081270
APA StyleMir, R., Elfaki, I., Elangeeb, M. E., Moawadh, M. S., Tayeb, F. J., Barnawi, J., Albalawi, I. A., Alharbi, A. A., Alhelali, M. H., & Alsaedi, B. S. O. (2023). Comprehensive Molecular Evaluation of HNF-1 Alpha, miR-27a, and miR-146 Gene Variants and Their Link with Predisposition and Progression in Type 2 Diabetes Patients. Journal of Personalized Medicine, 13(8), 1270. https://doi.org/10.3390/jpm13081270





