Sequence Alignment between TRIM33 Gene and Human Noncoding RNAs: A Potential Explanation for Paraneoplastic Dermatomyositis
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of Human ncRNAs Aligning to TRIM33 Gene Sequence
2.2. In Silico Characterization of Human ncRNAs Aligning to TRIM33 Gene Sequence
3. Results
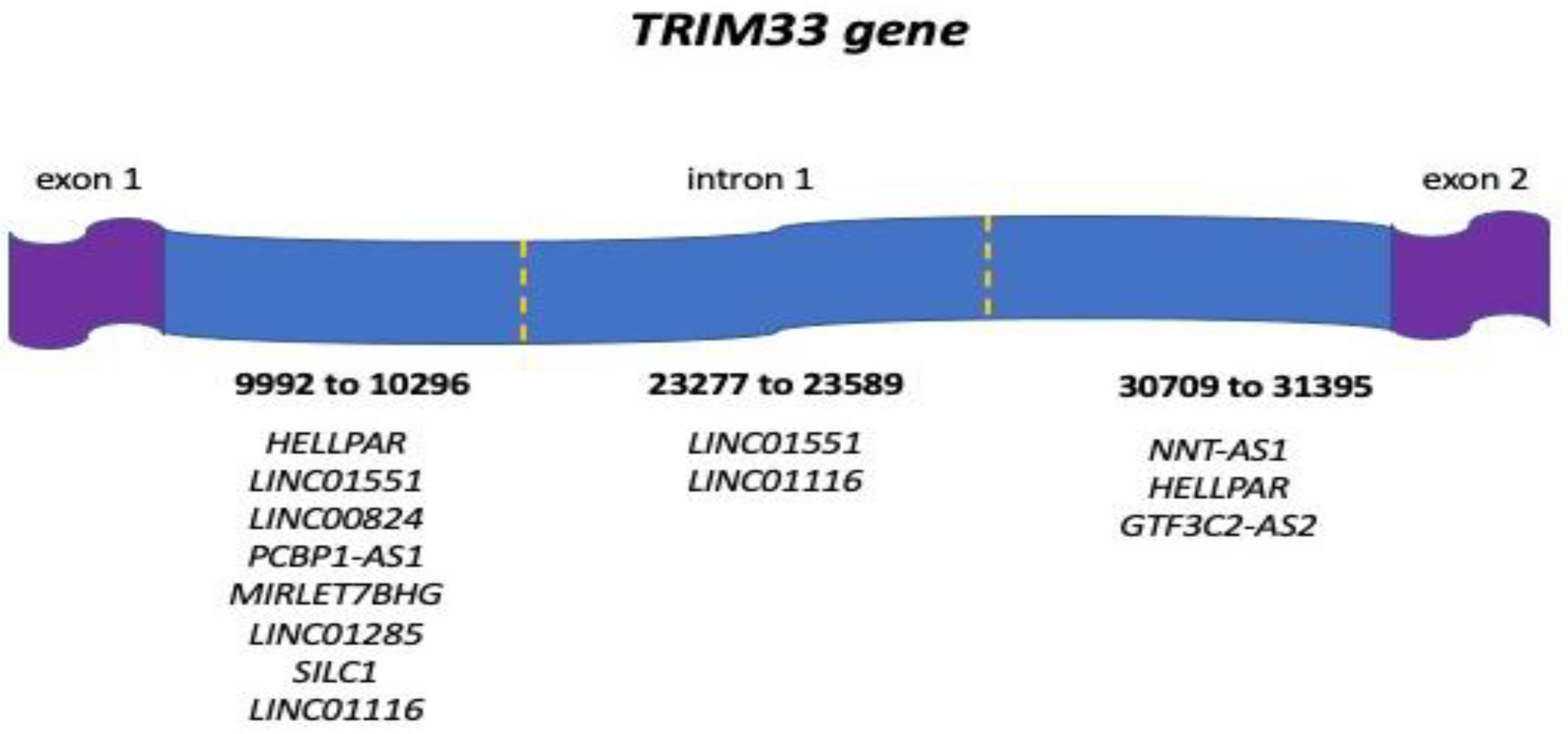
3.1. Human ncRNAs Aligning with the TRIM33 Gene
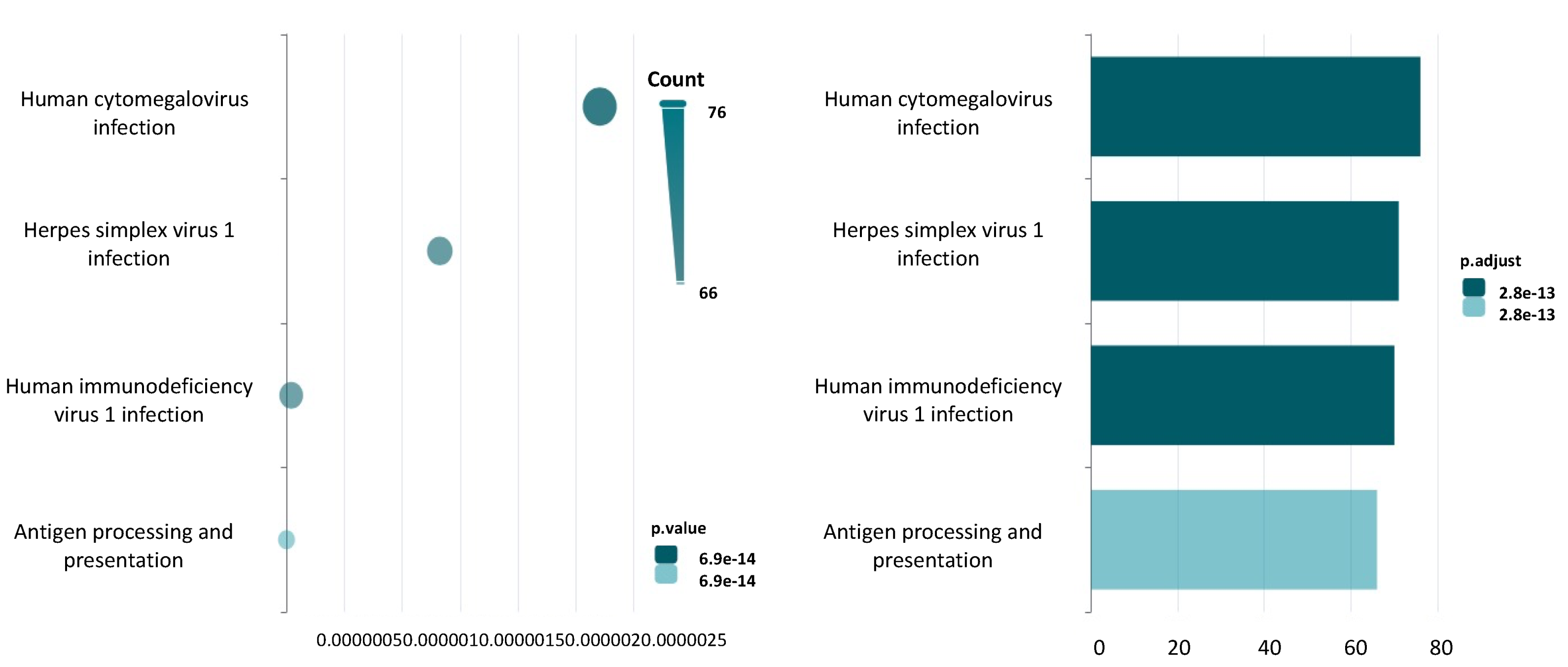
3.2. Human Diseases Associated with ncRNAs Aligning with the TRIM33 Gene
3.3. Regulatory Function of ncRNAs Aligning with the TRIM33 Gene
3.4. QmRLFS-Finder Analysis
3.5. Spliceator Analysis
3.6. MiR-142-3p as a Multiple Interactor with TRIM33 Gene and TRIM33 Gene-Aligned lncRNAs Dysregulated in Dermatomyositis
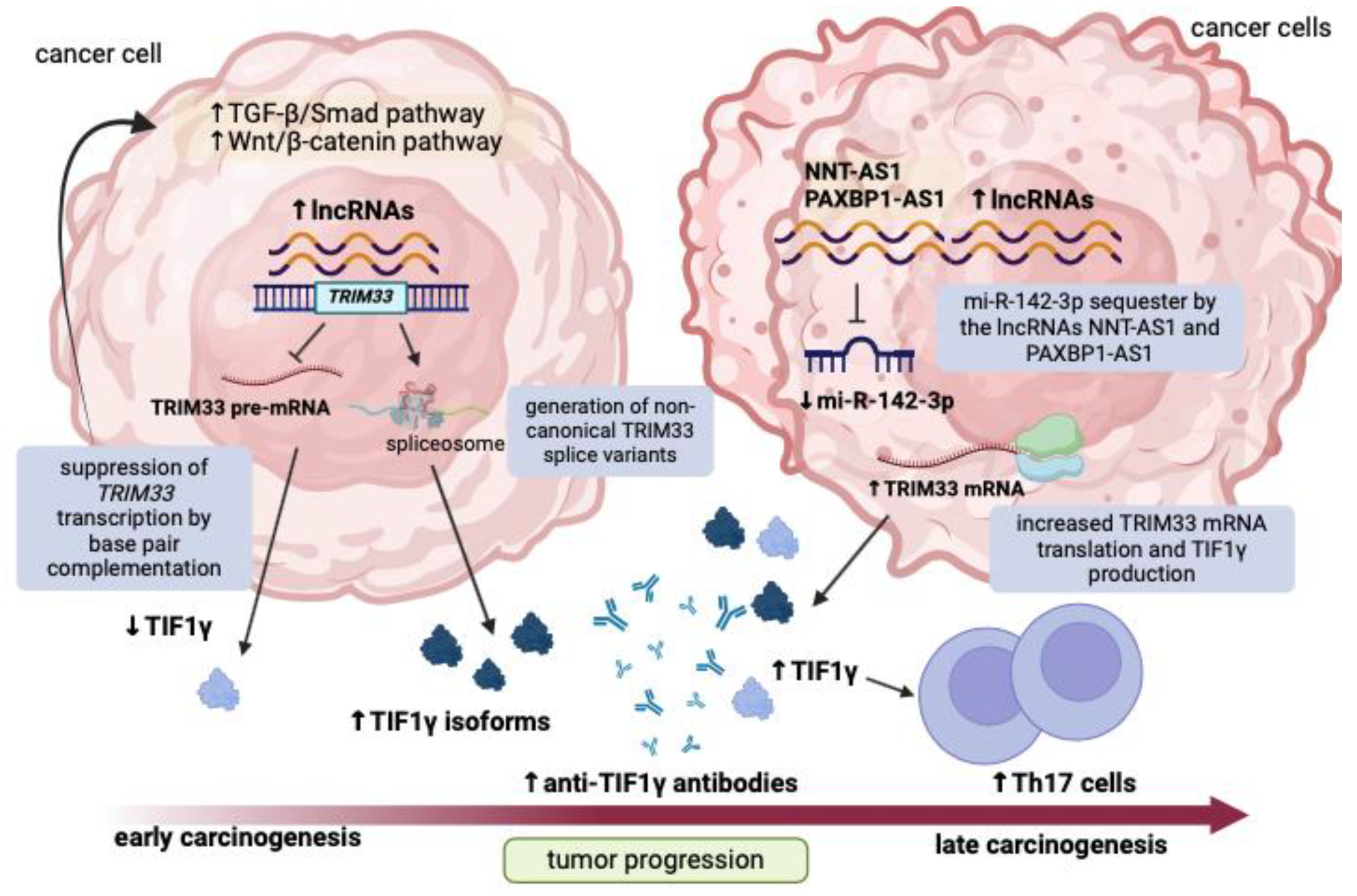
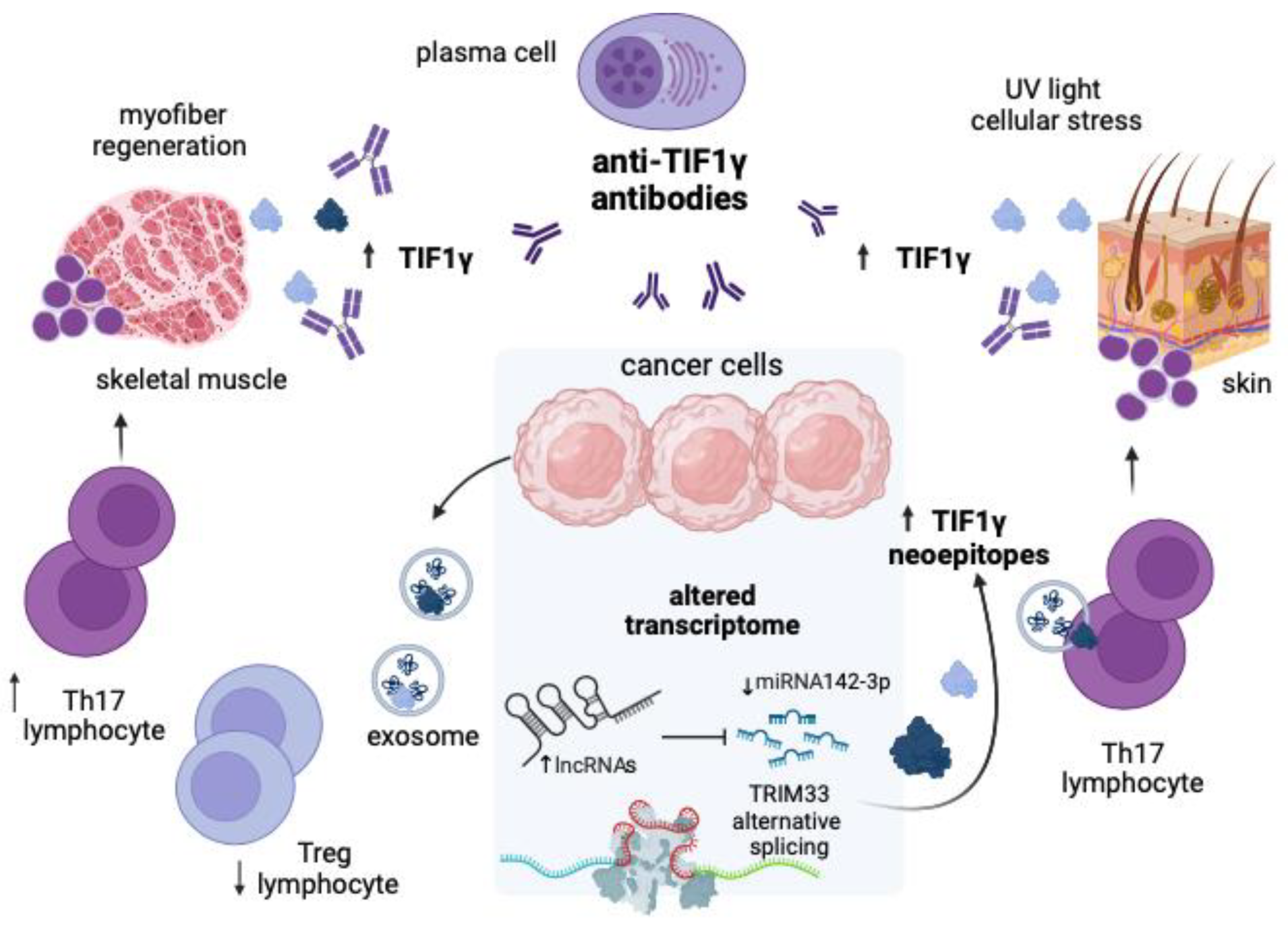
4. Discussion
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lundberg, I.E.; Tjärnlund, A.; Bottai, M.; Werth, V.P.; Pilkington, C.; de Visser, M.; Alfredsson, L.; Amato, A.A.; Barohn, R.J.; Liang, M.H.; et al. 2017 European League Against Rheumatism/American College of Rheumatology Classification Criteria for Adult and Juvenile Idiopathic Inflammatory Myopathies and Their Major Subgroups. Arthritis Rheumatol. 2017, 69, 2271–2282. [Google Scholar] [CrossRef] [PubMed]
- Patasova, K.; Lundberg, I.E.; Holmqvist, M. Genetic Influences in Cancer-Associated Myositis. Arthritis Rheumatol. 2023, 75, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Dourado, E.; Bottazzi, F.; Cardelli, C.; Conticini, E.; Schmidt, J.; Cavagna, L.; Barsotti, S. Idiopathic Inflammatory Myopathies: One Year in Review 2022. Clin. Exp. Rheumatol. 2023, 41, 199–213. [Google Scholar] [CrossRef]
- Carstens, P.O.; Schmidt, J. Diagnosis, Pathogenesis and Treatment of Myositis: Recent Advances. Clin. Exp. Immunol. 2014, 175, 349–358. [Google Scholar] [CrossRef]
- Malik, A.; Hayat, G.; Kalia, J.S.; Guzman, M.A. Idiopathic Inflammatory Myopathies: Clinical Approach and Management. Front. Neurol. 2016, 7, 64. [Google Scholar] [CrossRef] [PubMed]
- Ernste, F.C.; Reed, A.M. Idiopathic Inflammatory Myopathies: Current Trends in Pathogenesis, Clinical Features, and Up-to-Date Treatment Recommendations. Mayo Clin. Proc. 2013, 88, 83–105. [Google Scholar] [CrossRef] [PubMed]
- Dalakas, M.C. Inflammatory Muscle Diseases. N. Engl. J. Med. 2015, 372, 1734–1747. [Google Scholar] [CrossRef] [PubMed]
- Fiorentino, D.F.; Chung, L.S.; Christopher-Stine, L.; Zaba, L.; Li, S.; Mammen, A.L.; Rosen, A.; Casciola-Rosen, L. Most Patients with Cancer-Associated Dermatomyositis Have Antibodies to Nuclear Matrix Protein NXP-2 or Transcription Intermediary Factor 1γ. Arthritis Rheum. 2013, 65, 2954–2962. [Google Scholar] [CrossRef] [PubMed]
- Fiorentino, D.F.; Kuo, K.; Chung, L.; Zaba, L.; Li, S.; Casciola-Rosen, L. Distinctive Cutaneous and Systemic Features Associated with Antitranscriptional Intermediary Factor-1γ Antibodies in Adults with Dermatomyositis. J. Am. Acad. Dermatol. 2015, 72, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Kotobuki, Y.; Tonomura, K.; Fujimoto, M. Transcriptional Intermediary Factor 1 (TIF1) and Anti-TIF1γ Antibody-Positive Dermatomyositis. Immunol. Med. 2020, 44, 23–29. [Google Scholar] [CrossRef]
- De Vooght, J.; Vulsteke, J.B.; De Haes, P.; Bossuyt, X.; Lories, R.; De Langhe, E. Anti-TIF1-Γautoantibodies: Warning Lights of a Tumour Autoantigen. Rheumatology 2020, 59, 469–477. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Ding, Z.; Liang, H.; Zhang, B.; Chen, X. The Roles of TIF1γ in Cancer. Front. Oncol. 2019, 9, 979. [Google Scholar] [CrossRef] [PubMed]
- Kusy, S.; Gault, N.; Ferri, F.; Lewandowski, D.; Barroca, V.; Jaracz-Ros, A.; Losson, R.; Romeo, P.H. Adult Hematopoiesis Is Regulated by TIF1γ, a Repressor of TAL1 and PU.1 Transcriptional Activity. Cell Stem Cell 2011, 8, 412–425. [Google Scholar] [CrossRef]
- Cordel, N.; Derambure, C.; Coutant, S.; Mariette, X.; Jullien, D.; Debarbieux, S.; Chosidow, O.; Meyer, A.; Bessis, D.; Joly, P.; et al. TRIM33 Gene Somatic Mutations Identified by next Generation Sequencing in Neoplasms of Patients with Anti-TIF1γpositive Cancer-Associated Dermatomyositis. Rheumatology 2021, 60, 5863–5867. [Google Scholar] [CrossRef] [PubMed]
- Pinal-Fernandez, I.; Ferrer-Fabregas, B.; Trallero-Araguas, E.; Balada, E.; Martínez, M.A.; Milisenda, J.C.; Aparicio-Español, G.; Labrador-Horrillo, M.; Garcia-Patos, V.; Grau-Junyent, J.M.; et al. Tumour TIF1 Mutations and Loss of Heterozygosity Related to Cancer-Associated Myositis. Rheumatology 2018, 57, 388–396. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Wang, X.; Dong, C. Molecular Mechanisms of T Helper 17 Cell Differentiation: Emerging Roles for Transcription Cofactors. Adv. Immunol. 2019, 144, 121–153. [Google Scholar] [CrossRef]
- Ikeda, N.; Yamaguchi, Y.; Kanaoka, M.; Ototake, Y.; Akita, A.; Watanabe, T.; Aihara, M. Clinical Significance of Serum Levels of Anti-Transcriptional Intermediary Factor 1-γ Antibody in Patients with Dermatomyositis. J. Dermatol. 2020, 47, 490–496. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A Pathology Atlas of the Human Cancer Transcriptome. Science 2017, 357, 2507. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.Y.; Liu, Q.; Ren, Y.; Alam, S.K.; Wang, L.; Zhu, Z.; Hoeppner, L.H.; Dehm, S.M.; Cao, Q.; Yang, R. A Pan-Cancer Transcriptome Analysis of Exitron Splicing Identifies Novel Cancer Driver Genes and Neoepitopes. Mol. Cell 2021, 81, 2246–2260.e12. [Google Scholar] [CrossRef] [PubMed]
- Tsimberidou, A.M.; Fountzilas, E.; Bleris, L.; Kurzrock, R. Transcriptomics and Solid Tumors: The next Frontier in Precision Cancer Medicine. Semin. Cancer Biol. 2022, 84, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.L.; Zhang, Y.M.; Yang, H.B.; Shu, X.M.; Lu, X.; Wang, G.C. Transcriptomic Profiling of Long Non-Coding RNAs in Dermatomyositis by Microarray Analysis. Sci. Rep. 2016, 6, 32818. [Google Scholar] [CrossRef]
- Gao, S.; Zhang, H.; Zuo, X.; Xiao, Y.; Liu, D.; Zhu, H.; Luo, H. Integrated Comparison of the MiRNAome and MRNAome in Muscles of Dermatomyositis and Polymyositis Reveals Common and Specific MiRNA-MRNAs. Epigenomics 2019, 11, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Yates, A.D.; Achuthan, P.; Akanni, W.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; et al. Ensembl 2020. Nucleic Acids Res. 2019, 48, D682–D688. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Iny Stein, T.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 2016, 1.30.1–1.30.33. [Google Scholar] [CrossRef] [PubMed]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The Mutational Constraint Spectrum Quantified from Variation in 141,456 Humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Liu, T.; Cui, T.; Wang, Z.; Zhang, Y.; Tan, P.; Huang, Y.; Yu, J.; Wang, D. RNAInter in 2020: RNA Interactome Repository with Increased Coverage and Annotation. Nucleic Acids Res. 2020, 48, D189–D197. [Google Scholar] [CrossRef] [PubMed]
- Jenjaroenpun, P.; Wongsurawat, T.; Yenamandra, S.P.; Kuznetsov, V.A. QmRLFS-Finder: A Model, Web Server and Stand-Alone Tool for Prediction and Analysis of R-Loop Forming Sequences: Table 1. Nucleic Acids Res. 2015, 43, W527–W534. [Google Scholar] [CrossRef] [PubMed]
- Scalzitti, N.; Kress, A.; Orhand, R.; Weber, T.; Moulinier, L.; Jeannin-Girardon, A.; Collet, P.; Poch, O.; Thompson, J.D. Spliceator: Multi-Species Splice Site Prediction Using Convolutional Neural Networks. BMC Bioinform. 2021, 22, 561. [Google Scholar] [CrossRef]
- Kyrchanova, O.; Georgiev, P. Mechanisms of Enhancer-Promoter Interactions in Higher Eukaryotes. Int. J. Mol. Sci. 2021, 22, 671. [Google Scholar] [CrossRef] [PubMed]
- Petermann, E.; Lan, L.; Zou, L. Sources, Resolution and Physiological Relevance of R-Loops and RNA–DNA Hybrids. Nat. Rev. Mol. Cell Biol. 2022, 23, 521–540. [Google Scholar] [CrossRef]
- Elsakrmy, N.; Cui, H. R-Loops and R-Loop-Binding Proteins in Cancer Progression and Drug Resistance. Int. J. Mol. Sci. 2023, 24, 7064. [Google Scholar] [CrossRef]
- Catalanotto, C.; Cogoni, C.; Zardo, G. MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions. Int. J. Mol. Sci. 2016, 17, 1712. [Google Scholar] [CrossRef]
- Kazimierczyk, M.; Kasprowicz, M.K.; Kasprzyk, M.E.; Wrzesinski, J. Human Long Noncoding RNA Interactome: Detection, Characterization and Function. Int. J. Mol. Sci. 2020, 21, 1027. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Tang, L.; Zhang, L.; Ren, Y.; Peng, H.; Xiao, Y.; Xu, J.; Mao, D.; Liu, L.; Liu, L. Identification of Biomarkers Associated with CD4+ T-Cell Infiltration with Gene Coexpression Network in Dermatomyositis. Front. Immunol. 2022, 13, 854848. [Google Scholar] [CrossRef]
- Franco, C.; Giannella, A.; Gasparotto, M.; Zanatta, E.; Ghirardello, A.; Pettorossi, F.; Rahmè, Z.; Depascale, R.; Ragno, D.; Bevilacqua, G.; et al. Circulating Extracellular Vesicles and Small Non-Coding RNAs Cargo in Idiopathic Inflammatory Myopathies Reveal Differences across Myositis Subsets. J. Autoimmun. 2024, 147, 103255. [Google Scholar] [CrossRef]
- Li, L.; Zuo, X.; Liu, D.; Luo, H.; Zhang, H.; Peng, Q.; Wang, G.; Zhu, H. Plasma Exosomal RNAs Have Potential as Both Clinical Biomarkers and Therapeutic Targets of Dermatomyositis. Rheumatology 2022, 61, 2672–2681. [Google Scholar] [CrossRef] [PubMed]
- Hirai, T.; Ikeda, K.; Tsushima, H.; Fujishiro, M.; Hayakawa, K.; Yoshida, Y.; Morimoto, S.; Yamaji, K.; Takasaki, Y.; Takamori, K.; et al. Circulating Plasma MicroRNA Profiling in Patients with Polymyositis/Dermatomyositis before and after Treatment: MiRNA May Be Associated with Polymyositis/Dermatomyositis. Inflamm. Regen. 2018, 38, 1. [Google Scholar] [CrossRef]
- Muñoz-Braceras, S.; Pinal-Fernandez, I.; Casal-Dominguez, M.; Pak, K.; Milisenda, J.C.; Lu, S.; Gadina, M.; Naz, F.; Gutierrez-Cruz, G.; Dell’Orso, S.; et al. Identification of Unique MicroRNA Profiles in Different Types of Idiopathic Inflammatory Myopathy. Cells 2023, 12, 2198. [Google Scholar] [CrossRef]
- Georgantas, R.W.; Streicher, K.; Greenberg, S.A.; Greenlees, L.M.; Zhu, W.; Brohawn, P.Z.; Higgs, B.W.; Czapiga, M.; Morehouse, C.A.; Amato, A.; et al. Inhibition of Myogenic MicroRNAs 1, 133, and 206 by Inflammatory Cytokines Links Inflammation and Muscle Degeneration in Adult Inflammatory Myopathies. Arthritis Rheumatol. 2014, 66, 1022–1033. [Google Scholar] [CrossRef] [PubMed]
- Pinal-Fernandez, I.; Casal-Dominguez, M.; Mammen, A.L. Immune-Mediated Necrotizing Myopathy. Curr. Rheumatol. Rep. 2018, 20, 21. [Google Scholar] [CrossRef]
- Jain, S.; Singhal, S.; Francis, F.; Hajdu, C.; Wang, J.H.; Suriawinata, A.; Wang, Y.Q.; Zhang, M.; Weinshel, E.H.; Francois, F.; et al. Association of Overexpression of TIF1γ with Colorectal Carcinogenesis and Advanced Colorectal Adenocarcinoma. World J. Gastroenterol. 2011, 17, 3994–4000. [Google Scholar] [CrossRef]
- Ding, Z.Y.; Jin, G.N.; Wang, W.; Chen, W.X.; Wu, Y.H.; Ai, X.; Chen, L.; Zhang, W.G.; Liang, H.F.; Laurence, A.; et al. Reduced Expression of Transcriptional Intermediary Factor 1 Gamma Promotes Metastasis and Indicates Poor Prognosis of Hepatocellular Carcinoma. Hepatology 2014, 60, 1620–1636. [Google Scholar] [CrossRef] [PubMed]
- Kassem, L.; Deygas, M.; Fattet, L.; Lopez, J.; Goulvent, T.; Lavergne, E.; Chabaud, S.; Carrabin, N.; Chopin, N.; Bachelot, T.; et al. TIF1γ Interferes with TGFβ1/SMAD4 Signaling to Promote Poor Outcome in Operable Breast Cancer Patients. BMC Cancer 2015, 15, 453. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Song, K.S.; Sohn, E.H.; Kang, S.W.; Yoo, I.S.; Shim, S.C.; Yoo, S.J.; Kim, J. Anti-TIF1γ Antibody and the Expression of TIF1γ in Idiopathic Inflammatory Myopathies. Int. J. Rheum. Dis. 2019, 22, 314–320. [Google Scholar] [CrossRef] [PubMed]
- Harada, Y.; Tominaga, M.; Iitoh, E.; Kaieda, S.; Koga, T.; Fujimoto, K.; Chikasue, T.; Obara, H.; Kakuma, T.; Ida, H.; et al. Clinical Characteristics of Anti-TIF-1γ Antibody-Positive Dermatomyositis Associated with Malignancy. J. Clin. Med. 2022, 11, 1925. [Google Scholar] [CrossRef] [PubMed]
- Scholtissek, B.; Ferring-Schmitt, S.; Maier, J.; Wenzel, J. Expression of the Autoantigen TRIM33/TIF1γ in Skin and Muscle of Patients with Dermatomyositis Is Upregulated, Together with Markers of Cellular Stress. Clin. Exp. Dermatol. 2017, 42, 659–662. [Google Scholar] [CrossRef]
- Parkes, J.E.; Rothwell, S.; Oldroyd, A.; Chinoy, H.; Lamb, J.A.; Lundberg, I.E.; Miller, F.W.; Cooper, R.G.; Ollier, W.E.; Gregersen, P.K.; et al. Genetic Background May Contribute to the Latitude-Dependent Prevalence of Dermatomyositis and Anti-TIF1-γ Autoantibodies in Adult Patients with Myositis. Arthritis Res. Ther. 2018, 20, 117. [Google Scholar] [CrossRef] [PubMed]
- Mohassel, P.; Rosen, P.; Casciola-Rosen, L.; Pak, K.; Mammen, A.L. Expression of the Dermatomyositis Autoantigen Transcription Intermediary Factor 1γ in Regenerating Muscle. Arthritis Rheumatol. 2015, 67, 266–272. [Google Scholar] [CrossRef]
- Li, C.H.; Chen, Y. Insight into the Role of Long Noncoding RNA in Cancer Development and Progression. Int. Rev. Cell Mol. Biol. 2016, 326, 33–65. [Google Scholar] [CrossRef]
- Wu, G.C.; Pan, H.F.; Leng, R.X.; Wang, D.G.; Li, X.P.; Li, X.M.; Ye, D.Q. Emerging Role of Long Noncoding RNAs in Autoimmune Diseases. Autoimmun. Rev. 2015, 14, 798–805. [Google Scholar] [CrossRef]
- Ye, L.; Zuo, Y.; Yang, H.; Li, W.; Peng, Q.; Lu, X.; Wang, G.; Shu, X. Specific Autoantibodies and Clinical Phenotypes Correlate with the Aberrant Expression of Immune-Related MicroRNAs in Dermatomyositis. J. Immunol. Res. 2019, 2019, 2927061. [Google Scholar] [CrossRef]
- El-Sheikh, N.M.; Abulsoud, A.I.; Wasfey, E.F.; Hamdy, N.M. Insights on the Potential Oncogenic Impact of Long Non-Coding RNA Nicotinamide Nucleotide Transhydrogenase Antisense RNA 1 in Different Cancer Types; Integrating Pathway(s) and Clinical Outcome(s) Association. Pathol. Res. Pract. 2022, 240, 154183. [Google Scholar] [CrossRef]
- Lu, Q.; Liu, L.; Wang, S.; Zhang, Q.; Li, L. Comprehensive Analysis of M5C-Related LncRNAs in the Prognosis and Immune Landscape of Hepatocellular Carcinoma. Front. Genet. 2022, 13, 990594. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-Y.; Yang, W.; Zheng, F.-S.; Wang, Y.; Lu, J.-B. Long Non-Coding RNA SNHG1 Regulates Zinc Finger E-Box Binding Homeobox 1 Expression by Interacting with TAp63 and Promotes Cell Metastasis and Invasion in Lung Squamous Cell Carcinoma. Biomed. Pharmacother. 2017, 90, 650–658. [Google Scholar] [CrossRef] [PubMed]
- Alsaleem, M.A.; Ball, G.; Toss, M.S.; Raafat, S.; Aleskandarany, M.; Joseph, C.; Ogden, A.; Bhattarai, S.; Rida, P.C.G.; Khani, F.; et al. A Novel Prognostic Two-Gene Signature for Triple Negative Breast Cancer. Mod. Pathol. 2020, 33, 2208–2220. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, X.; Lin, C.; Jia, X.; Zhu, H.; Song, J.; Zhang, Y. Noncoding RNAs Regulate Alternative Splicing in Cancer. J. Exp. Clin. Cancer Res. 2021, 40, 11. [Google Scholar] [CrossRef]
- Ren, P.; Lu, L.; Cai, S.; Chen, J.; Lin, W.; Han, F. Alternative Splicing: A New Cause and Potential Therapeutic Target in Autoimmune Disease. Front. Immunol. 2021, 12, 713540. [Google Scholar] [CrossRef] [PubMed]
- Grabski, D.F.; Broseus, L.; Kumari, B.; Rekosh, D.; Hammarskjold, M.L.; Ritchie, W. Intron Retention and Its Impact on Gene Expression and Protein Diversity: A Review and a Practical Guide. Wiley Interdiscip. Rev. RNA 2021, 12, e1631. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, S.A. Dermatomyositis and Type 1 Interferons. Curr. Rheumatol. Rep. 2010, 12, 198–203. [Google Scholar] [CrossRef]
- Leylek, R.; Idoyaga, J. The Versatile Plasmacytoid Dendritic Cell: Function, Heterogeneity, and Plasticity. Int. Rev. Cell Mol. Biol. 2019, 349, 177–211. [Google Scholar] [CrossRef]
- Sharma, S. Immunomodulation: A Definitive Role of MicroRNA-142. Dev. Comp. Immunol. 2017, 77, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, A.; Mukhametshina, R.T.; Taghizadeh, S.; Vásquez-Pacheco, E.; Cabrera-Fuentes, H.; Rizvanov, A.; Mari, B.; Carraro, G.; Bellusci, S. MicroRNA-142 Is a Multifaceted Regulator in Organogenesis, Homeostasis, and Disease. Dev. Dyn. 2017, 246, 285–290. [Google Scholar] [CrossRef]
- Kinder, T.B.; Heier, C.R.; Tully, C.B.; Van der Muelen, J.H.; Hoffman, E.P.; Nagaraju, K.; Fiorillo, A.A. Muscle Weakness in Myositis: MicroRNA-Mediated Dystrophin Reduction in a Myositis Mouse Model and Human Muscle Biopsies. Arthritis Rheumatol. 2020, 72, 1170–1183. [Google Scholar] [CrossRef]
- Li, L.; Zuo, X.; Liu, D.; Luo, H.; Zhu, H. The Functional Roles of RNAs Cargoes Released by Neutrophil-Derived Exosomes in Dermatomyositis. Front. Pharmacol. 2021, 12, 727901. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y. Architecture of RNA–RNA Interactions. Curr. Opin. Genet. Dev. 2022, 72, 138–144. [Google Scholar] [CrossRef] [PubMed]
| Gene | Type | Tissue Expression | Subcellular Localization | Associated Disease | No. of Matched Transcripts | Alignment Length (bp) | %ID | Hit on a Phenotype-Associated Variant |
|---|---|---|---|---|---|---|---|---|
| NNT-AS1 | lncRNA | almost ubiquitous | nucleus, cytoskeleton, mitochondria, extracellular, plasma membrane | cervical cancer, ovarian cancer, osteogenic sarcoma, hepatocellular cancer, colorectal cancer | 1 | 676 | 91.8 | no |
| HELLPAR | lncRNA | skeletal muscle tissue, liver | nucleus | HELLP syndrome | 3 | 403 ± 170.6 | 93.9 ± 0.5 | no |
| GTF3C2-AS2 | lncRNA | almost ubiquitous | / | leukoencephalopathy with vanishing white matter; retinitis pigmentosa | 1 | 671 | 92.1 | no |
| LINC01551 | lncRNA | overexpressed in the brain | / | hypertropia | 4 | 308.5 ± 0.5 | 96.7 ± 3.3 | no |
| LINC01116 | lncRNA | almost ubiquitous (overexpressed in the kidney) | nucleus, cytoskeleton, extracellular | breast cancer, glioblastoma, glioma, lung cancer, ovarian squamous cell carcinoma | 2 | 310.5 ± 2.1 | 95.8 ± 4.1 | no |
| LINC00824 | lncRNA | overexpressed in whole blood, intestine and spleen | / | primary spontaneous pneumothorax | 2 | 297 ± 0 | 98.9 ± 0 | no |
| PCBP1-AS1 | lncRNA | almost ubiquitous | nucleus, extracellular | retinitis pigmentosa | 1 | 279 | 96.7 | no |
| MIRLET7BHG | lncRNA | almost ubiquitous | nucleus, extracellular | brachydactyly type b2 | 1 | 304 | 94.7 | no |
| LINC01285 | lncRNA | almost ubiquitous | / | monkeypox virus infection | 1 | 305 | 94.7 | no |
| SILC1 | lncRNA | overexpressed in the brain | nucleus | gastric cancer | 1 | 267 | 97 | no |
| Gene | Transcript | Type | Tissue Expression | Localization | Type of Regulatory Region | R-Loops |
|---|---|---|---|---|---|---|
| ZNF584-DT | ENST00000593393.1 | lncRNA | testis, endometrium | / | enhancer, CTCF | 0 |
| MKLN1-AS | ENST00000416220.7 | lncRNA | almost ubiquitous | nucleus | enhancer | 0 |
| ENSG00000254258 | ENST00000522373.1 | lncRNA | almost ubiquitous | / | enhancer | 0 |
| ARHGEF35-AS1 | ENST00000650250.1 | lncRNA | almost ubiquitous | / | enhancer | 0 |
| ENSG00000284294 | ENST00000638287.1 | lncRNA | sural nerve, colonic epithelium, bone marrow cell | / | enhancer | 0 |
| ENSG00000284294 | ENST00000640084.1 | lncRNA | sural nerve, colonic epithelium, bone marrow cell | / | enhancer | 0 |
| ENSG00000287839 | ENST00000668003.2 | lncRNA | whole blood, intestine | / | enhancer | 0 |
| ENSG00000285658 | ENST00000648317.1 | lncRNA | bone marrow cell, skeletal muscle tissue, colonic epithelium | / | TFB | 0 |
| OR2A1-AS1 | ENST00000468575.1 | lncRNA | almost ubiquitous | / | enhancer | 0 |
| MRPL20-DT | ENST00000607307.1 | lncRNA | almost ubiquitous | / | promoter | 0 |
| ANXA2R-OT1 | ENST00000702244.1 | lncRNA | almost ubiquitous | cytosol | enhancer | 0 |
| ENSG00000215014 | ENST00000366221.3 | lncRNA | overexpressed in testis | / | promoter | 0 |
| XACT | ENST00000674361.1 | lncRNA | testis, colon, nervous system | nucleolus | CTCF | 0 |
| ENSG00000267353 | ENST00000588717.1 | lncRNA | myometrium, endometrium | / | promoter | 0 |
| LINC00501 | ENST00000425388.1 | lncRNA | almost ubiquitous | extracellular | promoter | 0 |
| ARIH2OS | ENST00000647812.1 | lncRNA | testis, ovary | nucleus, plasma membrane, extracellular | promoter | 0 |
| ENSG00000287277 | ENST00000664549.1 | lncRNA | colonic epithelium, bone marrow cell, sural nerve | / | enhancer | 0 |
| ENSG00000287277 | ENST00000669799.1 | lncRNA | colonic epithelium, bone marrow cell, sural nerve | / | enhancer | 0 |
| ENSG00000287277 | ENST00000666057.1 | lncRNA | colonic epithelium, bone marrow cell, sural nerve | / | enhancer | 0 |
| ENSG00000287277 | ENST00000665216.1 | lncRNA | colonic epithelium, bone marrow cell, sural nerve | / | enhancer | 0 |
| PAXBP1-AS1 | ENST00000691027.1 | lncRNA | almost ubiquitous | / | promoter | 0 |
| PAXBP1-AS1 | ENST00000653345.1 | lncRNA | almost ubiquitous | / | promoter | 0 |
| PAXBP1-AS1 | ENST00000660366.1 | lncRNA | almost ubiquitous | / | promoter | 0 |
| PAXBP1-AS1 | ENST00000440052.5 | lncRNA | almost ubiquitous | / | promoter | 0 |
| PAXBP1-AS1 | ENST00000458479.1 | lncRNA | almost ubiquitous | / | promoter | 0 |
| PAXBP1-AS1 | ENST00000665654.1 | lncRNA | almost ubiquitous | / | promoter | 0 |
| PAXBP1-AS1 | ENST00000662524.1 | lncRNA | almost ubiquitous | / | promoter | 0 |
| PAXBP1-AS1 | ENST00000665120.1 | lncRNA | almost ubiquitous | / | promoter | 0 |
| miRNA | RNAInter Score | Results from Other Studies | Methods | Reference |
|---|---|---|---|---|
| hsa-miR-142-3p | 0.4564 | downregulated in plasma exosomes of patients with IIMs compared to controls | NGS | [35] |
| hsa-miR-3613-5p | 0.4528 | downregulated in plasma exosomes of patients with IIMs compared to controls | NGS | [35] |
| hsa-miR-196a-5p | 0.4457 | downregulated in PM muscle biopsy samples but upregulated in HSkMM after exposure to 20% PM serum | miRNAome analysis | [22] |
| hsa-miR-21-5p | 0.4212 | upregulated in DM and ASyS muscle biopsy samples compared to controls | NanoString nCounter system | [38] |
| hsa-miR-30e-5p | 0.4139 | upregulated in HSkMM after stimulation with 20% PM serum | miRNAome analysis | [22] |
| hsa-miR-32-5p | 0.4086 | upregulated in plasma exosomes of patients with IIMs compared to controls | NGS | [35] |
| hsa-miR-491-5p | 0.4086 | upregulated in HSkMM after stimulation with 20% PM serum | miRNAome analysis | [22] |
| hsa-let-7f-5p | 0.4017 | upregulated in plasma exosomes of CAM patients compared to non-CAM IIM patients | NGS | [35] |
| hsa-let-7a-5p | 0.4017 | downregulated in plasma exosomes of patients with IIMs compared to controls | NGS | [35] |
| hsa-miR-133b | 0.3649 | upregulated and downregulated in IIM muscle biopsy samples compared to controls according to two different studies | NanoString nCounter system;miRNA microarray and qPCR | [38,39] |
| hsa-miR-30d-5p | 0.3649 | downregulated in PM muscle biopsy samples but upregulated in HSkMM after exposure to 20% PM serum | miRNAome analysis | [22] |
| hsa-miR-30a-5p | 0.3649 | downregulated in PM muscle biopsy samples but upregulated in HSkMM after exposure to 20% PM serum | miRNAome analysis | [22] |
| hsa-miR-133a-3p | 0.3649 | downregulated in IIM muscle biopsy samples comparedto controls | miRNA microarray and qPCR | [39] |
| hsa-miR-30e-3p | 0.3579 | downregulated in muscle biopsy samples from DM patients and correlated with the IFN-I signature | NanoString nCounter system | [38] |
| hsa-miR-30a-3p | 0.3579 | downregulated in muscle biopsy samples from DM patients and correlated with the IFN-I signature | NanoString nCounter system | [38] |
| hsa-miR-498-5p | 0.3097 | upregulated in plasma samples from PM/DM patients after immunosuppressive treatment | miRNA microarrayand qRT-PCR | [37] |
| hsa-miR-495-3p | 0.2986 | upregulated in IBM muscle biopsy samples compared to DMand controls | NanoString nCounter system | [38] |
| hsa-miR-130a-3p | 0.2129 | upregulated in HSkMM after stimulation with 20% PM serum | miRNAome analysis | [22] |
| hsa-miR-543 | 0.2129 | upregulated in IBM muscle biopsy samples compared to DMand controls | NanoString nCounter system | [38] |
| hsa-miR-143-3p | 0.2129 | downregulated in plasma exosomes of CAM patients compared to non-CAM patients | NGS | [35] |
| hsa-miR-302d-3p | 0.2129 | downregulated in muscle biopsy samples from IIM patients compared to controls | NanoString nCounter system | [38] |
| hsa-miR-382-5p | 0.2129 | upregulated in IIM muscle biopsy samples compared to controls | NanoString nCounter system | [38] |
| hsa-miR-411-5p | 0.2129 | upregulated in IBM muscle biopsy samples compared to DMand controls | NanoString nCounter system | [38] |
| hsa-miR-27a-3p | 0.1862 | downregulated in plasma exosomes of DM patients compared to PM/ASyS patients | NGS | [35] |
| hsa-miR-26a-5p | 0.1862 | upregulated in HSkMM after stimulation with 20% PM serum | miRNAome analysis | [22] |
| hsa-miR-320e | 0.1619 | uregulated in DM muscle biopsy samples compared to controls | NanoString nCounter system | [38] |
| hsa-miR-361-3p | 1E-10 | upregulated in DM muscle biopsy samples and associated with IFN-I signature | NanoString nCounter system | [38] |
| hsa-miR-221-3p | 1E-10 | upregulated in HSkMM after stimulation with 20% PM serum; differentially expressed in muscle biopsy samples of IIM patients compared to controls (mainly upregulated) | miRNAome analysis NanoString nCounter system | [22,38] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Talotta, R. Sequence Alignment between TRIM33 Gene and Human Noncoding RNAs: A Potential Explanation for Paraneoplastic Dermatomyositis. J. Pers. Med. 2024, 14, 628. https://doi.org/10.3390/jpm14060628
Talotta R. Sequence Alignment between TRIM33 Gene and Human Noncoding RNAs: A Potential Explanation for Paraneoplastic Dermatomyositis. Journal of Personalized Medicine. 2024; 14(6):628. https://doi.org/10.3390/jpm14060628
Chicago/Turabian StyleTalotta, Rossella. 2024. "Sequence Alignment between TRIM33 Gene and Human Noncoding RNAs: A Potential Explanation for Paraneoplastic Dermatomyositis" Journal of Personalized Medicine 14, no. 6: 628. https://doi.org/10.3390/jpm14060628
APA StyleTalotta, R. (2024). Sequence Alignment between TRIM33 Gene and Human Noncoding RNAs: A Potential Explanation for Paraneoplastic Dermatomyositis. Journal of Personalized Medicine, 14(6), 628. https://doi.org/10.3390/jpm14060628






