The Role of the Gut Microbiome in Inflammatory Bowel Disease: The Middle East Perspective
Abstract
1. Introduction
1.1. Background
1.2. Objectives
1.3. Methods
2. Normal Gut Microbiome
2.1. Composition
2.2. Functions
3. Alterations in the Gut Microbiome in IBD
3.1. Dysbiosis
3.2. Functional Changes
3.3. Microbial Interactions
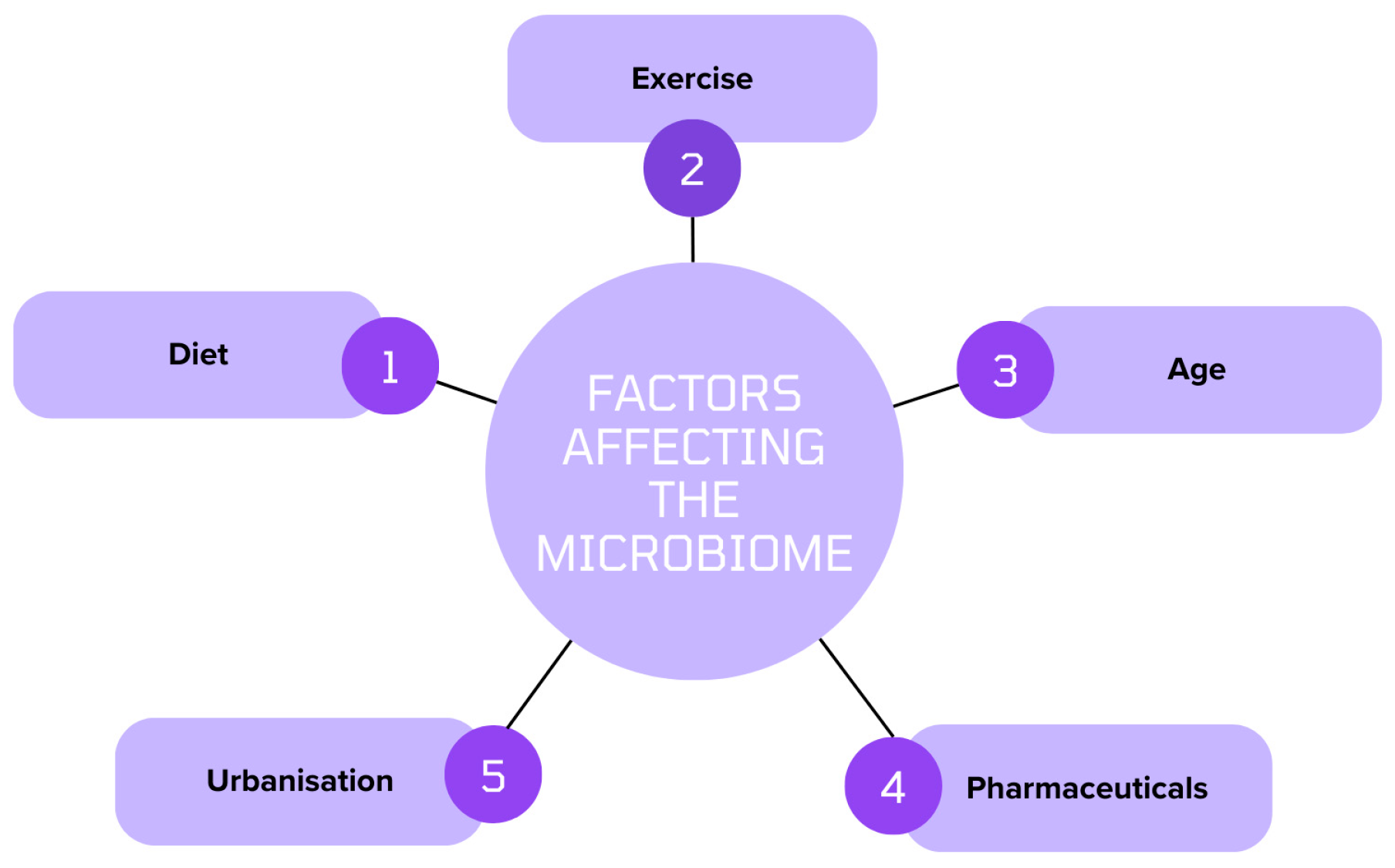
4. Factors Affecting the Middle Eastern Microbiome
4.1. Diet and Nutrition
4.2. Genetics
4.3. Environmental Factors
5. Therapeutic Interventions
5.1. Probiotics and Prebiotics
5.2. Diet Modification
5.3. Faecal Microbiota Transplantation (FMT)
6. Future Directions
6.1. Precision Medicine
6.2. Microbiome Modulation
6.3. Population Studies
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Piovani, D.; Danese, S.; Peyrin-Biroulet, L.; Nikolopoulos, G.K.; Lytras, T.; Bonovas, S. Environmental Risk Factors for Inflammatory Bowel Diseases: An Umbrella Review of Meta-analyses. Gastroenterology 2019, 157, 647–659.e4. [Google Scholar] [CrossRef]
- Mosli, M.; Alawadhi, S.; Hasan, F.; Rached, A.A.; Sanai, F.; Danese, S. Incidence, Prevalence, and clinical epidemiology of inflammatory bowel disease in the Arab World: A systematic review and meta-analysis. Inflamm. Intest. Dis. 2021, 6, 123–131. [Google Scholar] [CrossRef]
- Olfatifar, M.; Zali, M.R.; Pourhoseingholi, M.A.; Balaii, H.; Ghavami, S.B.; Ivanchuk, M.; Ivanchuk, P.; Nazari, S.H.; Shahrokh, S.; Sabour, S.; et al. The emerging epidemic of inflammatory bowel disease in Asia and Iran by 2035: A modeling study. BMC Gastroenterol. 2021, 21, 204. [Google Scholar] [CrossRef]
- Hall, A.B.; Tolonen, A.C.; Xavier, R.J. Human genetic variation and the gut microbiome in disease. Nat. Rev. Genet. 2017, 18, 690–699. [Google Scholar] [CrossRef]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the human intestinal microbial flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef]
- Wegorzewska, M.M.; Glowacki, R.W.P.; Hsieh, S.A.; Donermeyer, D.L.; Hickey, C.A.; Horvath, S.C.; Martens, E.C.; Stappenbeck, T.S.; Allen, P.M. Diet modulates colonic T cell responses by regulating the expression of a Bacteroides thetaiotaomicron antigen. Sci. Immunol. 2019, 4, eaau9079. [Google Scholar] [CrossRef]
- Khan, I.; Ullah, N.; Zha, L.; Bai, Y.; Khan, A.; Zhao, T.; Che, T.; Zhang, C. Alteration of Gut Microbiota in Inflammatory Bowel Disease (IBD): Cause or Consequence? IBD Treatment Targeting the Gut Microbiome. Pathogens 2019, 8, 126. [Google Scholar] [CrossRef]
- Fujimoto, T.; Imaeda, H.; Takahashi, K.; Kasumi, E.; Bamba, S.; Fujiyama, Y.; Andoh, A. Decreased abundance of Faecalibacterium prausnitzii in the gut microbiota of Crohn’s disease. J. Gastroenterol. Hepatol. 2013, 28, 613–619. [Google Scholar] [CrossRef]
- Alharbi, R.S.; Shaik, N.A.; Almahdi, H.; ElSokary, H.A.; Jamalalail, B.A.; Mosli, M.H.; Alsufyani, H.A.; Al-Aama, J.Y.; Elango, R.; Saadah, O.I.; et al. Genetic association study of NOD2 and IL23R amino acid substitution polymorphisms in Saudi Inflammatory Bowel Disease patients. J. King Saud Univ. Sci. 2022, 34, 101726. [Google Scholar] [CrossRef]
- Zou, Y.; Xue, W.; Luo, G.; Deng, Z.; Qin, P.; Guo, R.; Xiao, L. 1520 reference genomes from cultivated human gut bacteria enable functional microbiome analyses. Nat. Biotechnol. 2019, 37, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Sender, R.; Fuchs, S.; Milo, R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell 2016, 164, 337–340. [Google Scholar] [CrossRef]
- Wu, X.; Wu, Y.; He, L.; Wu, L.; Wang, X.; Liu, Z. Effects of the intestinal microbial metabolite butyrate on the development of colorectal cancer. J. Cancer 2018, 9, 2510–2517. [Google Scholar] [CrossRef]
- Silva, J.P.B.; Navegantes-Lima, K.C.; de Oliveira, A.L.B.; Rodrigues, D.V.S.; Gaspar, S.L.F.; Monteiro, V.V.S.; Moura, D.P.; Monteiro, M.C. Protective Mechanisms of Butyrate on Inflammatory Bowel Disease. Curr. Pharm. Des. 2018, 24, 4154–4166. [Google Scholar] [CrossRef]
- Laursen, M.F.; Sinha, A.K.; Pedersen, M.; Roager, H.M. Key bacterial taxa determine longitudinal dynamics of aromatic amino acid catabolism in infants’ gut. Gut Microbes 2023, 15, 2221426. [Google Scholar] [CrossRef]
- Revilla-Guarinos, A.; Gebhard, S.; Mascher, T.; Zúñiga, M. Antimicrobial peptide resistance in Firmicutes. Environ. Microbiol. 2014, 16, 1225–1237. [Google Scholar] [CrossRef]
- Munoz, R.; Rosselló-Móra, R.; Amann, R. Revised phylogeny of Bacteroidetes and proposal of sixteen new taxa and two new combinations including Rhodothermaeota phyl. nov. Syst. Appl. Microbiol. 2016, 39, 281–296. [Google Scholar] [CrossRef]
- Rakoff-Nahoum, S.; Coyne, M.J.; Comstock, L.E. An ecological network of polysaccharide utilization among human intestinal symbionts. Curr. Biol. 2014, 24, 40–49. [Google Scholar] [CrossRef]
- Chatzidaki-Livanis, M.; Coyne, M.J.; Roelofs, K.G.; Gentyala, R.R.; Caldwell, J.M.; Comstock, L.E. Gut symbiont Bacteroides fragilis secretes a eukaryotic-like ubiquitin protein that mediates intraspecies antagonism. mBio 2017, 8, e01902-17. [Google Scholar] [CrossRef]
- Yoshida, N.; Emoto, T.; Yamashita, T.; Watanabe, H.; Hayashi, T.; Tabata, T.; Hoshi, N.; Hatano, N.; Ozawa, G.; Sasaki, N.; et al. Bacteroides vulgatus and Bacteroides dorei reduce gut microbial lipopolysaccharide production and inhibit atherosclerosis. Circulation 2018, 138, 2486–2498. [Google Scholar] [CrossRef]
- O’Toole, P.W.; Jeffery, I.B. Gut microbiota and aging. Science 2015, 350, 1214–1215. [Google Scholar] [CrossRef]
- Milani, C.; Duranti, S.; Bottacini, F.; Casey, E.; Turroni, F.; Mahony, J.; Belzer, C.; Delgado Palacio, S.; Arboleya Montes, S.; Mancabelli, L.; et al. The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiol. Mol. Biol. Rev. 2017, 81, e00036-17. [Google Scholar] [CrossRef]
- Penders, J.; Thijs, C.; Vink, C.; Stelma, F.F.; Snijders, B.; Kummeling, I.; Van den Brandt, P.A.; Stobberingh, E.E. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 2006, 118, 511–521. [Google Scholar] [CrossRef]
- Park, S.-H.; Kim, K.-A.; Ahn, Y.-T.; Jeong, J.-J.; Huh, C.-S.; Kim, D.-H. Comparative analysis of gut microbiota in elderly people of urbanized towns and longevity villages. BMC Microbiol. 2015, 15, 49. [Google Scholar] [CrossRef]
- Brooks, C.N.; Wight, M.E.; Azeez, O.E.; Bleich, R.M.; Zwetsloot, K.A. Growing old together: What we know about the influence of diet and exercise on the aging host’s gut microbiome. Front. Sports Act. Living 2023, 5, 1168731. [Google Scholar] [CrossRef]
- Lutgendorff, F.; Akkermans, L.M.; Soderholm, J.D. The role of microbiota and probiotics in stress-induced gastrointestinal damage. Curr. Mol. Med. 2008, 8, 282–298. [Google Scholar] [CrossRef]
- Zhang, J.; Guo, Z.; Xue, Z.; Sun, Z.; Zhang, M.; Wang, L.; Wang, G.; Wang, F.; Xu, J.; Cao, H.; et al. A phylo-functional core of gut microbiota in healthy young Chinese cohorts across lifestyles, geography and ethnicities. ISME J. 2015, 9, 1979–1990. [Google Scholar] [CrossRef]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef]
- Clarke, S.F.; Murphy, E.F.; O’Sullivan, O.; Lucey, A.J.; Humphreys, M.; Hogan, A.; Hayes, P.; O’Reilly, M.; Jeffery, I.B.; Wood-Martin, R.; et al. Exercise and associated dietary extremes impact on gut microbial diversity. Gut 2014, 63, 1913–1920. [Google Scholar] [CrossRef]
- Patangia, D.V.; Ryan, C.A.; Dempsey, E.; Ross, R.P.; Stanton, C. Impact of antibiotics on the human microbiome and consequences for host health. Microbiologyopen 2022, 11, e1260. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Weaver, C.M. Diet, gut microbiome, and bone health. Curr. Osteoporos. Rep. 2015, 13, 125–130. [Google Scholar] [CrossRef]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef]
- Yang, Q.; Liang, Q.; Balakrishnan, B.; Belobrajdic, D.P.; Feng, Q.-J.; Zhang, W. Role of Dietary Nutrients in the Modulation of Gut Microbiota: A Narrative Review. Nutrients 2020, 12, 381. [Google Scholar] [CrossRef]
- Bielik, V.; Kolisek, M. Bioaccessibility and Bioavailability of Minerals in Relation to a Healthy Gut Microbiome. Int. J. Mol. Sci. 2021, 22, 6803. [Google Scholar] [CrossRef]
- Zaky, A.; Glastras, S.J.; Wong, M.Y.W.; Pollock, C.A.; Saad, S. The Role of the Gut Microbiome in Diabetes and Obesity-Related Kidney Disease. Int. J. Mol. Sci. 2021, 22, 9641. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Soto-Martin, E.C.; Warnke, I.; Farquharson, F.M.; Christodoulou, M.; Horgan, G.; Derrien, M.; Faurie, J.-M.; Flint, H.J.; Duncan, S.H.; Louis, P. Vitamin Biosynthesis by Human Gut Butyrate-Producing Bacteria and Cross-Feeding in Synthetic Microbial Communities. mBio 2020, 11, e00886-20. [Google Scholar] [CrossRef]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef]
- Magnúsdóttir, S.; Ravcheev, D.; de Crécy-Lagard, V.; Thiele, I. Systematic genome assessment of B-vitamin biosynthesis suggests co-operation among gut microbes. Front. Genet. 2015, 6, 148. [Google Scholar] [CrossRef]
- Rowley, C.A.; Kendall, M.M. To B12 or not to B12: Five questions on the role of cobalamin in host-microbial interactions. PLoS Pathog. 2019, 15, e1007479. [Google Scholar] [CrossRef]
- Subramanian, V.S.; Sabui, S.; Moradi, H.; Marchant, J.S.; Said, H.M. Inhibition of intestinal ascorbic acid uptake by lipopolysaccharide is mediated via transcriptional mechanisms. Biochim. Biophys. Acta Biomembr. 2018, 1860, 556–565. [Google Scholar] [CrossRef]
- Pham, V.T.; Fehlbaum, S.; Seifert, N.; Richard, N.; Bruins, M.J.; Sybesma, W.; Rehman, A.; Steinert, R.E. Effects of colon-targeted vitamins on the composition and metabolic activity of the human gut microbiome—A pilot study. Gut Microbes 2021, 13, 1875774. [Google Scholar] [CrossRef]
- Allen, J.; Sears, C.L. Impact of the gut microbiome on the genome and epigenome of colon epithelial cells: Contributions to colorectal cancer development. Genome Med. 2019, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.; Hao, S.; Sears, C.L.; Timp, W. Epigenetic Changes Induced by Bacteroides fragilis Toxin. Infect. Immun. 2019, 87, e00447-18. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rodriguez-Sillke, Y.; Visekruna, A.; Glauben, R.; Siegmund, B.; Steinhoff, U. Recognition of food antigens by the mucosal and systemic immune system: Consequences for intestinal development and homeostasis. Int. J. Med. Microbiol. 2021, 311, 151493. [Google Scholar] [CrossRef]
- Pittayanon, R.; Lau, J.T.; Leontiadis, G.I.; Tse, F.; Yuan, Y.; Surette, M.; Moayyedi, P. Differences in Gut Microbiota in Patients with vs without Inflammatory Bowel Diseases: A Systematic Review. Gastroenterology 2020, 158, 930–946.e1. [Google Scholar] [CrossRef]
- Garay, J.G.; Turpin, W.T.; Lee, S.-H.; Smith, M.I.; Goethel, A.; Griffiths, A.M.; Moayyedi, P.; Espin-Garcia, O.; Abreu, M.; Aumais, G.L.; et al. Gut Microbiome Composition Is Associated with Future Onset of Crohn’s Disease in Healthy First-Degree Relatives. Gastroenterology 2023, 165, 670–681. [Google Scholar] [CrossRef] [PubMed]
- Dörffel, Y.; Swidsinski, A.; Loening-Baucke, V.; Wiedenmann, B.; Pavel, M. Common biostructure of the colonic microbiota in neuroendocrine tumors and Crohn’s disease and the effect of therapy. Inflamm. Bowel Dis. 2012, 18, 1663–1671. [Google Scholar] [CrossRef]
- Quévrain, E.; Maubert, M.A.; Michon, C.; Chain, F.; Marquant, R.; Tailhades, J.; Miquel, S.; Carlier, L.; Bermúdez-Humarán, L.G.; Pigneur, B.; et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut 2016, 65, 415–425. [Google Scholar] [CrossRef]
- Joossens, M.; Huys, G.; Cnockaert, M.; De Preter, V.; Verbeke, K.; Rutgeerts, P.; Vandamme, P.; Vermeire, S. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut 2011, 60, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Ning, L.; Zhou, Y.-L.; Sun, H.; Zhang, Y.; Shen, C.; Wang, Z.; Xuan, B.; Zhao, Y.; Ma, Y.; Yan, Y.; et al. Microbiome and metabolome features in inflammatory bowel disease via multi-omics integration analyses across cohorts. Nat. Commun. 2023, 14, 7135. [Google Scholar] [CrossRef]
- Russo, E.; Giudici, F.; Fiorindi, C.; Ficari, F.; Scaringi, S.; Amedei, A. Immunomodulating Activity and Therapeutic Effects of Short Chain Fatty Acids and Tryptophan Post-biotics in Inflammatory Bowel Disease. Front. Immunol. 2019, 10, 2754. [Google Scholar] [CrossRef]
- Henke, M.T.; Kenny, D.J.; Cassilly, C.D.; Vlamakis, H.; Xavier, R.J.; Clardy, J. Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn’s disease, produces an inflammatory polysaccharide. Proc. Natl. Acad. Sci. USA 2019, 116, 12672–12677. [Google Scholar] [CrossRef] [PubMed]
- Li, X.V.; Leonardi, I.; Putzel, G.G.; Semon, A.; Fiers, W.D.; Kusakabe, T.; Lin, W.-Y.; Gao, I.H.; Doron, I.; Gutierrez-Guerrero, A.; et al. Immune regulation by fungal strain diversity in inflammatory bowel disease. Nature 2022, 603, 672–678. [Google Scholar] [CrossRef] [PubMed]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Swidsinski, A.; Loening-Baucke, V.; Herber, A. Mucosal flora in Crohn’s disease and ulcerative colitis—An overview. J. Physiol. Pharmacol. 2009, 60, 61–71. [Google Scholar]
- Swidsinski, A.; Weber, J.; Loening-Baucke, V.; Hale, L.P.; Lochs, H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J. Clin. Microbiol. 2005, 43, 3380–3389. [Google Scholar] [CrossRef] [PubMed]
- Prudent, V.; Demarre, G.; Vazeille, E.; Wery, M.; Quenech’du, N.; Ravet, A.; Dauverd-Girault, J.; van Dijk, E.; Bringer, M.-A.; Descrimes, M.; et al. The Crohn’s disease-related bacterial strain LF82 assembles biofilm-like communities to protect itself from phagolysosomal attack. Commun. Biol. 2021, 4, 627. [Google Scholar] [CrossRef]
- De La Fuente, M.; Franchi, L.; Araya, D.; Díaz-Jiménez, D.; Olivares, M.; Álvarez-Lobos, M.; Golenbock, D.; González, M.-J.; López-Kostner, F.; Quera, R.; et al. Escherichia coli isolates from inflammatory bowel diseases patients survive in macrophages and activate NLRP3 inflammasome. Int. J. Med. Microbiol. 2014, 304, 384–392. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, G.; Quintero, M.A.; Deo, S.K.; Abreu, M.T.; Daunert, S. Bacterial Quorum-Sensing Molecules in Serum: A Potential Tool for Crohn’s Disease Management. Clin. Transl. Gastroenterol. 2022, 13, e00547. [Google Scholar] [CrossRef] [PubMed]
- Fahed, A.C.; El-Hage-Sleiman, A.-K.M.; Farhat, T.I.; Nemer, G.M. Diet, Genetics, and Disease: A Focus on the Middle East and North Africa Region. J. Nutr. Metab. 2012, 2012, 109037. [Google Scholar] [CrossRef]
- Musaiger, A.O. Diet and Prevention of Coronary Heart Disease in the Arab Middle East Countries. Med. Princ. Pract. 2002, 11 (Suppl. S2), 9–16. [Google Scholar] [CrossRef]
- Dominianni, C.; Sinha, R.; Goedert, J.J.; Pei, Z.; Yang, L.; Hayes, R.B.; Ahn, J. Sex, Body Mass Index, and Dietary Fiber Intake Influence the Human Gut Microbiome. PLoS ONE 2015, 10, e0124599. [Google Scholar] [CrossRef]
- Leeuwendaal, N.K.; Stanton, C.; O’toole, P.W.; Beresford, T.P. Fermented Foods, Health and the Gut Microbiome. Nutrients 2022, 14, 1527. [Google Scholar] [CrossRef]
- Taylor, B.C.; Lejzerowicz, F.; Poirel, M.; Shaffer, J.P.; Jiang, L.; Aksenov, A.; Litwin, N.; Humphrey, G.; Martino, C.; Miller-Montgomery, S.; et al. Consumption of Fermented Foods Is Associated with Systematic Differences in the Gut Microbiome and Metabolome. mSystems 2020, 5, 10–1128. [Google Scholar] [CrossRef]
- Imhann, F.; Vila, A.V.; Bonder, M.J.; Fu, J.; Gevers, D.; Visschedijk, M.C.; Spekhorst, L.M.; Alberts, R.; Franke, L.; van Dullemen, H.M.; et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut 2018, 67, 108–119. [Google Scholar] [CrossRef]
- Molendijk, I.; van der Marel, S.; Maljaars, P.W.J. Towards a Food Pharmacy: Immunologic Modulation through Diet. Nutrients 2019, 11, 1239. [Google Scholar] [CrossRef]
- Al-Shamali, M.A.; Kalaoui, M.; Patty, I.; Hasan, F.; Khajah, A.; Al-Nakib, B. Ulcerative colitis in Kuwait: A review of 90 cases. Digestion 2003, 67, 218–224. [Google Scholar] [CrossRef]
- Al-Ghamdi, A.S.; Al-Mofleh, I.A.; Al-Rashed, R.S.; Al-Amri, S.M.; Aljebreen, A.M.; Isnani, A.C.; El-Badawi, R. Epidemiology and outcome of Crohn’s disease in a teaching hospital in Riyadh. World J. Gastroenterol. 2004, 10, 1341–1344. [Google Scholar] [CrossRef]
- Abdulla, M.; Al Saeed, M.; Fardan, R.H.; Alalwan, H.F.; Almosawi, Z.S.A.; Almahroos, A.F.; Al Qamish, J. Inflammatory bowel disease in Bahrain: Single-center experience. Clin. Exp. Gastroenterol. 2017, 10, 133–145. [Google Scholar] [CrossRef]
- Radhakrishnan, S.; Zubaidi, G.; Daniel, M.; Sachdev, G.K.; Mohan, A.N. Ulcerative colitis in Oman. A prospective study of the incidence and disease pattern from 1987 to 1994. Digestion 1997, 58, 266–270. [Google Scholar] [CrossRef]
- Abdul-Baki, H.; ElHajj, I.; El-Zahabi, L.M.; Azar, C.; Aoun, E.; Zantout, H.; Nasreddine, W.; Ayyach, B.; Mourad, F.H.; Soweid, A.; et al. Clinical epidemiology of inflammatory bowel disease in Lebanon. Inflamm. Bowel Dis. 2007, 13, 475–480. [Google Scholar] [CrossRef]
- Manolio, T.A. Genomewide association studies and assessment of the risk of disease. N. Engl. J. Med. 2010, 363, 166–176. [Google Scholar] [CrossRef]
- Wijmenga, C.; Zhernakova, A. The importance of cohort studies in the post-GWAS era. Nat. Genet. 2018, 50, 322–328. [Google Scholar] [CrossRef]
- Vermeire, S.; Wild, G.; Kocher, K.; Cousineau, J.; Dufresne, L.; Bitton, A.; Langelier, D.; Pare, P.; Lapointe, G.; Cohen, A.; et al. CARD15 genetic variation in a Quebec population: Prevalence, genotype-phenotype relationship, and haplotype structure. Am. J. Hum. Genet. 2002, 71, 74–83. [Google Scholar] [CrossRef]
- Abdelnaby, H.; Ndiaye, N.C.; D’Amico, F.; Fouad, A.M.; Hassan, S.; Elshafey, A.; Al Hashash, W.; Faisal, M.; Alshamali, Y.; Al-Taweel, T.; et al. NOD2/CARD15 polymorphisms (P268S, IVS8+158, G908R, L1007fs, R702W) among Kuwaiti patients with Crohn’s disease: A case-control study. Saudi J. Gastroenterol. 2021, 27, 249–256. [Google Scholar] [CrossRef]
- Sohail, M.U.; Hume, M.E.; Byrd, J.A.; Nisbet, D.J.; Shabbir, M.Z.; Ijaz, A.; Rehman, H. Molecular analysis of the caecal and tracheal microbiome of heat-stressed broilers supplemented with prebiotic and probiotic. Avian Pathol. 2015, 44, 67–74. [Google Scholar] [CrossRef]
- Chen, S.; Hu, Z.; Tang, J.; Zhu, H.; Zheng, Y.; Xiao, J.; Xu, Y.; Wang, Y.; Luo, Y.; Mo, X.; et al. High temperature and humidity in the environment disrupt bile acid metabolism, the gut microbiome, and GLP-1 secretion in mice. Commun. Biol. 2024, 7, 465. [Google Scholar] [CrossRef]
- Zuo, T.; Kamm, M.A.; Colombel, J.-F.; Ng, S.C. Urbanization and the gut microbiota in health and inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 440–452. [Google Scholar] [CrossRef]
- Feeney, M.A.; Murphy, F.; Clegg, A.J.; Trebble, T.M.; Sharer, N.M.; Snook, J.A. A case–control study of childhood environmental risk factors for the development of inflammatory bowel disease. Eur. J. Gastroenterol. Hepatol. 2002, 14, 529–534. [Google Scholar] [CrossRef]
- Card, T.; Logan, R.F.A.; Rodrigues, L.C.; Wheeler, J.G. Antibiotic use and the development of Crohn’s disease. Gut 2004, 53, 246–250. [Google Scholar] [CrossRef]
- Al-Ali, D.; Ahmed, A.; Shafiq, A.; McVeigh, C.; Chaari, A.; Zakaria, D.; Bendriss, G. Fecal microbiota transplants: A review of emerging clinical data on applications, efficacy, and risks (2015–2020). Qatar Med. J. 2021, 2021, 5. [Google Scholar] [CrossRef]
- Senok, A.C. Probiotics in the Arabian Gulf Region. Food Nutr. Res. 2009, 53, 1842. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fedorak, R.; Demeria, D. Probiotic bacteria in the prevention and the treatment of inflammatory bowel disease. Gastroenterol. Clin. N. Am. 2012, 41, 821–842. [Google Scholar] [CrossRef] [PubMed]
- Cheifetz, A.S.; Gianotti, R.; Luber, R.; Gibson, P.R. Complementary and alternative medicines used by patients with inflammatory bowel diseases. Gastroenterology 2017, 152, 415–429.e15. [Google Scholar] [CrossRef] [PubMed]
- Guslandi, M. Role of probiotics in Crohn’s disease and in Pouchitis. J. Clin. Gastroenterol. 2015, 49 (Suppl. S1), S46–S49. [Google Scholar] [CrossRef] [PubMed]
- Gionchetti, P.; Rizzello, F.; Helwig, U.; Venturi, A.; Lammers, K.M.; Brigidi, P.; Vitali, B.; Poggioli, G.; Miglioli, M.; Campieri, M. Prophylaxis of pouchitis onset with probiotic therapy: A double-blind, placebo-controlled trial. Gastroenterology 2003, 124, 1202–1209. [Google Scholar] [CrossRef] [PubMed]
- Mimura, T.; Rizzello, F.; Helwig, U.; Poggioli, G.; Schreiber, S.; Talbot, I.C.; Nicholls, R.J.; Gionchetti, P.; Campieri, M.; Kamm, M.A. Once daily high dose probiotic therapy (VSL#3) for maintaining remission in recurrent or refractory pouchitis. Gut 2004, 53, 108–114. [Google Scholar] [CrossRef]
- Abraham, B.P.; Quigley, E.M.M. Probiotics in inflammatory bowel disease. Gastroenterol. Clin. N. Am. 2017, 46, 769–782. [Google Scholar] [CrossRef]
- Sood, A.; Midha, V.; Makharia, G.K.; Ahuja, V.; Singal, D.; Goswami, P.; Tandon, R.K. The probiotic preparation, VSL#3 induces remission in patients with mild-to-moderately active ulcerative colitis. Clin. Gastroenterol. Hepatol. 2009, 7, 1202–1209.e1. [Google Scholar] [CrossRef]
- Lin, S.C.; Cheifetz, A.S. The Use of Complementary and Alternative Medicine in Patients with Inflammatory Bowel Disease. Gastroenterol. Hepatol. 2018, 14, 415–425. [Google Scholar]
- Miele, E.; Pascarella, F.; Giannetti, E.; Quaglietta, L.; Baldassano, R.N.; Staiano, A. Effect of a probiotic preparation (VSL#3) on induction and maintenance of remission in children with ulcerative colitis. Am. J. Gastroenterol. 2009, 104, 437–443. [Google Scholar] [CrossRef]
- Shen, J.; Zuo, Z.-X.; Mao, A.-P. Effect of probiotics on inducing remission and maintaining therapy in ulcerative colitis, crohn’s disease, and pouchitis. Inflamm. Bowel Dis. 2014, 20, 21–35. [Google Scholar] [CrossRef] [PubMed]
- Derwa, Y.; Gracie, D.J.; Hamlin, P.J.; Ford, A.C. Systematic review with meta-analysis: The efficacy of probiotics in inflammatory bowel disease. Aliment. Pharmacol. Ther. 2017, 46, 389–400. [Google Scholar] [CrossRef]
- Orel, R. Intestinal microbiota, probiotics and prebiotics in inflammatory bowel disease. World J. Gastroenterol. 2014, 20, 11505. [Google Scholar] [CrossRef] [PubMed]
- Kanauchi, O.; Matsumoto, Y.; Matsumura, M.; Fukuoka, M.; Bamba, T. The beneficial effects of microflora, especially obligate anaerobes, and their products on the colonic environment in inflammatory bowel disease. Curr. Pharm. Des. 2005, 11, 1047–1053. [Google Scholar] [CrossRef] [PubMed]
- Quigley, E.M.M. Prebiotics and Probiotics. Nutr. Clin. Pract. 2011, 27, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Reddavide, R.; Rotolo, O.; Caruso, M.G.; Stasi, E.; Notarnicola, M.; Miraglia, C.; Nouvenne, A.; Meschi, T.; Angelis, G.L.D.; Di Mario, F.; et al. The role of diet in the prevention and treatment of Inflammatory Bowel Diseases. Acta Biomed. 2018, 89, 60–75. [Google Scholar] [CrossRef] [PubMed]
- Eeckhaut, V.; Ducatelle, R.; Sas, B.; Vermeire, S.; Van Immerseel, F. Progress towards butyrate-producing pharmabiotics: Butyricicoccus pullicaecorum capsule and efficacy in TNBS models in comparison with therapeutics. Gut 2014, 63, 367. [Google Scholar] [CrossRef]
- Calder, P.C. n−3 Polyunsaturated fatty acids, inflammation, and inflammatory diseases. Am. J. Clin. Nutr. 2006, 83 (Suppl. S6), 1505S–1519S. [Google Scholar] [CrossRef]
- Mozaffari, H.; Daneshzad, E.; Larijani, B.; Bellissimo, N.; Azadbakht, L. Dietary intake of fish, n-3 polyunsaturated fatty acids, and risk of inflammatory bowel disease: A systematic review and meta-analysis of observational studies. Eur. J. Nutr. 2020, 59, 1–17. [Google Scholar] [CrossRef]
- Cabré, E.; Mañosa, M.; Gassull, M.A. Omega-3 fatty acids and inflammatory bowel diseases—A systematic review. Br. J. Nutr. 2012, 107, S240–S252. [Google Scholar] [CrossRef]
- Dziąbowska-Grabias, K.; Sztanke, M.; Zając, P.; Celejewski, M.; Kurek, K.; Szkutnicki, S.; Korga, P.; Bulikowski, W.; Sztanke, K. Antioxidant therapy in inflammatory bowel diseases. Antioxidants 2021, 10, 412. [Google Scholar] [CrossRef] [PubMed]
- Shahinfar, H.; Payandeh, N.; ElhamKia, M.; Abbasi, F.; Alaghi, A.; Djafari, F.; Eslahi, M.; Gohari, N.S.F.; Ghorbaninejad, P.; Hasanzadeh, M.; et al. Administration of dietary antioxidants for patients with inflammatory bowel disease: A systematic review and meta-analysis of randomized controlled clinical trials. Complement. Ther. Med. 2021, 63, 102787. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Wang, H.; Zhang, X.; Li, X.; Yu, J. Ascorbic acid ameliorates oxidative stress and inflammation in dextran sulfate sodium-induced ulcerative colitis in mice. Int. J. Clin. Exp. Med. 2015, 8, 20245–20253. [Google Scholar] [PubMed]
- Seidner, D.L.; Lashner, B.A.; Brzezinski, A.; Banks, P.L.; Goldblum, J.; Fiocchi, C.; Katz, J.; Lichtenstein, G.R.; Anton, P.A.; Kam, L.Y.; et al. An oral supplement enriched with fish oil, soluble fiber, and antioxidants for Corticosteroid sparing in ulcerative colitis: A randomized controlled trial. Clin. Gastroenterol. Hepatol. 2005, 3, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Ordovas, J.M.; Ferguson, L.R.; Tai, E.S.; Mathers, J.C. Personalised nutrition and health. BMJ 2018, 361, bmj.k2173. [Google Scholar] [CrossRef] [PubMed]
- Biesiekierski, J.R.; Jalanka, J.; Staudacher, H.M. Can gut microbiota composition predict response to dietary treatments? Nutrients 2019, 11, 1134. [Google Scholar] [CrossRef]
- Bennet, S.M.P.; Bohn, L.; Storsrud, S.; Liljebo, T.; Collin, L.; Lindfors, P.; Tornblom, H.; Ohman, L.; Simren, M. Multivariate modelling of faecal bacterial profiles of patients with IBS predicts responsiveness to a diet low in FODMAPs. Gut 2018, 67, 872–881. [Google Scholar] [CrossRef]
- Levine, A.; Wine, E.; Assa, A.; Boneh, R.S.; Shaoul, R.; Kori, M.; Cohen, S.; Peleg, S.; Shamaly, H.; On, A.; et al. Crohn’s Disease Exclusion Diet Plus Partial Enteral Nutrition Induces Sustained Remission in a Randomized Controlled Trial. Gastroenterology 2019, 157, 440–450.e8. [Google Scholar] [CrossRef] [PubMed]
- Svolos, V.; Hansen, R.; Nichols, B.; Quince, C.; Ijaz, U.Z.; Papadopoulou, R.T.; Edwards, C.A.; Watson, D.; Alghamdi, A.; Brejnrod, A.; et al. Treatment of Active Crohn’s Disease with an Ordinary Food-based Diet That Replicates Exclusive Enteral Nutrition. Gastroenterology 2019, 156, 1354–1367.e6. [Google Scholar] [CrossRef]
- Wellens, J.; Vissers, E.; Matthys, C.; Vermeire, S.; Sabino, J. Personalized dietary regimens for inflammatory bowel disease: Current knowledge and future perspectives. Pharmacogenomics Pers. Med. 2023, 16, 15–27. [Google Scholar] [CrossRef]
- Weingarden, A.R.; Vaughn, B.P. Intestinal microbiota, fecal microbiota transplantation, and inflammatory bowel disease. Gut Microbes 2017, 8, 238–252. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, M.J.; Weingarden, A.R.; Sadowsky, M.J.; Khoruts, A. Standardized frozen preparation for transplantation of fecal microbiota for recurrent clostridium difficile infection. Am. J. Gastroenterol. 2012, 107, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Van Nood, E.; Vrieze, A.; Nieuwdorp, M.; Fuentes, S.; Zoetendal, E.G.; De Vos, W.M.; Visser, C.E.; Kuijper, E.J.; Bartelsman, J.F.W.M.; Tijssen, J.G.P.; et al. Duodenal infusion of donor feces for recurrent clostridium difficile. N. Engl. J. Med. 2013, 368, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Bennet, J.D.; Brinkman, M. Treatment of ulcerative colitis by implantation of normal colonic flora. Lancet 1989, 333, 164. [Google Scholar] [CrossRef] [PubMed]
- Cui, B.; Feng, Q.; Wang, H.; Wang, M.; Peng, Z.; Li, P.; Huang, G.; Liu, Z.; Wu, P.; Fan, Z.; et al. Fecal microbiota transplantation through mid-gut for refractory Crohn’s disease: Safety, feasibility, and efficacy trial results. J. Gastroenterol. Hepatol. 2014, 30, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Hsu, M.; Tun, K.M.; Batra, K.; Haque, L.; Vongsavath, T.; Hong, A.S. Safety and Efficacy of Fecal Microbiota Transplantation in Treatment of Inflammatory Bowel Disease in the Pediatric Population: A Systematic Review and Meta-Analysis. Microorganisms 2023, 11, 1272. [Google Scholar] [CrossRef] [PubMed]
- Colman, R.J.; Rubin, D.T. Fecal microbiota transplantation as therapy for inflammatory bowel disease: A systematic review and meta-analysis. J. Crohn’s Colitis 2014, 8, 1569–1581, Erratum in J. Crohn’s Colitis 2023, 17, 149. [Google Scholar] [CrossRef] [PubMed]
- Tariq, R.M.; Syed, T.; Yadav, D.M.; Prokop, L.J.M.; Singh, S.M.; Loftus, E.V.J.; Pardi, D.S.; Khanna, S.M. Outcomes of fecal microbiota transplantation for C. difficile infection in inflammatory bowel disease. J. Clin. Gastroenterol. 2021, 57, 285–293. [Google Scholar] [CrossRef]
- Fischer, M.; Kao, D.; Kelly, C.; Kuchipudi, A.; Jafri, S.-M.; Blumenkehl, M.; Rex, D.; Mellow, M.; Kaur, N.; Sokol, H.; et al. Fecal Microbiota Transplantation is Safe and Efficacious for Recurrent or Refractory Clostridium difficile Infection in Patients with Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2016, 22, 2402–2409. [Google Scholar] [CrossRef]
- Olesen, S.W.; Panchal, P.; Chen, J.; Budree, S.; Osman, M. Global disparities in faecal Microbiota Transplantation Research. Lancet Gastroenterol. Hepatol. 2020, 5, 241. [Google Scholar] [CrossRef]
- Moossavi, S.; Salimzadeh, H.; Katoonizadeh, A.; Mojarrad, A.; Merat, D.; Ansari, R.; Vahedi, H.; Merat, S.; Malekzadeh, R. Physicians’ Knowledge and Attitude towards Fecal Microbiota Transplant in Iran. Middle East J. Dig. Dis. 2015, 7, 155–160. [Google Scholar] [PubMed]
- Bendriss, G.; Al-Ali, D.; Shafiq, A.; Laswi, I.; Mhaimeed, N.; Salameh, M.; Burney, Z.; Pillai, K.; Chaari, A.; Zakaria, D.; et al. Targeting the gut microbiome: A brief report on the awareness, practice, and readiness to engage in clinical interventions in Qatar. Qatar Med. J. 2021, 2020, 47. [Google Scholar] [CrossRef]
- Al-Bakri, A.G.; Akour, A.A.; Al-Delaimy, W.K. Knowledge, attitudes, ethical and social perspectives towards fecal microbiota transplantation (FMT) among Jordanian healthcare providers. BMC Med. Ethics 2021, 22, 19. [Google Scholar] [CrossRef] [PubMed]
- Pesce, M.; Seguella, L.; Del Re, A.; Lu, J.; Palenca, I.; Corpetti, C.; Rurgo, S.; Sanseverino, W.; Sarnelli, G.; Esposito, G. Next-Generation Probiotics for Inflammatory Bowel Disease. Int. J. Mol. Sci. 2022, 23, 5466. [Google Scholar] [CrossRef]
- Thomas, J.P.; Modos, D.; Korcsmaros, T.; Brooks-Warburton, J. Network Biology Approaches to Achieve Precision Medicine in Inflammatory Bowel Disease. Front. Genet. 2021, 12, 760501. [Google Scholar] [CrossRef] [PubMed]
- Borg-Bartolo, S.P.; Boyapati, R.K.; Satsangi, J.; Kalla, R. Precision medicine in inflammatory bowel disease: Concept, progress and challenges. F1000Research 2020, 9, 54. [Google Scholar] [CrossRef] [PubMed]
- Mishima, Y.; Sartor, R.B. Manipulating resident microbiota to enhance regulatory immune function to treat inflammatory bowel diseases. J. Gastroenterol. 2020, 55, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Emencheta, S.C.; Olovo, C.V.; Eze, O.C.; Kalu, C.F.; Berebon, D.P.; Onuigbo, E.B.; Vila, M.M.D.C.; Balcão, V.M.; Attama, A.A. The Role of Bacteriophages in the Gut Microbiota: Implications for Human Health. Pharmaceutics 2023, 15, 2416. [Google Scholar] [CrossRef] [PubMed]
- Kernbauer, E.; Ding, Y.; Cadwell, K. An enteric virus can replace the beneficial function of commensal bacteria. Nature 2014, 516, 94–98. [Google Scholar] [CrossRef]
- Blum-Oehler, G.; Oswald, S.; Eiteljörge, K.; Sonnenborn, U.; Schulze, J.; Kruis, W.; Hacker, J. Development of strain-specific PCR reactions for the detection of the probiotic Escherichia coli strain Nissle 1917 in fecal samples. Res. Microbiol. 2003, 154, 59–66. [Google Scholar] [CrossRef]
- Sonnenborn, U.; Schulze, J. The non-pathogenic Escherichia coli strain Nissle 1917–features of a versatile probiotic. Microb. Ecol. Health Dis. 2009, 21, 122–158. [Google Scholar]
- Wang, L.; Liao, Y.; Yang, R.; Zhu, Z.; Zhang, L.; Wu, Z.; Sun, X. An engineered probiotic secreting Sj16 ameliorates colitis via Ruminococcaceae/butyrate/retinoic acid axis. Bioeng. Transl. Med. 2021, 6, e10219. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Liu, X.-Y.; Zhang, D.; Zhang, Y.-D.; Li, Z.-H.; Liu, X.; Wu, F.; Chen, G.-Q. Construction of a sustainable 3-hydroxybutyrate-producing probiotic Escherichia coli for treatment of colitis. Cell. Mol. Immunol. 2021, 18, 2344–2357. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Liu, Y.; Lan, X.; Xu, X.; Zhang, X.; Li, X.; Zhao, Y.; Li, G.; Du, C.; Lu, S.; et al. Oral Escherichia coli expressing IL-35 meliorates experimental colitis in mice. J. Transl. Med. 2018, 16, 71. [Google Scholar] [CrossRef]
- Anomaly, J. The Future of Phage: Ethical Challenges of Using Phage Therapy to Treat Bacterial Infections. Public Health Ethics 2020, 13, 82–88. [Google Scholar] [CrossRef]

| ‘Inflammatory bowel disease’, ‘IBD’, ‘Ulcerative Colitis’, ‘UC’, ‘Crohn’s Disease’, ‘CD’, ‘Microbiome’, ‘Firmucutes’, ‘Bacteroidetes’, ‘Dysbiosis’, ‘Middle East’, ‘Arab’, ‘Diet’, ‘NOD’, ‘Probiotics’, ‘Prebiotics’, ‘Faecal Microbiota Transplantation’, |
| Phylum | Role in Gut Microbiome | Genera |
|---|---|---|
| Firmicutes | Fermentation: Degradation of dietary fibres into short-chain fatty acids (SCFAs) such as butyrate, an energy source for colonocytes. Gut Barrier: Supports the integrity of the intestinal barrier. Immune Regulation: Mediates inflammatory reactions and the immune response, such as colonic Treg cells. | Lactobacillus Clostridium Enterococcus |
| Bacteroidetes | Carbohydrate Digestion: Produces SCFAs such as acetate and propionate by breaking down complex carbohydrates. Metabolic Regulation: Expresses bile salt hydrolases, important in bile acid metabolism. Immune Regulation: Mediates inflammatory reactions and the immune response, such as promotion of CD4+ T cell differentiation. | Bacteroides Prevotella |
| Actinobacteria | Metabolic Regulation: Produces acetate, a co-substrate for butyrate, an energy source for colonocytes. Immune Regulation: Maintains gut barrier homeostasis and induces colonic Treg cells. | Bifidobacterium Corynebacterium |
| Proteobacteria | Pathogenicity: When overgrown, associated with dysbiosis and intestinal inflammation, increasing risk of metabolic syndrome and IBD. Metabolism: Fixation of nitrogen. | Escherichia Salmonella Helicobacter |
| Verrucomicrobia | Mucin Degradation: Helps to break down mucin, producing metabolites such as SCFAs. Immune Regulation: Produces antimicrobial peptides. | Akkermansia |
| Fusobacteria | Amino Acid Metabolism: Contributes to the metabolism of peptides and amino acids. Pathogenicity: Associated with dysbiosis and inflammation in the gastrointestinal tract, as well as colorectal cancer. | Fusobacterium |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El-Sayed, A.; Kapila, D.; Taha, R.S.I.; El-Sayed, S.; Mahen, M.R.A.; Taha, R.; Alrubaiy, L. The Role of the Gut Microbiome in Inflammatory Bowel Disease: The Middle East Perspective. J. Pers. Med. 2024, 14, 652. https://doi.org/10.3390/jpm14060652
El-Sayed A, Kapila D, Taha RSI, El-Sayed S, Mahen MRA, Taha R, Alrubaiy L. The Role of the Gut Microbiome in Inflammatory Bowel Disease: The Middle East Perspective. Journal of Personalized Medicine. 2024; 14(6):652. https://doi.org/10.3390/jpm14060652
Chicago/Turabian StyleEl-Sayed, Ahmed, Diya Kapila, Rama Sami Issa Taha, Sherif El-Sayed, Mohd Rafiw Ahmed Mahen, Roa’a Taha, and Laith Alrubaiy. 2024. "The Role of the Gut Microbiome in Inflammatory Bowel Disease: The Middle East Perspective" Journal of Personalized Medicine 14, no. 6: 652. https://doi.org/10.3390/jpm14060652
APA StyleEl-Sayed, A., Kapila, D., Taha, R. S. I., El-Sayed, S., Mahen, M. R. A., Taha, R., & Alrubaiy, L. (2024). The Role of the Gut Microbiome in Inflammatory Bowel Disease: The Middle East Perspective. Journal of Personalized Medicine, 14(6), 652. https://doi.org/10.3390/jpm14060652






