Starvation and Inflammation Modulate Adipose Mesenchymal Stromal Cells’ Molecular Signature
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Methods
2.1.1. Ethics
2.1.2. ASC Isolation and Culture
- (i)
- “F”: 48 h in a complete medium;
- (ii)
- “F+IL”: as “F” supplemented with 1 ng/mL interleukin 1 beta (IL1β);
- (iii)
- “S”: 48 h without serum (starvation), after 48 h in a complete medium;
- (iv)
- “S+IL”: as “S” supplemented with 1 ng/mL IL1β;
- (v)
- “F+IL/S”: 48 h without serum (starvation), after 48 h in a complete medium supplemented with 1 ng/mL IL1β.
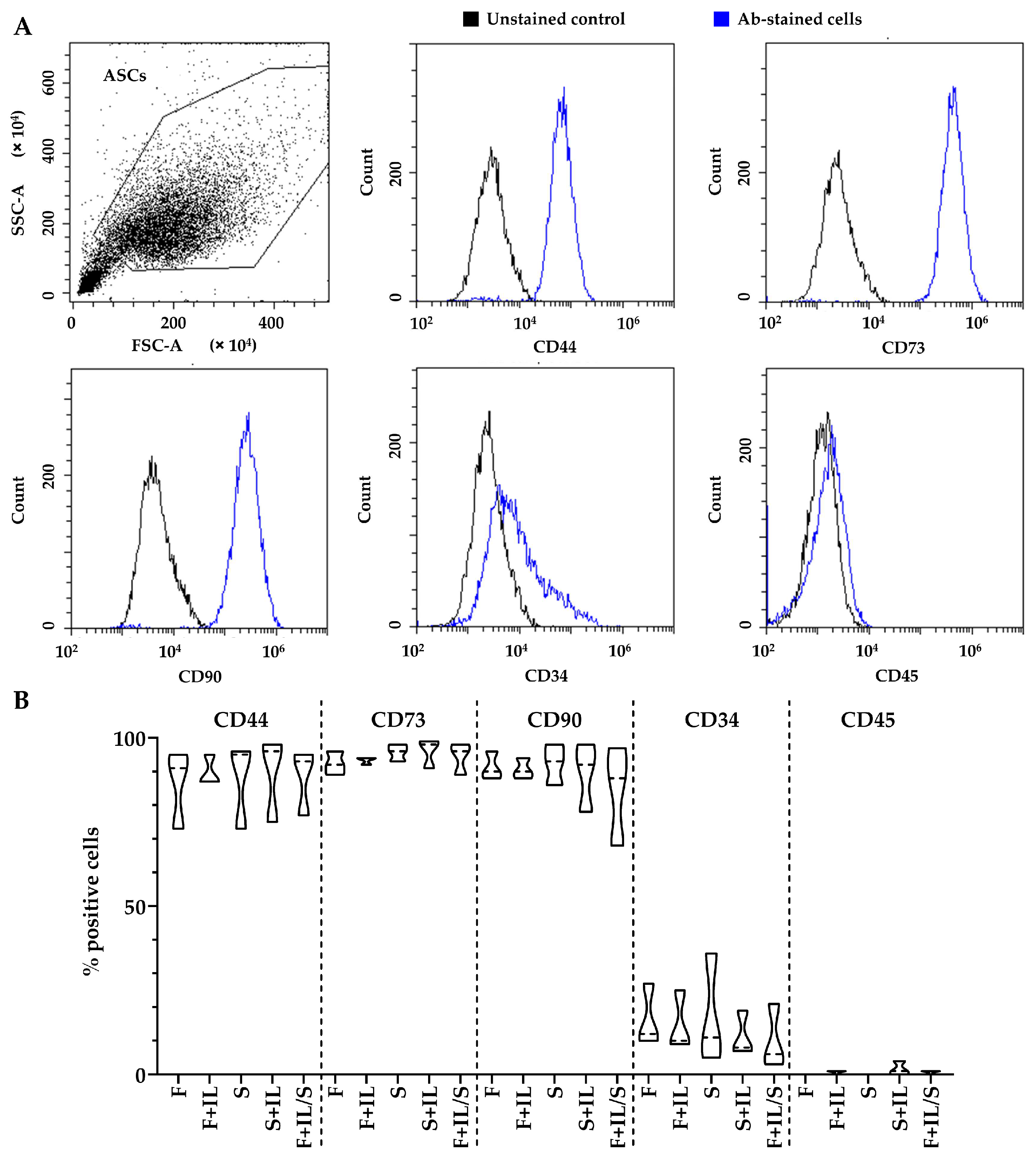
2.1.3. Flow Cytometry
2.1.4. Gene Expression Analysis with PCR Arrays
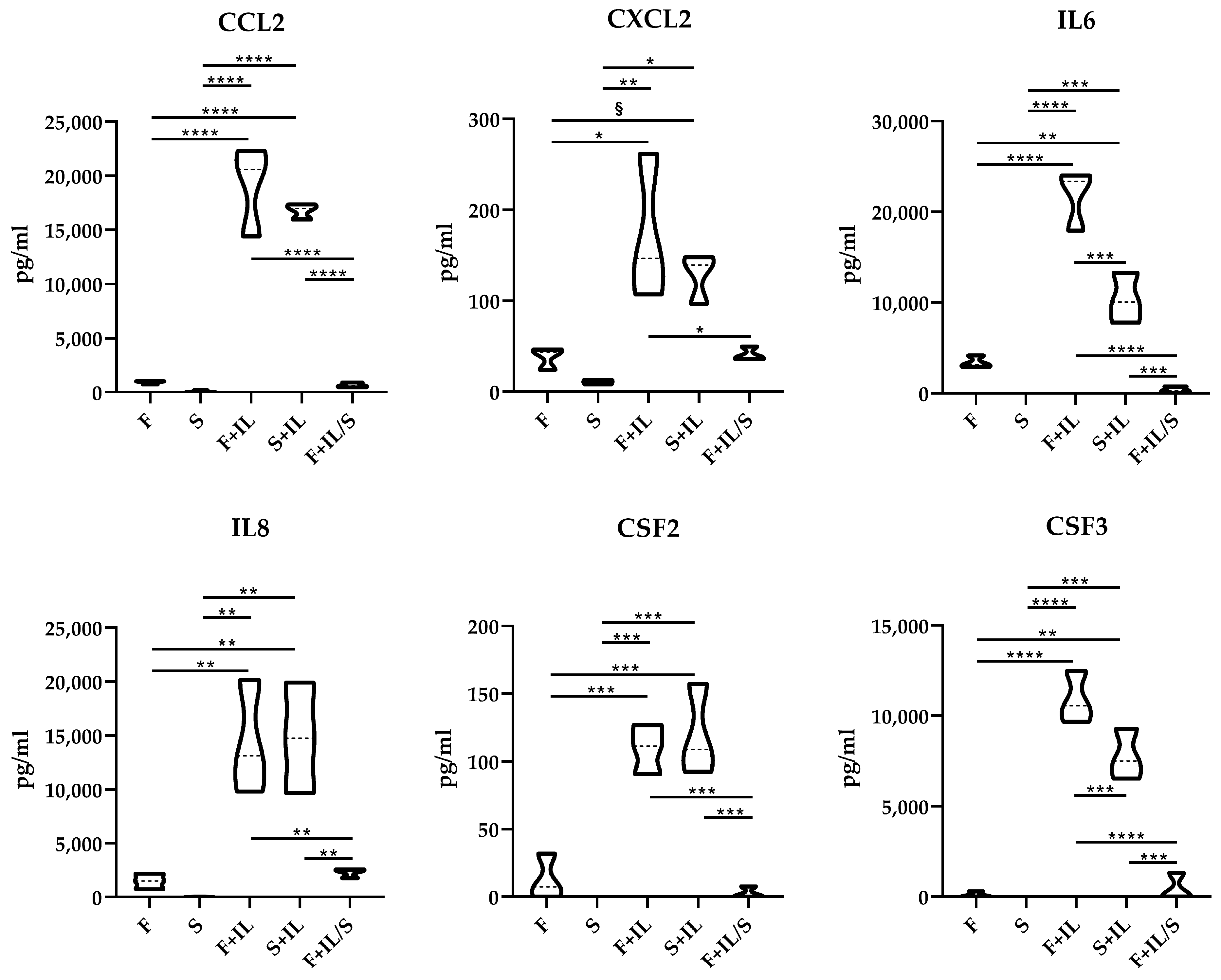
2.1.5. ELISA Assays
2.1.6. Functional Experiment on Chondrocytes
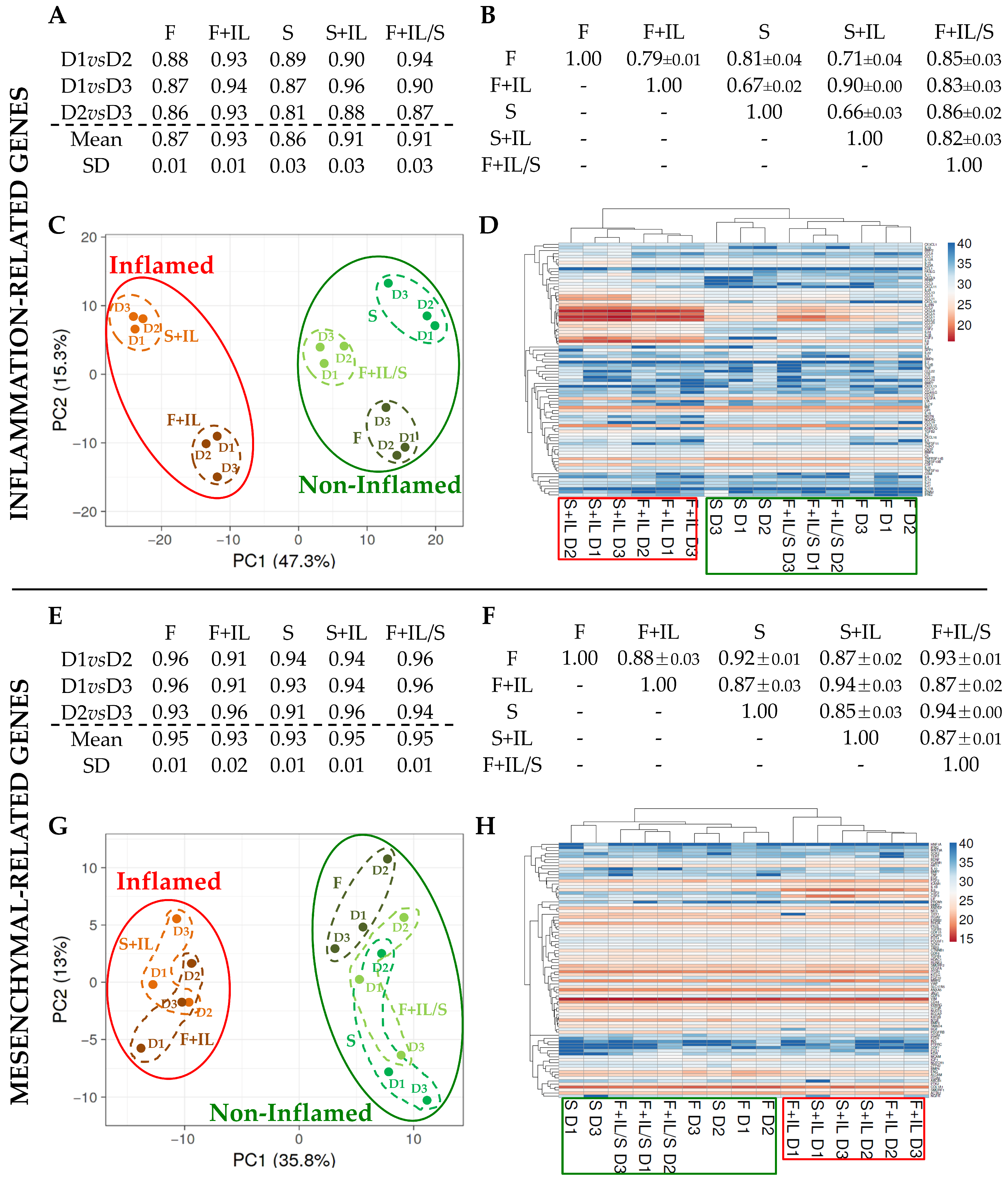
2.1.7. Hierarchical Clustering
2.1.8. Statistical Analyses
3. Results
3.1. Results
3.1.1. ASCs’ Immunophenotype
3.1.2. Inflammation and Starvation Affect ASCs’ Molecular Signature Related to Inflammation Markers
3.1.3. Inflammation and Starvation Affect ASCs’ Molecular Signature Related to Mesenchymal Markers
3.1.4. Genes Affected by Inflammation and Whose Modulation Is Abolished by Sequential Starvation
3.1.5. ELISA Confirmation of Gene Modulation
3.1.6. Secretomes’ Effect on Chondrocytes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Merimi, M.; El-Majzoub, R.; Lagneaux, L.; Moussa Agha, D.; Bouhtit, F.; Meuleman, N.; Fahmi, H.; Lewalle, P.; Fayyad-Kazan, M.; Najar, M. The Therapeutic Potential of Mesenchymal Stromal Cells for Regenerative Medicine: Current Knowledge and Future Understandings. Front. Cell Dev. Biol. 2021, 9, 661532. [Google Scholar] [CrossRef]
- Han, Y.; Yang, J.; Fang, J.; Zhou, Y.; Candi, E.; Wang, J.; Hua, D.; Shao, C.; Shi, Y. The secretion profile of mesenchymal stem cells and potential applications in treating human diseases. Signal Transduct. Target. Ther. 2022, 7, 92. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.B.; Moncivais, K.; Caplan, A.I. Mesenchymal stem cells: Environmentally responsive therapeutics for regenerative medicine. Exp. Mol. Med. 2013, 45, 54. [Google Scholar] [CrossRef] [PubMed]
- Miceli, V.; Zito, G.; Bulati, M.; Gallo, A.; Busà, R.; Iannolo, G.; Conaldi, P.G. Different priming strategies improve distinct therapeutic capabilities of mesenchymal stromal/stem cells: Potential implications for their clinical use. World J. Stem Cells 2023, 15, 400–420. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Nogués, C.; O’Brien, T. Current good manufacturing practice considerations for mesenchymal stromal cells as therapeutic agents. Biomater. Biosyst. 2021, 2, 100018. [Google Scholar] [CrossRef]
- Eleuteri, S.; Fierabracci, A. Insights into the Secretome of Mesenchymal Stem Cells and Its Potential Applications. Int. J. Mol. Sci. 2019, 20, 4597. [Google Scholar] [CrossRef]
- Partan, R.U.; Putra, K.M.; Kusuma, N.F.; Darma, S.; Reagan, M.; Muthia, P.; Radiandina, A.S.; Saleh, M.I.; Salim, E.M. Umbilical Cord Mesenchymal Stem Cell Secretome Improves Clinical Outcomes and Changes Biomarkers in Knee Osteoarthritis. J. Clin. Med. 2023, 12, 7138. [Google Scholar] [CrossRef]
- Lee, J.Y.; Kang, M.H.; Jang, J.E.; Lee, J.E.; Yang, Y.; Choi, J.Y.; Kang, H.S.; Lee, U.; Choung, J.W.; Jung, H.; et al. Comparative analysis of mesenchymal stem cells cultivated in serum free media. Sci. Rep. 2022, 12, 8620. [Google Scholar] [CrossRef]
- Chouaib, B.; Haack-Sørensen, M.; Chaubron, F.; Cuisinier, F.; Collart-Dutilleul, P. Towards the Standardization of Mesenchymal Stem Cell Secretome-Derived Product Manufacturing for Tissue Regeneration. Int. J. Mol. Sci. 2023, 24, 12594. [Google Scholar] [CrossRef]
- Giannasi, C.; Niada, S.; Della Morte, E.; Casati, S.R.; De Palma, C.; Brini, A.T. Serum starvation affects mitochondrial metabolism of adipose-derived stem/stromal cells. Cytotherapy 2023, 25, 704–711. [Google Scholar] [CrossRef]
- Paschalidis, T.; Bakopoulou, A.; Papa, P.; Leyhausen, G.; Geurtsen, W.; Koidis, P. Dental pulp stem cells’ secretome enhances pulp repair processes and compensates TEGDMA-induced cytotoxicity. Dent. Mater. 2014, 30, 405–418. [Google Scholar] [CrossRef] [PubMed]
- Stanco, D.; Viganò, M.; Perucca Orfei, C.; Di Giancamillo, A.; Peretti, G.M.; Lanfranchi, L.; de Girolamo, L. Multidifferentiation potential of human mesenchymal stem cells from adipose tissue and hamstring tendons for musculoskeletal cell-based therapy. Regen. Med. 2015, 10, 729–743. [Google Scholar] [CrossRef] [PubMed]
- Ragni, E.; Piccolo, S.; Taiana, M.; Visconte, C.; Grieco, G.; de Girolamo, L. Inflammation and Starvation Affect Housekeeping Gene Stability in Adipose Mesenchymal Stromal Cells. Curr. Issues Mol. Biol. 2024, 46, 842–855. [Google Scholar] [CrossRef] [PubMed]
- Scherberich, A.; Di Maggio, N.D.; McNagny, K.M. A familiar stranger: CD34 expression and putative functions in SVF cells of adipose tissue. World J. Stem Cells 2013, 5, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Műzes, G.; Sipos, F. Mesenchymal Stem Cell-Derived Secretome: A Potential Therapeutic Option for Autoimmune and Immune-Mediated Inflammatory Diseases. Cells 2022, 11, 2300. [Google Scholar] [CrossRef] [PubMed]
- Galderisi, U.; Peluso, G.; Di Bernardo, G. Clinical Trials Based on Mesenchymal Stromal Cells are Exponentially Increasing: Where are We in Recent Years? Stem Cell Rev. Rep. 2022, 18, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.P.; Ha, P.; Xiao, C.Y.; Kim, S.Y.; Jensen, A.R.; Easley, J.; Yao, Q.; Zhang, X. Updates on mesenchymal stem cell therapies for articular cartilage regeneration in large animal models. Front. Cell Dev. Biol. 2022, 10, 982199. [Google Scholar] [CrossRef]
- D’Arrigo, D.; Roffi, A.; Cucchiarini, M.; Moretti, M.; Candrian, C.; Filardo, G. Secretome and Extracellular Vesicles as New Biological Therapies for Knee Osteoarthritis: A Systematic Review. Clin. Med. 2019, 8, 1867. [Google Scholar] [CrossRef] [PubMed]
- Copp, G.; Robb, K.P.; Viswanathan, S. Culture-expanded mesenchymal stromal cell therapy: Does it work in knee osteoarthritis? A pathway to clinical success. Cell Mol. Immunol. 2023, 20, 626–650. [Google Scholar] [CrossRef]
- Redondo-Castro, E.; Cunningham, C.; Miller, J.; Martuscelli, L.; Aoulad-Ali, S.; Rothwell, N.J.; Kielty, C.M.; Allan, S.M.; Pinteaux, E. Interleukin-1 primes human mesenchymal stem cells towards an anti-inflammatory and pro-trophic phenotype in vitro. Stem Cell Res. Ther. 2017, 8, 79. [Google Scholar] [CrossRef]
- Colombini, A.; Libonati, F.; Cangelosi, D.; Lopa, S.; De Luca, P.; Coviello, D.A.; Moretti, M.; de Girolamo, L. Inflammatory priming with IL-1β promotes the immunomodulatory behavior of adipose derived stem cells. Front. Bioeng. Biotechnol. 2022, 10, 1000879. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Dai, H.; Li, J. Immunomodulatory properties of mesenchymal stromal/stem cells: The link with metabolism. Adv. Res. 2023, 45, 15–29. [Google Scholar] [CrossRef] [PubMed]
- Torres Palomino, D.C.; Cavalheiro Marti, L. Chemokines and immunity. Einstein 2015, 13, 469–473. [Google Scholar] [CrossRef] [PubMed]
- Willcockson, H.; Ozkan, H.; Chubinskaya, S.; Loeser, R.F.; Longobardi, L. CCL2 induces articular chondrocyte MMP expression through ERK and p38 signaling pathways. Osteoarthr. Cartil. Open 2021, 3, 100136. [Google Scholar] [CrossRef] [PubMed]
- Alaaeddine, N.; Antoniou, J.; Moussa, M.; Hilal, G.; Kreichaty, G.; Ghanem, I.; Abouchedid, W.; Saghbini, E.; Di Battista, J.A. The chemokine CCL20 induces proinflammatory and matrix degradative responses in cartilage. Inflamm. Res. 2015, 64, 721–731. [Google Scholar] [CrossRef]
- Edderkaoui, B. Chemokines in Cartilage Regeneration and Degradation: New Insights. Int. J. Mol. Sci. 2023, 25, 381. [Google Scholar] [CrossRef]
- Wang, E.C.Y.; Newton, Z.; Hayward, O.A.; Clark, S.R.; Collins, F.; Perks, W.V.; Singh, R.K.; Twohig, J.P.; Williams, A.S. Regulation of early cartilage destruction in inflammatory arthritis by death receptor 3. Arthritis Rheumatol. 2014, 66, 2762–2772. [Google Scholar] [CrossRef] [PubMed]
- Poo, Y.S.; Nakaya, H.; Gardner, J.; Larcher, T.; Schroder, W.A.; Le, T.T.; Major, L.D.; Suhrbier, A. CCR2 deficiency promotes exacerbated chronic erosive neutrophil-dominated chikungunya virus arthritis. J. Virol. 2014, 88, 6862–6872. [Google Scholar] [CrossRef]
- Onuora, S. Pain: Granulocyte-macrophage colony-stimulating factor required for inflammatory and arthritic pain. Nat. Rev. Rheumatol. 2012, 8, 499. [Google Scholar] [CrossRef]
- Jrad, A.I.S.; Trad, M.; Bzeih, W.; El Hasbani, G.; Uthman, I. Role of pro-inflammatory interleukins in osteoarthritis: A narrative review. Connect. Tissue Res. 2023, 64, 238–247. [Google Scholar] [CrossRef]
- Zhang, J.; An, J. Cytokines, inflammation, and pain. Int. Anesthesiol. Clin. 2007, 45, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Lotz, M.; Moats, T.; Villiger, P.M. Leukemia inhibitory factor is expressed in cartilage and synovium and can contribute to the pathogenesis of arthritis. J. Clin. Investig. 1992, 90, 888–896. [Google Scholar] [CrossRef] [PubMed]
- Grimaud, E.; Blanchard, F.; Charrier, C.; Gouin, F.; Redini, F.; Heymann, D. Leukaemia inhibitory factor (lif) is expressed in hypertrophic chondrocytes and vascular sprouts during osteogenesis. Cytokine 2002, 20, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Boff, D.; Crijns, H.; Janssens, R.; Vanheule, V.; Menezes, G.B.; Macari, S.; Silva, T.A.; Amaral, F.A.; Proost, P. The chemokine fragment CXCL9(74-103) diminishes neutrophil recruitment and joint inflammation in antigen-induced arthritis. J. Leukoc. Biol. 2018, 104, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Lv, S.; Tong, P.; Yan, L.; Chen, Z.; Zhou, L.; Yuan, Q.; Guo, L.; Shan, L. Intra-Articular Injection of Adipose-Derived Stem Cells Ameliorates Pain and Cartilage Anabolism/Catabolism in Osteoarthritis: Preclinical and Clinical Evidences. Front. Pharmacol. 2022, 13, 54025. [Google Scholar] [CrossRef]
- Platas, J.; Guillén, M.I.; del Caz, M.D.; Gomar, F.; Mirabet, V.; Alcaraz, M.J. Conditioned media from adipose-tissue-derived mesenchymal stem cells downregulate degradative mediators induced by interleukin-1β in osteoarthritic chondrocytes. Mediat. Inflamm. 2013, 2013, 357014. [Google Scholar] [CrossRef]
- Niada, S.; Giannasi, C.; Gomarasca, M.; Stanco, D.; Casati, S.; Brini, A.T. 6 Adipose-derived stromal cell secretome reduces TNFα-induced hypertrophy and catabolic markers in primary human articular chondrocytes. Stem Cell Res. 2019, 38, 101463. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piccolo, S.; Grieco, G.; Visconte, C.; De Luca, P.; Taiana, M.; Zagra, L.; Ragni, E.; de Girolamo, L. Starvation and Inflammation Modulate Adipose Mesenchymal Stromal Cells’ Molecular Signature. J. Pers. Med. 2024, 14, 847. https://doi.org/10.3390/jpm14080847
Piccolo S, Grieco G, Visconte C, De Luca P, Taiana M, Zagra L, Ragni E, de Girolamo L. Starvation and Inflammation Modulate Adipose Mesenchymal Stromal Cells’ Molecular Signature. Journal of Personalized Medicine. 2024; 14(8):847. https://doi.org/10.3390/jpm14080847
Chicago/Turabian StylePiccolo, Simona, Giulio Grieco, Caterina Visconte, Paola De Luca, Michela Taiana, Luigi Zagra, Enrico Ragni, and Laura de Girolamo. 2024. "Starvation and Inflammation Modulate Adipose Mesenchymal Stromal Cells’ Molecular Signature" Journal of Personalized Medicine 14, no. 8: 847. https://doi.org/10.3390/jpm14080847
APA StylePiccolo, S., Grieco, G., Visconte, C., De Luca, P., Taiana, M., Zagra, L., Ragni, E., & de Girolamo, L. (2024). Starvation and Inflammation Modulate Adipose Mesenchymal Stromal Cells’ Molecular Signature. Journal of Personalized Medicine, 14(8), 847. https://doi.org/10.3390/jpm14080847








